Lineage Replacement and Genetic Changes of Four HR-HPV Types during the Period of Vaccine Coverage: A Six-Year Retrospective Study in Eastern China
Abstract
1. Introduction
2. Methods
2.1. Study Population and Sample Collection
2.2. Viral DNA Amplification and L1 Gene Sequencing
2.3. Phylogenetic Analysis and Sequence Alignment
2.4. Homology Modeling
2.5. Statistical Analysis
3. Results
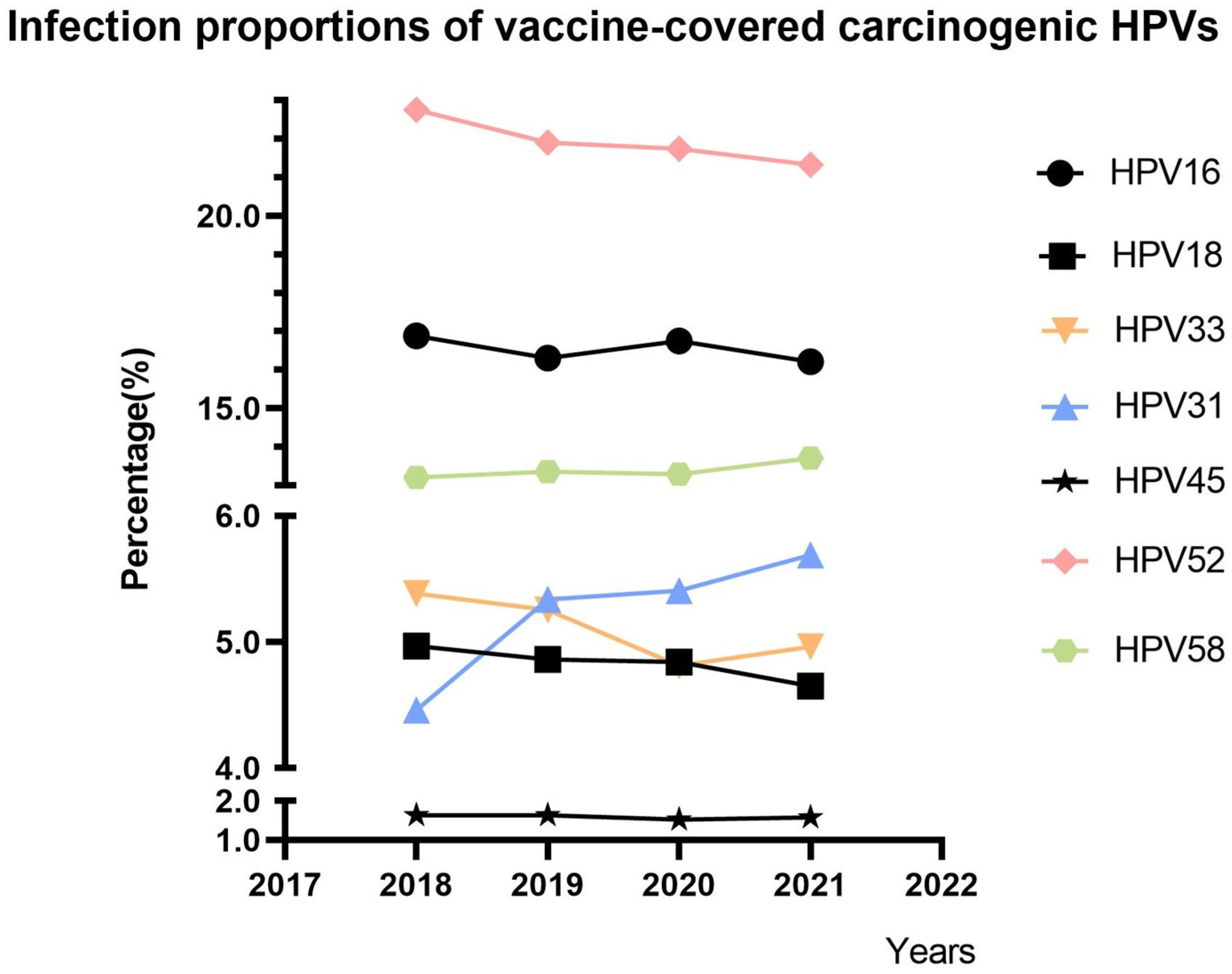
3.1. Trends in the Proportions of Vaccine-Covered Carcinogenic HPVs
3.2. Characteristics of HPV31-, 33-, 52-, and 58-Positive Patients
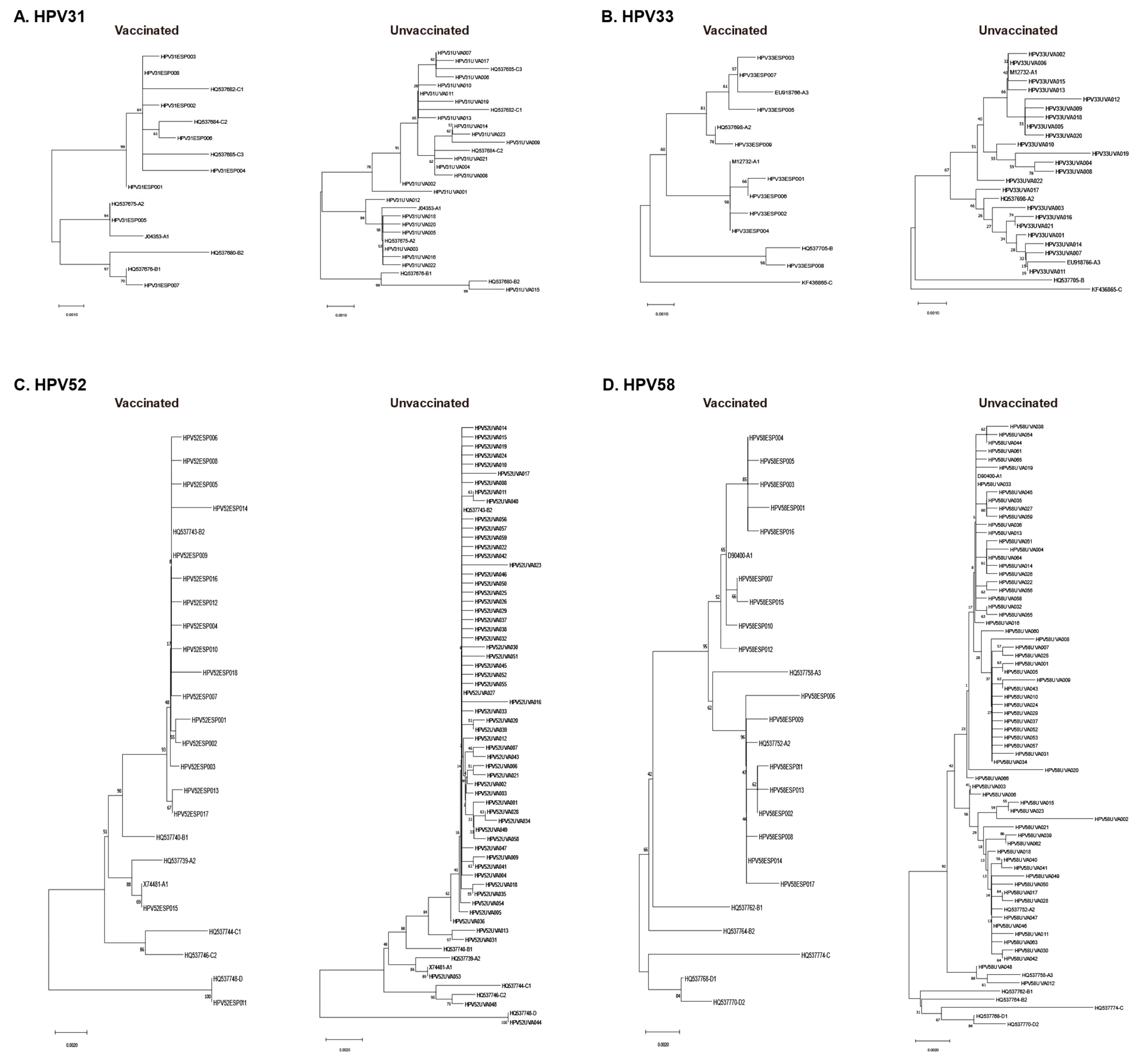
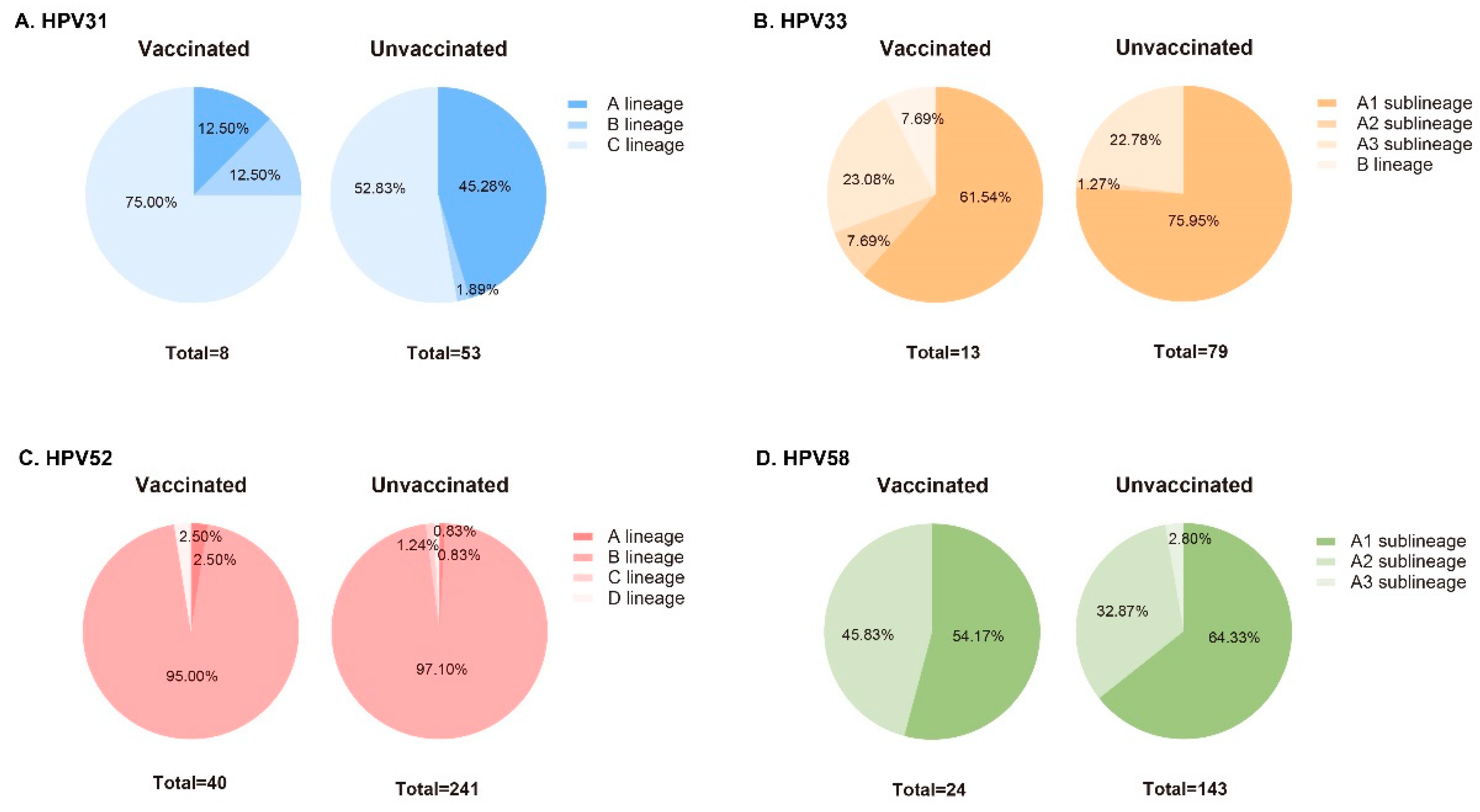
3.3. Phylogenetic Analysis of L1 Sequences
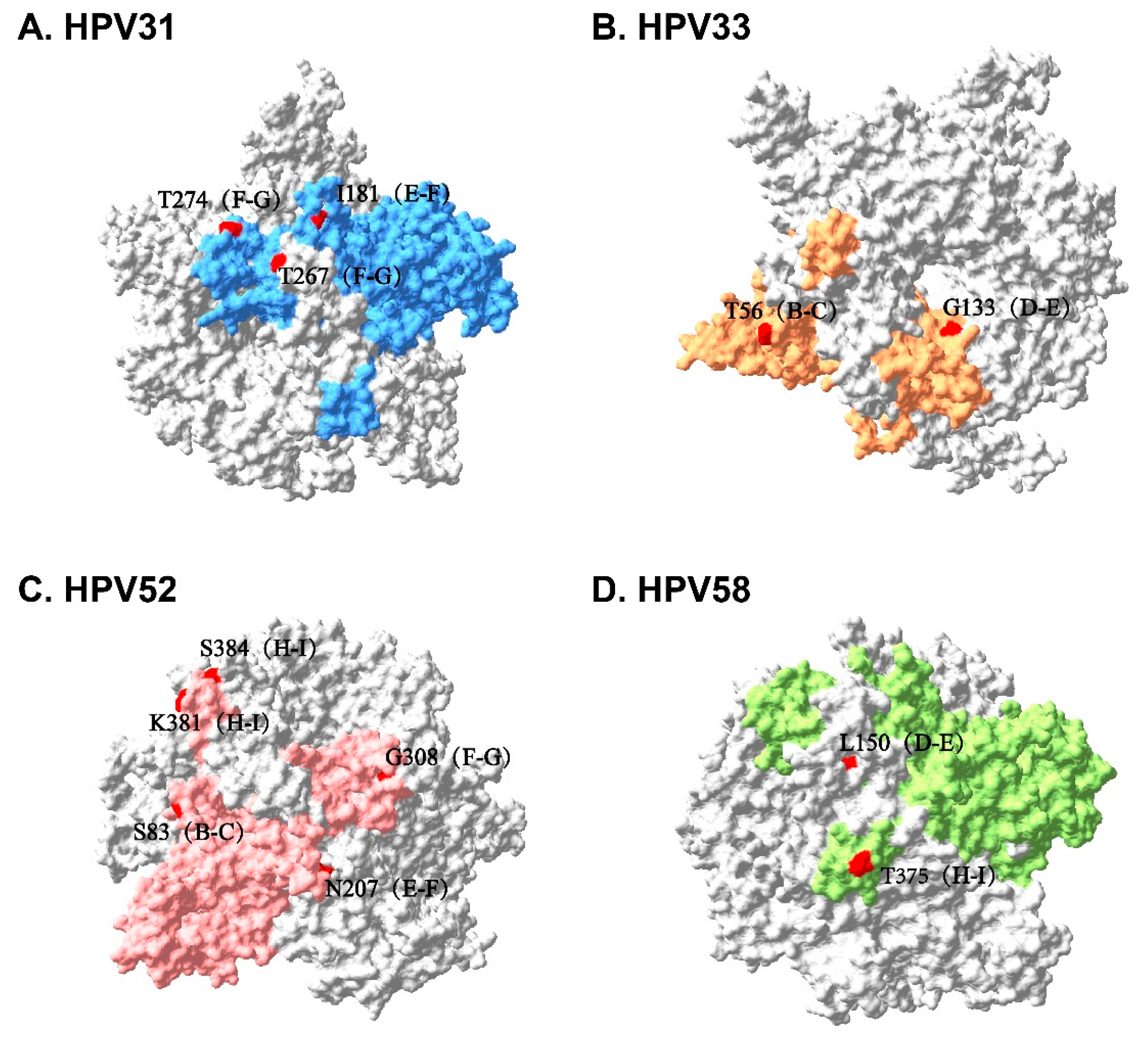
3.4. Sequence Alignment and Protein Structure Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [PubMed]
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Collado, J.J.; Gómez, D.; Muñoz, J.; Bosch, F.X.; de Sanjosé, S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in the World; Summary Report; ICO/IARC HPV Information Centre: Barcelona, Spain, 22 October 2021. [Google Scholar]
