Advancing Reversed-Phase Chromatography Analytics of Influenza Vaccines Using Machine Learning Approaches on a Diverse Range of Antigens and Formulations
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Preparation
2.3. Conditioning and Separation
2.4. Data Processing
2.5. Machine Learning
3. Results
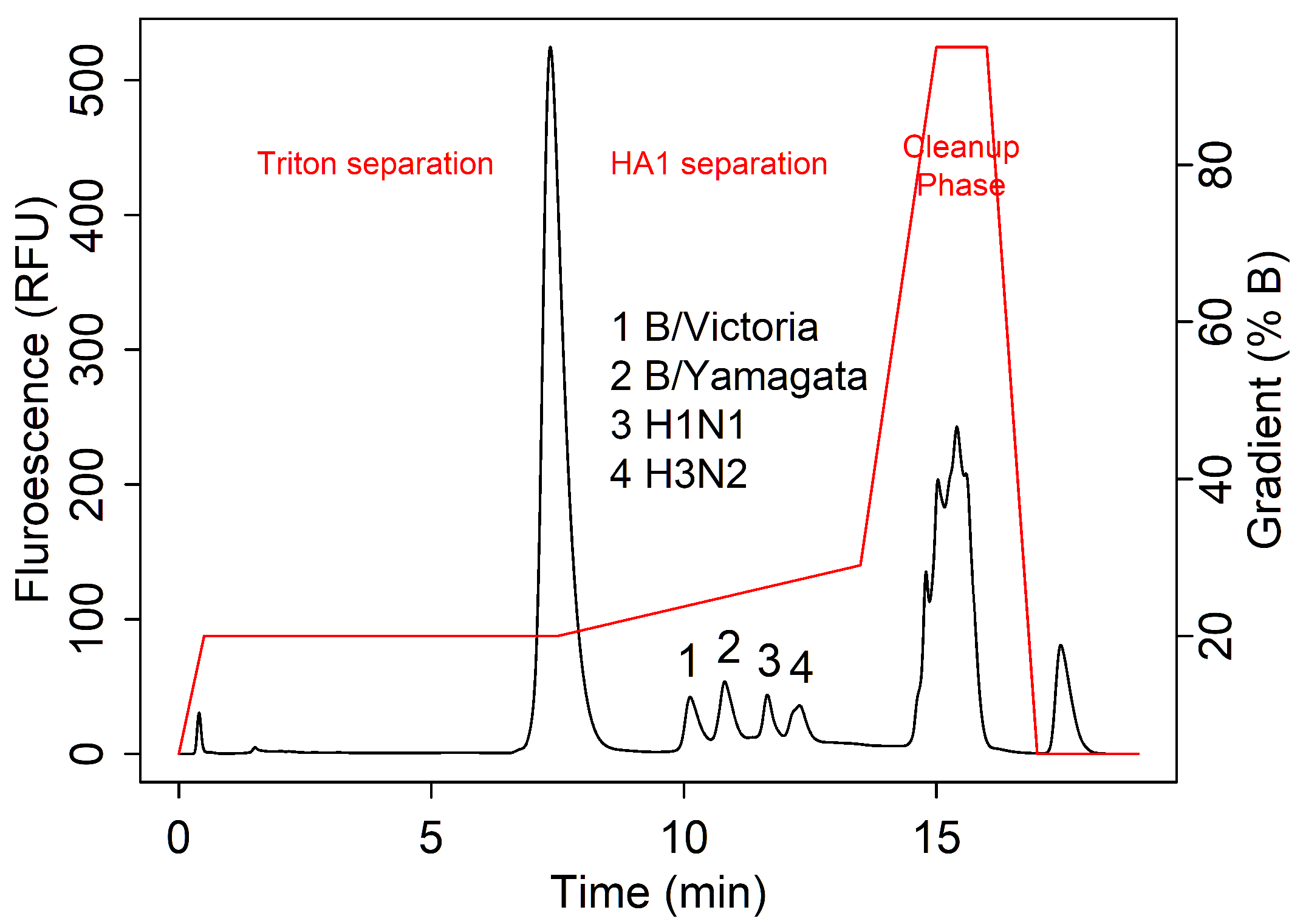
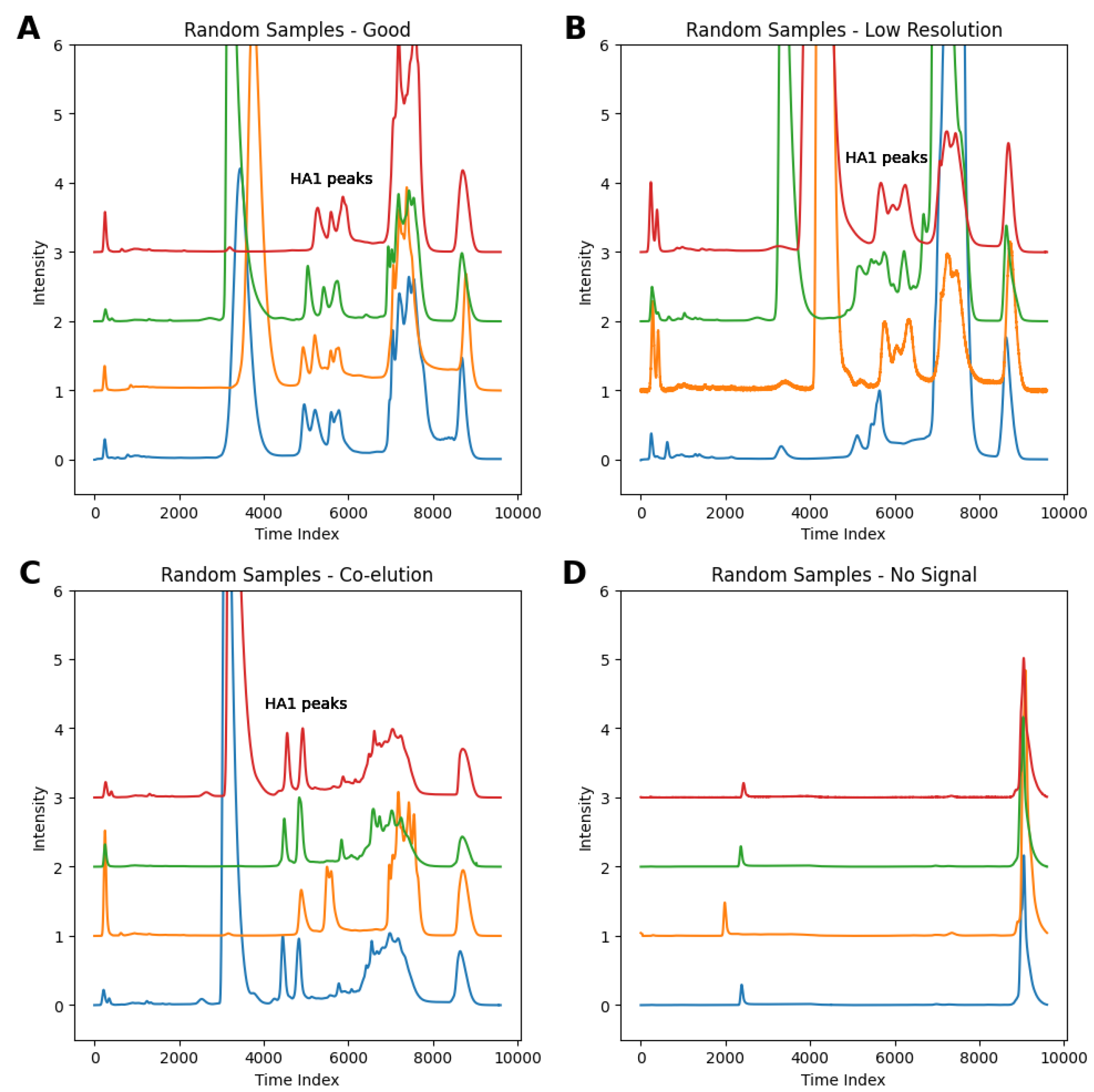
3.1. Data Collection and Key Features
3.2. Examples of Challenging Separations
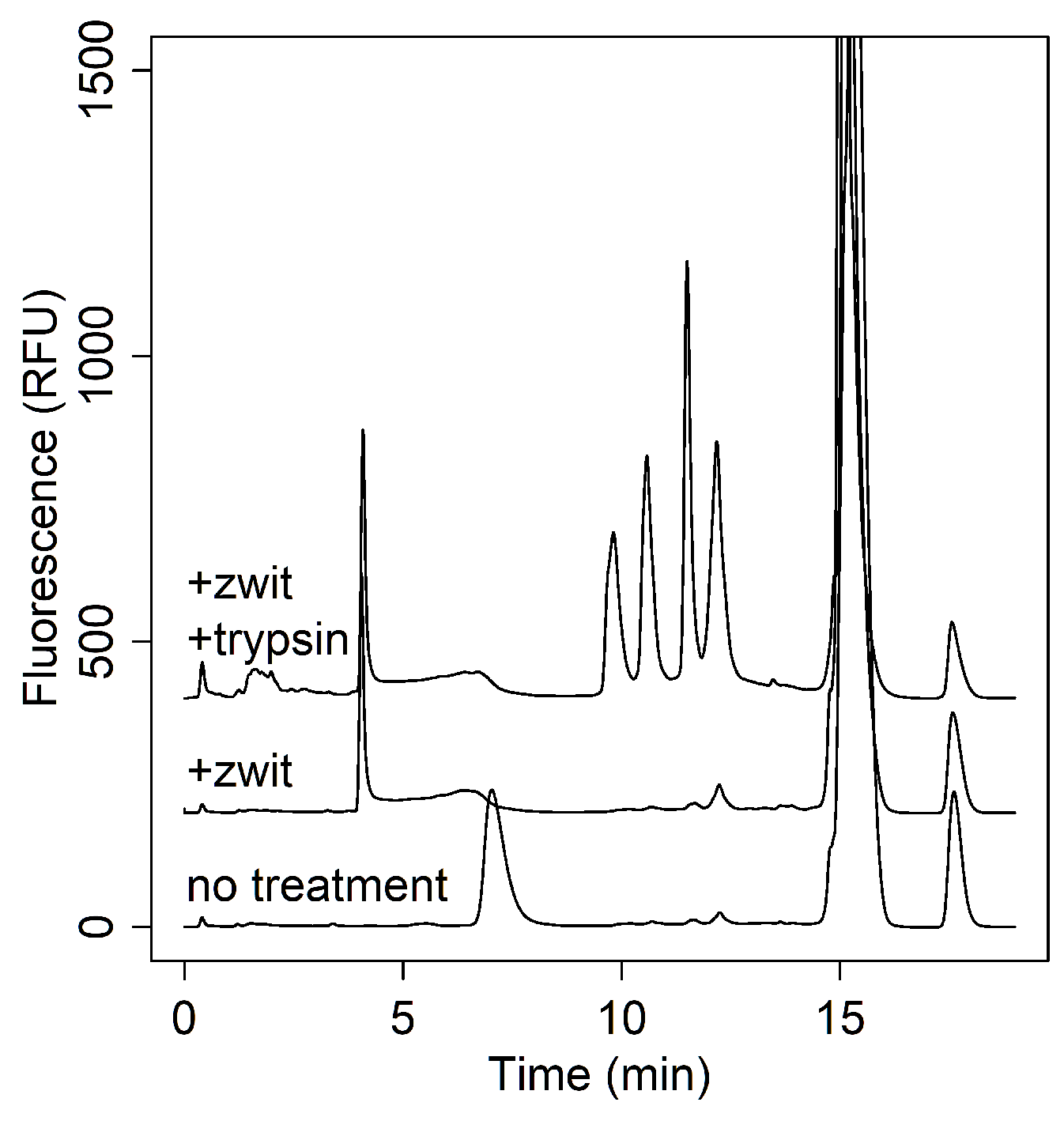
3.2.1. Excipient Interference
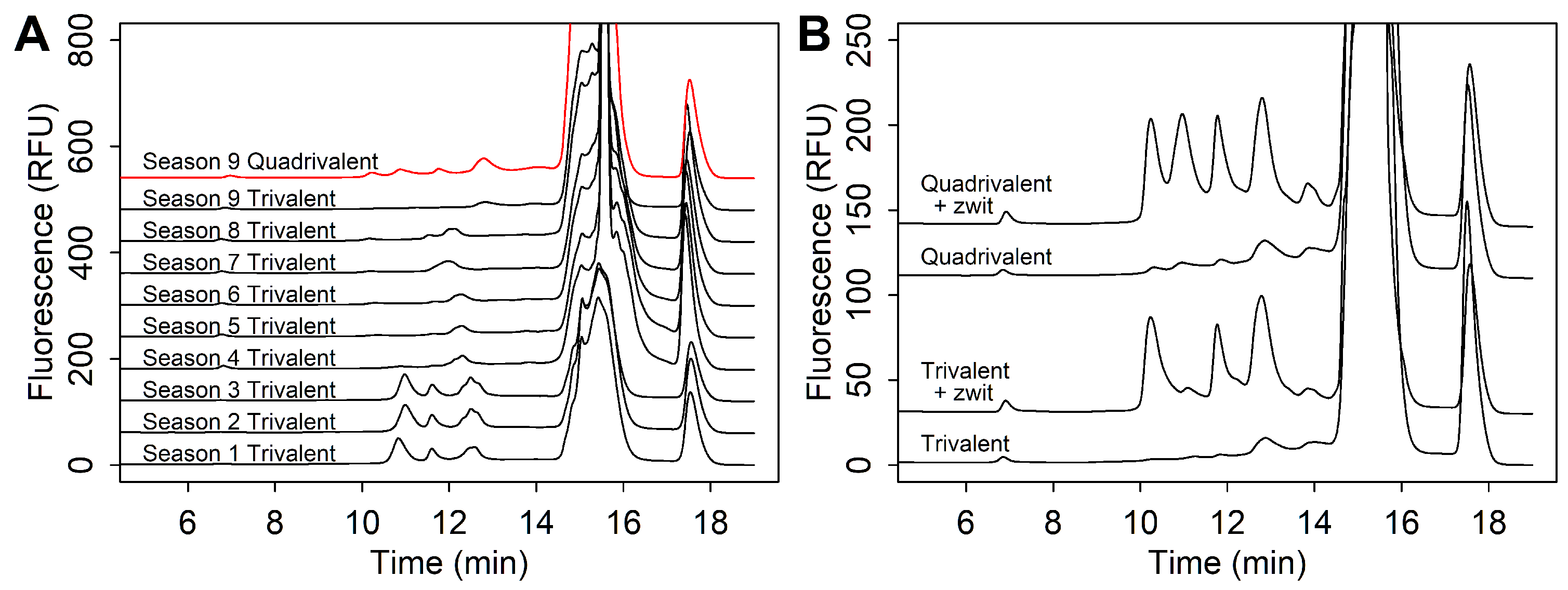