- Williamson, A.L. Recent Developments in Human Papillomavirus (HPV) Vaccinology. Viruses 2023, 15, 1440. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.U.; Burk, R.D.; Chen, Z.; Van Doorslaer, K.; Zur Hausen, H.; De Villiers, E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010, 401, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.D.; Harari, A.; Chen, Z. Human papillomavirus genome variants. Virology 2013, 445, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Sui, L.; Li, Y. Vaccine escape challenges virus prevention: The example of two vaccine-preventable oncogenic viruses. J. Med. Virol. 2023, 95, e29184. [Google Scholar] [CrossRef] [PubMed]
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Collado, J.J.; Gómez, D.; Muñoz, J.; Bosch, F.X.; de Sanjosé, S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in China; Summary Report; ICO/IARC HPV Information Centre: Barcelona, Spain, 22 October 2021. [Google Scholar]
- Wang, R.; Pan, W.; Jin, L.; Huang, W.; Li, Y.; Wu, D.; Gao, C.; Ma, D.; Liao, S. Human papillomavirus vaccine against cervical cancer: Opportunity and challenge. Cancer Lett. 2020, 471, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.T.; Lowy, D.R. Papillomavirus-like particles and HPV vaccine development. Semin. Cancer Biol. 1996, 7, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Scherer, E.M.; Smith, R.A.; Simonich, C.A.; Niyonzima, N.; Carter, J.J.; Galloway, D.A. Characteristics of memory B cells elicited by a highly efficacious HPV vaccine in subjects with no pre-existing immunity. PLoS Pathog. 2014, 10, e1004461. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nygård, M.; Saah, A.; Munk, C.; Tryggvadottir, L.; Enerly, E.; Hortlund, M.; Sigurdardottir, L.G.; Vuocolo, S.; Kjaer, S.K.; Dillner, J. Evaluation of the Long-Term Anti-Human Papillomavirus 6 (HPV6), 11, 16, and 18 Immune Responses Generated by the Quadrivalent HPV Vaccine. Clin. Vaccine Immunol. CVI 2015, 22, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Roteli-Martins, C.M.; Naud, P.; De Borba, P.; Teixeira, J.C.; De Carvalho, N.S.; Zahaf, T.; Sanchez, N.; Geeraerts, B.; Descamps, D. Sustained immunogenicity and efficacy of the HPV-16/18 AS04-adjuvanted vaccine: Up to 8.4 years of follow-up. Hum. Vaccines Immunother. 2012, 8, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Bishop, B.; Dasgupta, J.; Klein, M.; Garcea, R.L.; Christensen, N.D.; Zhao, R.; Chen, X.S. Crystal structures of four types of human papillomavirus L1 capsid proteins: Understanding the specificity of neutralizing monoclonal antibodies. J. Biol. Chem. 2007, 282, 31803–31811. [Google Scholar] [CrossRef] [PubMed]