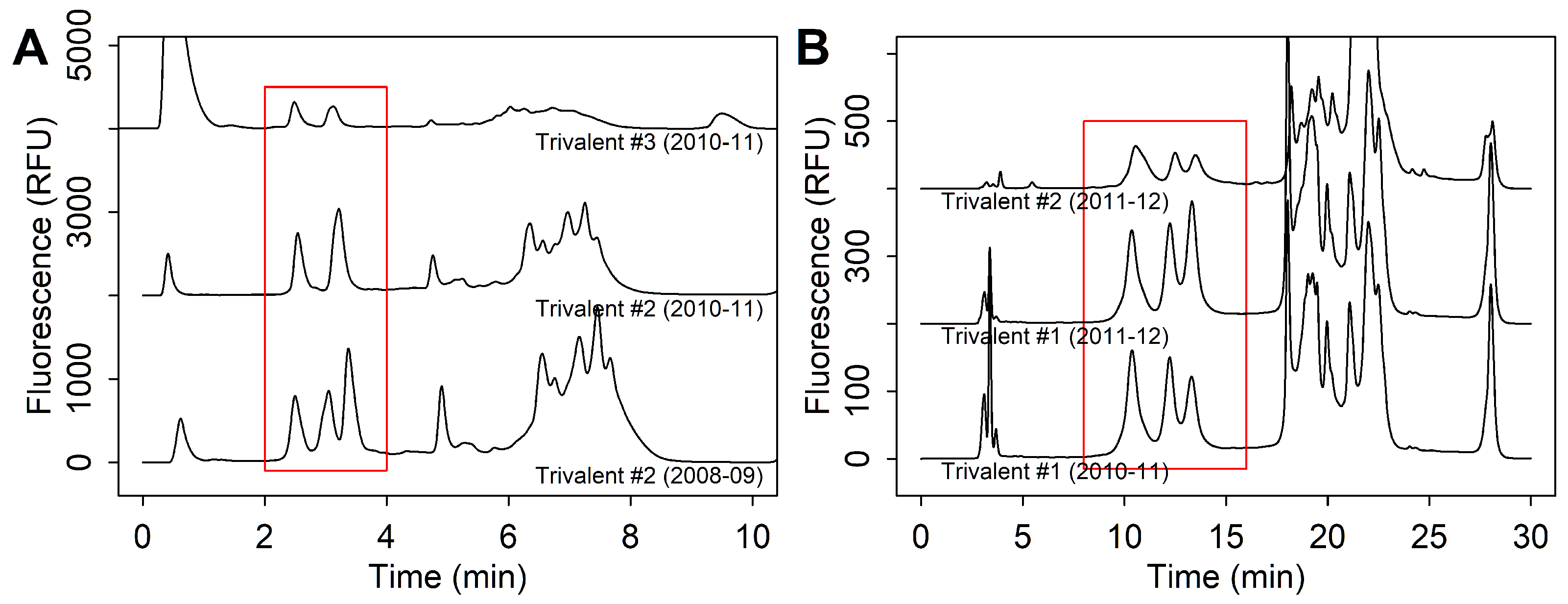
3.2.2. Strain Co-Elution
3.2.3. Recombinant Modalities
3.3. Machine Learning for Chromatographic Classification in Vaccine Quality Assessment
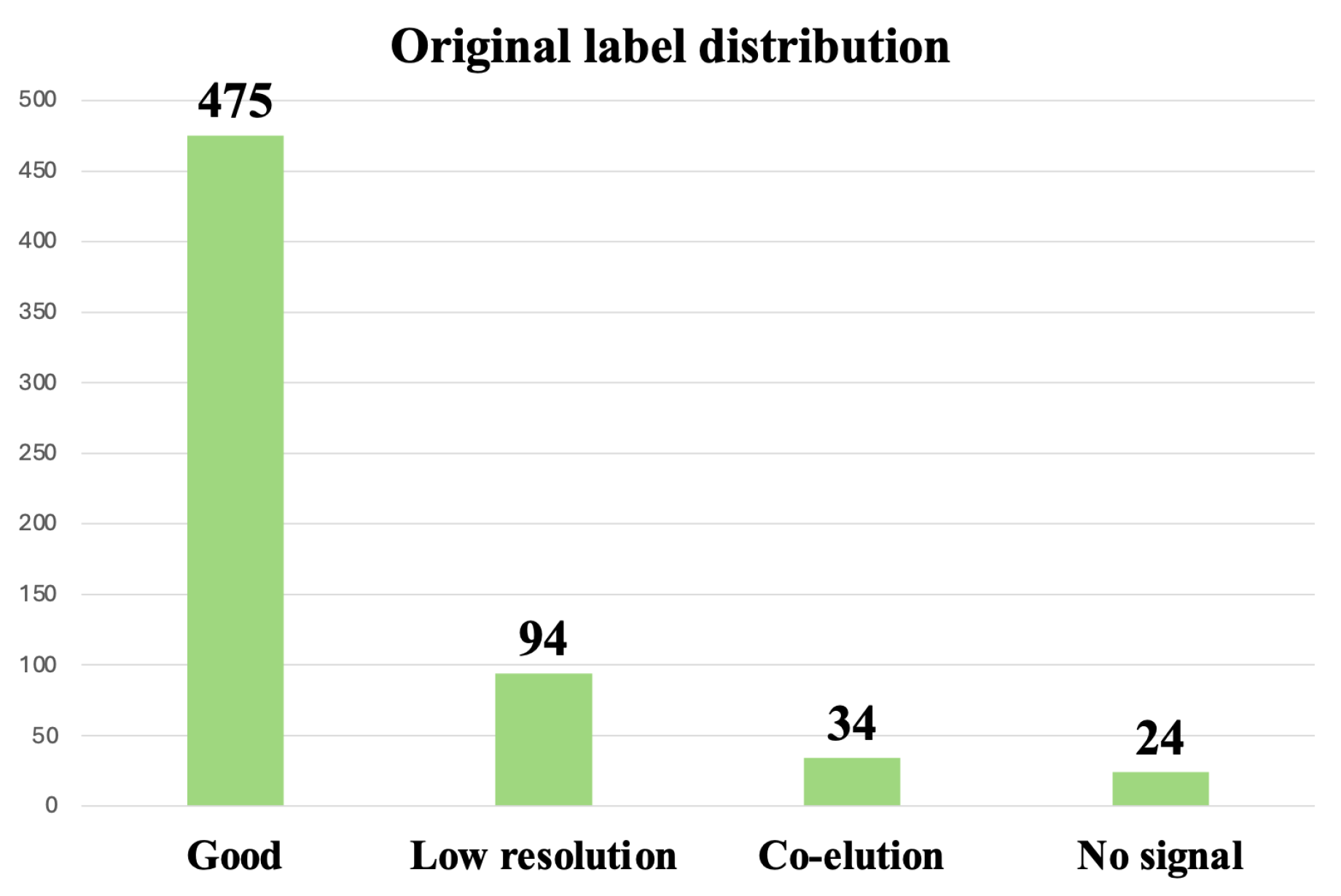
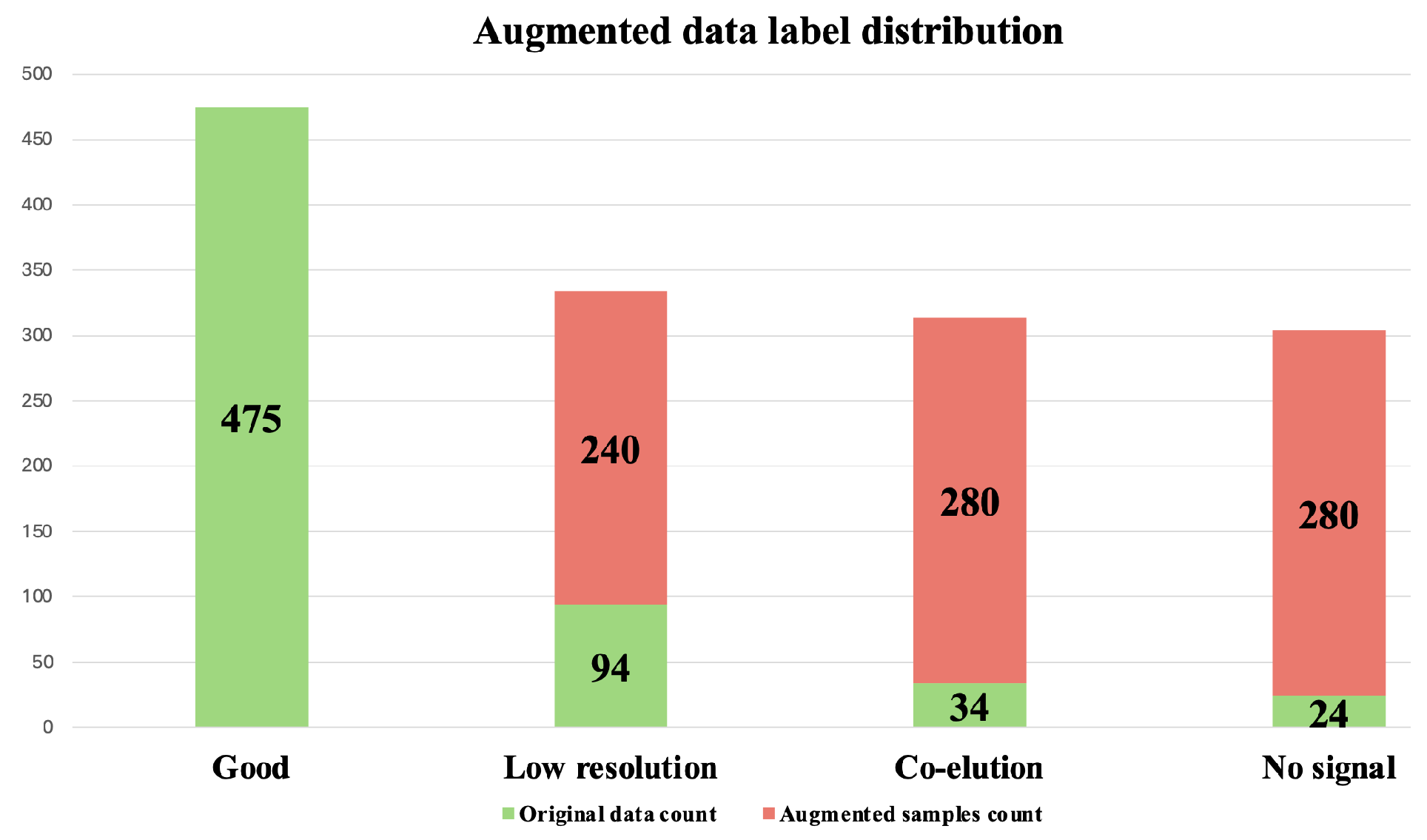