- Naud, P.S.; Roteli-Martins, C.M.; De Carvalho, N.S.; Teixeira, J.C.; De Borba, P.C.; Sanchez, N.; Zahaf, T.; Catteau, G.; Geeraerts, B.; Descamps, D. Sustained efficacy, immunogenicity, and safety of the HPV-16/18 AS04-adjuvanted vaccine: Final analysis of a long-term follow-up study up to 9.4 years post-vaccination. Hum. Vaccines Immunother. 2014, 10, 2147–2162. [Google Scholar] [CrossRef] [PubMed]
- Porras, C.; Tsang, S.H.; Herrero, R.; Guillén, D.; Darragh, T.M.; Stoler, M.H.; Hildesheim, A.; Wagner, S.; Boland, J.; Lowy, D.R.; et al. Efficacy of the bivalent HPV vaccine against HPV 16/18-associated precancer: Long-term follow-up results from the Costa Rica Vaccine Trial. Lancet Oncol. 2020, 21, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Bruni, L.; Saura-Lázaro, A.; Montoliu, A.; Brotons, M.; Alemany, L.; Diallo, M.S.; Afsar, O.Z.; LaMontagne, D.S.; Mosina, L.; Contreras, M.; et al. HPV vaccination introduction worldwide and WHO and UNICEF estimates of national HPV immunization coverage 2010–2019. Prev. Med. 2021, 144, 106399. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Ploner, A.; Elfström, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundström, K.; Dillner, J.; Sparén, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Drolet, M.; Bénard, E.; Boily, M.