3.3.1. Oversampling and Undersampling in Addressing Class Imbalance
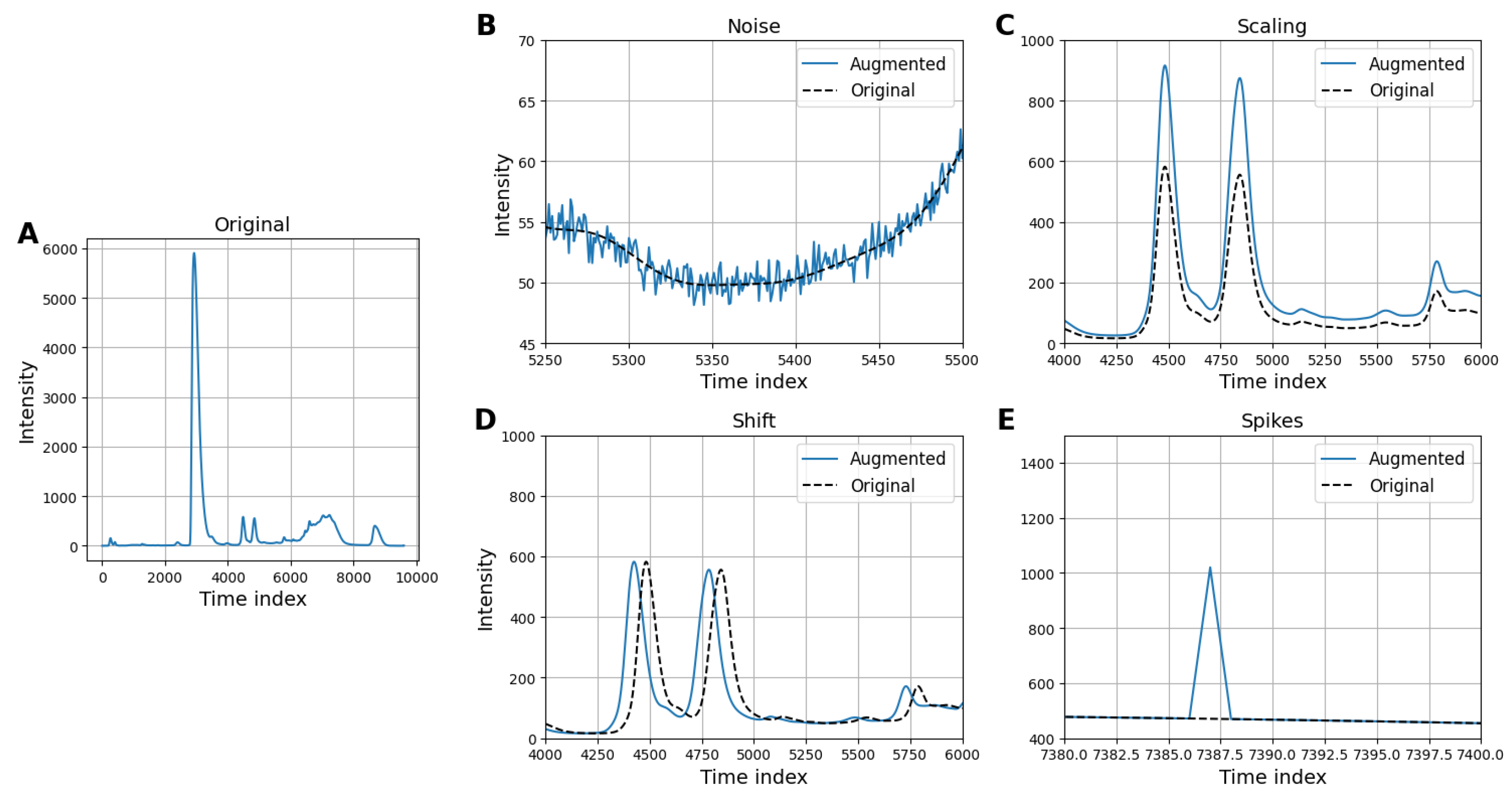
3.3.2. Data Augmentation for Addressing Class Imbalance
- Gaussian noise: Adds random noise sampled from a Gaussian distribution to simulate signal variability and improve model generalization.
- Intensity scaling: Randomly scales the intensity of the chromatogram to mimic variations in signal strength and enhance model robustness.
- Time shifting: Shifts the chromatogram along the time axis by a random amount to account for temporal variations in retention times.
- Peak spiking: Distorts the intensity of a randomly selected point in the chromatogram to simulate signal irregularities.
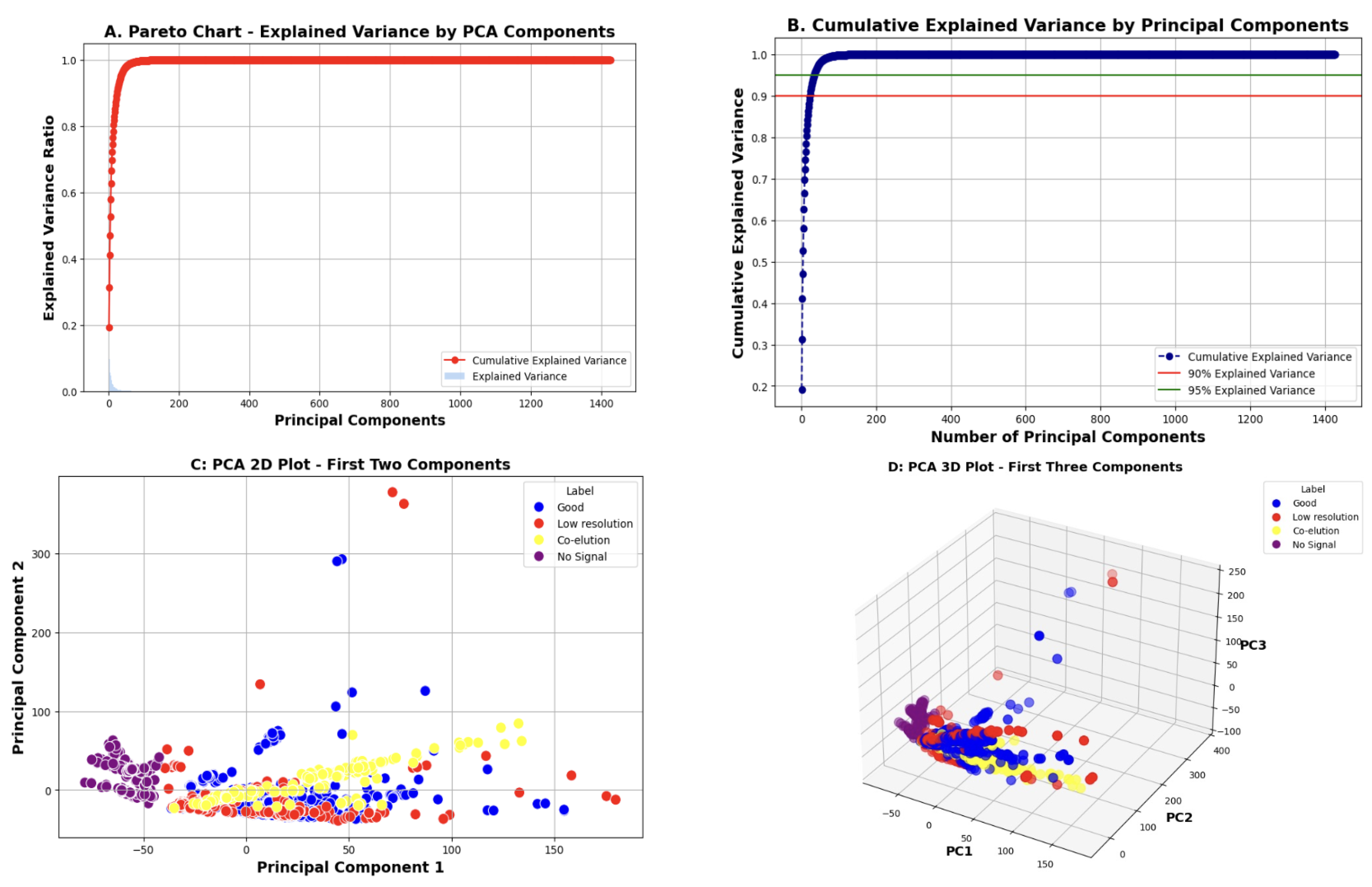
3.3.3. PCA for Dimensionality Reduction
- The Pareto chart displays the amount of variance explained by each principal component, which helps determine which components are most informative.
- The cumulative explained variance plot shows how variance accumulates as more components are added. This helps select the optimal number of components to retain while balancing accuracy and complexity.
- The 2D and 3D PCA plots project the data onto the first respective two and three principal components to visualize class separability.
3.3.4. Classification, Model Performance, and Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACN | Acetonitrile |
| DTT | Dithiothreitol |
| HA | Hemagglutinin |
| HPLC | High-Performance Liquid Chromatography |
| ML | Machine Learning |
| PCA | Principal Component Analysis |
| RPLC | Reversed-Phase Liquid Chromatography |
| RP | Reversed-Phase |
| SRID | Single Radial Immunodiffusion |
| TFA | trifluoroacetic acid |
| TPCK | L-1-tosylamido-2-phenylethyl chloromethyl ketone |
References
- Weir, J.P.; Gruber, M.F. An overview of the regulation of influenza vaccines in the United States. Influenza Other Respir. Viruses 2016, 10, 354–360. [Google Scholar] [CrossRef]
- Harding, A.T.; Heaton, N.S. Efforts to Improve the Seasonal Influenza Vaccine. Vaccines 2018, 6, 19. [Google Scholar] [CrossRef]
- Chen, J.R.; Liu, Y.M.; Tseng, Y.C.; Ma, C. Better influenza vaccines: An industry perspective. J. Biomed. Sci. 2020, 27, 33. [Google Scholar] [CrossRef]
- Sampath, S.; Khedr, A.; Qamar, S.; Tekin, A.; Singh, R.; Green, R.; Kashyap, R. Pandemics Throughout the History. Cureus 2021, 13, e18136. [Google Scholar] [CrossRef]
- Ekimov, A.; Arunachalam, A.B.; Blake, T.; Bodle, J.; Couzens, L.; Dubey, S.; Eichelberger, M.; Engelhardt, O.G.; Gubinelli, F.; Joshi, M.; et al. Assessing the stability-indicating properties of alternative potency assays for inactivated influenza vaccine. Vaccine 2023, 41, 4639–4647. [Google Scholar] [CrossRef]
- Wen, Y.; Han, L.; Palladino, G.; Ferrari, A.; Xie, Y.; Carfi, A.; Dormitzer, P.R.; Settembre, E.C. Conformationally selective biophysical assay for influenza vaccine potency determination. Vaccine 2015, 33, 5342–5349. [Google Scholar] [CrossRef]
- Pierce, C.L.; Williams, T.L.; Santana, W.I.; Levine, M.; Chen, L.M.; Cooper, H.C.; Solano, M.I.; Woolfitt, A.R.; Marasco, W.A.; Fang, H.; et al. Immunocapture isotope dilution mass spectrometry in response to a pandemic influenza threat. Vaccine 2017, 35, 5011–5018. [Google Scholar] [CrossRef]
- Serafin, B.; Kamen, A.; de Crescenzo, G.; Henry, O. Antibody-independent surface plasmon resonance assays for influenza vaccine quality control. Appl. Microbiol. Biotechnol. 2024, 108, 307. [Google Scholar] [CrossRef]
- Bodle, J.; Vandenberg, K.; Laurie, K.; Barr, I.G.; Zhang, Y.; Rockman, S. An ELISA-based assay for determining haemagglutinin potency in egg, cell, or recombinant protein derived influenza vaccines. Front. Immunol. 2023, 14, 1147028. [Google Scholar] [CrossRef]
- Schmeisser, F.; Vodeiko, G.M.; Lugovtsev, V.Y.; Stout, R.R.; Weir, J.P. An alternative method for preparation of pandemic influenza strain-specific antibody for vaccine potency determination. Vaccine 2010, 28, 2442–2449. [Google Scholar] [CrossRef]
- Narayan, K.; Paduraru, C.; Blake, T.; Arunachalam, A.B. Rapid determination of influenza vaccine potency by an SPR-based method using subtype or lineage-specific monoclonal antibodies. Front. Immunol. 2023, 14, 1128683. [Google Scholar] [CrossRef]
- Garcia-Cañas, V.; Lorbetskie, B.; Girard, M. Rapid and selective characterization of influenza virus constituents in monovalent and multivalent preparations using non-porous reversed-phase high performance liquid chromatography columns. J. Chromatogr. A 2006, 1123, 225–232. [Google Scholar] [CrossRef]
- Lorbetskie, B.; Fortin, N.; Durno, L.; Wang, J.; Li, C.; Li, X.; Girard, M.; Sauvé, S. Fast and highly selective determination of hemagglutinin content in quadrivalent influenza vaccine by reversed-phase high-performance liquid chromatography method. J. Chromatogr. A 2017, 1528, 18–24. [Google Scholar] [CrossRef]
- Lorbetskie, B.; Cunningham, A.M.; Lemieux, M.; Durno, L.; Farnsworth, A.; Wang, J.; Li, C.; Li, X.; Gilbert, M.; Sauvé, S.; et al. Selective Capture and Determination of Receptor-Binding Hemagglutinin in Influenza Vaccine Preparations Using a Coupled Receptor-Binding/RP-HPLC Assay. Anal. Chem. 2019, 91, 8908–8917. [Google Scholar] [CrossRef]
- Manceur, A.P.; Kamen, A.A. Critical review of current and emerging quantification methods for the development of influenza vaccine candidates. Vaccine 2015, 33, 5913–5919. [Google Scholar] [CrossRef]
- Kretschmer, F.; Harrieder, E.M.; Hoffmann, M.A.; Böcker, S.; Witting, M. RepoRT: A comprehensive repository for small molecule retention times. Nat. Methods 2024, 21, 153–155. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef]
- D’Atri, V.; Barrientos, R.C.; Losacco, G.L.; Rudaz, S.; Delobel, A.; Regalado, E.L.; Guillarme, D. Trends in Pharmaceutical Analysis: The Evolving Role of Liquid Chromatography. Anal. Chem. 2025, 97, 4706–4727. [Google Scholar] [CrossRef]
- Edwards-Parton, S.; Thornhill, N.F.; Bracewell, D.G.; Liddell, J.M.; Titchener-Hooker, N.J. Principal Component Score Modeling for the Rapid Description of Chromatographic Separations. Biotechnol. Prog. 2008, 24, 202–208. [Google Scholar] [CrossRef]
- Stienstra, C.M.K.; Nazdrajić, E.; Hopkins, W.S. From Reverse Phase Chromatography to HILIC: Graph Transformers Power Method-Independent Machine Learning of Retention Times. Anal. Chem. 2025, 97, 4461–4472. [Google Scholar] [CrossRef]
- Zhao, R.H.; Yue, B.F.; Ni, J.Y.; Zhou, H.F.; Zhang, Y.K. Application of an artificial neural network in chromatography—retention behavior prediction and pattern recognition. Chemom. Intell. Lab. Syst. 1999, 45, 163–170. [Google Scholar] [CrossRef]
- Singh, Y.R.; Shah, D.B.; Kulkarni, M.; Patel, S.R.; Maheshwari, D.G.; Shah, J.S.; Shah, S. Current trends in chromatographic prediction using artificial intelligence and machine learning. Anal. Methods 2023, 15, 2785–2797. [Google Scholar] [CrossRef]
- Bosten, E.; Pardon, M.; Chen, K.; Koppen, V.; Van Herck, G.; Hellings, M.; Cabooter, D. Assisted Active Learning for Model-Based Method Development in Liquid Chromatography. Anal. Chem. 2024, 96, 13699–13709. [Google Scholar] [CrossRef]
- Marchetto, A.; Tirapelle, M.; Mazzei, L.; Sorensen, E.; Besenhard, M.O. In Silico High-Performance Liquid Chromatography Method Development via Machine Learning. Anal. Chem. 2025, 97, 6991–7001. [Google Scholar] [CrossRef]
- Reuschenbach, M.; Drees, F.; Leupold, M.S.; Tintrop, L.K.; Schmidt, T.C.; Renner, G. qPeaks: A linear regression-based asymmetric peak model for parameter-free automatized detection and characterization of chromatographic peaks in non-target screening data. Anal. Chem. 2024, 96, 7120–7129. [Google Scholar] [CrossRef]
- Lee, C.C.; Lin, Y.C.; Pan, T.Y.; Yang, C.H.; Li, P.H.; Chen, S.Y.; Gao, J.J.; Yang, C.; Chu, L.J.; Huang, P.J.; et al. HeapMS: An Automatic Peak-Picking Pipeline for Targeted Proteomic Data Powered by 2D Heatmap Transformation and Convolutional Neural Networks. Anal. Chem. 2023, 95, 15486–15496. [Google Scholar] [CrossRef]
- Toghi Eshghi, S.; Auger, P.; Mathews, W.R. Quality assessment and interference detection in targeted mass spectrometry data using machine learning. Clin. Proteom. 2018, 15, 33. [Google Scholar] [CrossRef]
- Lorbetskie, B.; Wang, J.; Gravel, C.; Allen, C.; Walsh, M.; Rinfret, A.; Li, X.; Girard, M. Optimization and qualification of a quantitative reversed-phase HPLC method for hemagglutinin in influenza preparations and its comparative evaluation with biochemical assays. Vaccine 2011, 29, 3377–3389. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, C.; Ke, L.; Hayes, N.; Zhu, Y.; Qiu, H.; Zhang, B.; Zhou, T.; Wei, G.W. A review of machine learning methods for imbalanced data challenges in chemistry. Chem. Sci. 2025, 16, 7637–7658. [Google Scholar] [CrossRef]
- Iglesias, G.; Talavera, E.; González-Prieto, Á.; Mozo, A.; Gómez-Canaval, S. Data augmentation techniques in time series domain: A survey and taxonomy. Neural Comput. Appl. 2023, 35, 10123–10145. [Google Scholar] [CrossRef]
- Cordella, C.B. PCA: The basic building block of chemometrics. Anal. Chem. 2012, 47, 567. [Google Scholar]
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4. [Google Scholar]
- Ghasriani, H.; Frahm, G.E.; Johnston, M.J.W.; Aubin, Y. Effects of Excipients on the Structure and Dynamics of Filgrastim Monitored by Thermal Unfolding Studies by CD and NMR Spectroscopy. ACS Omega 2020, 5, 31845–31857. [Google Scholar] [CrossRef] [PubMed]
- Barberis, I.; Myles, P.; Ault, S.; Bragazzi, N.; Martini, M. History and evolution of influenza control through vaccination: From the first monovalent vaccine to universal vaccines. J. Prev. Med. Hyg. 2016, 57, E115. [Google Scholar] [PubMed]
- Pate, M.; Thornhill, N.; Chandwani, R.; Hoare, M.; Titchener-Hooker, N. Pre-processing of chromatographic data for principal component analysis. Bioprocess Eng. 1998, 19, 297–305. [Google Scholar] [CrossRef]
- Wood, J.M.; Weir, J.P. Standardisation of inactivated influenza vaccines—Learning from history. Influenza Other Respir. Viruses 2018, 12, 195–201. [Google Scholar] [CrossRef]
| Variable | Value |
|---|---|
| Years | 2009–2023 |
| No. of products | 22 |
| Reference standard sources | NIBSC, TGA, CBER |
| No. injections * | 13,454 |
| No. influenza A strains analyzed | 18 |
| No. influenza B strains analyzed | 8 |
| Influenza A subtypes | H1N1; H3N2; H7N9; |
| Influenza B lineages | Victoria; Yamagata; |
| Columns used | 15 |
| Methods | 438 |
| Mobile phase A | Water, 0.04–0.1% TFA |
| Mobile phase B | 25% ACN, 75% 2-propanol, 0.03% to 0.1% TFA |
| Injection volumes | 0.5–150 µL |
| Column temps | 25–65 °C |
| Method | Accuracy | Precision | Recall | F1 Score |
|---|---|---|---|---|
| Random Forest | 0.968 | 0.961 | 0.960 | 0.960 |
| Support Vector Machine | 0.893 | 0.902 | 0.893 | 0.891 |
| Logistic Regression | 0.911 | 0.912 | 0.911 | 0.911 |
| K-Nearest Neighbors | 0.920 | 0.923 | 0.920 | 0.920 |
| Decision Tree | 0.921 | 0.922 | 0.921 | 0.921 |
| Naive Bayes | 0.798 | 0.812 | 0.798 | 0.790 |
| Gradient Boosting | 0.953 | 0.954 | 0.953 | 0.953 |
| XGBoost | 0.960 | 0.960 | 0.960 | 0.960 |
| AdaBoost | 0.876 | 0.881 | 0.876 | 0.877 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorbetskie, B.; Manouchehri, N.; Girard, M.; Sauvé, S.; Lu, H. Advancing Reversed-Phase Chromatography Analytics of Influenza Vaccines Using Machine Learning Approaches on a Diverse Range of Antigens and Formulations. Vaccines 2025, 13, 820. https://doi.org/10.3390/vaccines13080820
Lorbetskie B, Manouchehri N, Girard M, Sauvé S, Lu H. Advancing Reversed-Phase Chromatography Analytics of Influenza Vaccines Using Machine Learning Approaches on a Diverse Range of Antigens and Formulations. Vaccines. 2025; 13(8):820. https://doi.org/10.3390/vaccines13080820
Chicago/Turabian StyleLorbetskie, Barry, Narges Manouchehri, Michel Girard, Simon Sauvé, and Huixin Lu. 2025. "Advancing Reversed-Phase Chromatography Analytics of Influenza Vaccines Using Machine Learning Approaches on a Diverse Range of Antigens and Formulations" Vaccines 13, no. 8: 820. https://doi.org/10.3390/vaccines13080820
APA StyleLorbetskie, B., Manouchehri, N., Girard, M., Sauvé, S., & Lu, H. (2025). Advancing Reversed-Phase Chromatography Analytics of Influenza Vaccines Using Machine Learning Approaches on a Diverse Range of Antigens and Formulations. Vaccines, 13(8), 820. https://doi.org/10.3390/vaccines13080820