-C.; Ali, H.; Baandrup, L.; Bauer, H.; Beddows, S.; Brisson, J.; Brotherton, J.M.L.; Cummings, T.; et al. Population-level impact and herd effects following human papillomavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2015, 15, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. The logic of virus evolution. Cell Host Microbe 2022, 30, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Horvath, C.A.; Boulet, G.A.; Renoux, V.M.; Delvenne, P.O.; Bogers, J.P. Mechanisms of cell entry by human papillomaviruses: An overview. Virol. J. 2010, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.Y.; Chang, M.H.; Ni, Y.H.; Chen, H.L. Survey of hepatitis B surface variant infection in children 15 years after a nationwide vaccination programme in Taiwan. Gut 2004, 53, 1499–1503. [Google Scholar] [CrossRef] [PubMed]
- Konopleva, M.V.; Borisova, V.N.; Sokolova, M.V.; Semenenko, T.A.; Suslov, A.P. Recombinant HBsAg of the Wild-Type and the G145R Escape Mutant, included in the New Multivalent Vaccine against Hepatitis B Virus, Dramatically Differ in their Effects on Leukocytes from Healthy Donors In Vitro. Vaccines 2022, 10, 235. [Google Scholar] [CrossRef] [PubMed]
- Gurgel, A.P.A.D.; Chagas, B.S.; Amaral, C.M.D.; Nascimento, K.C.G.; Leal, L.R.S.; Neto, J.d.C.S.; Muniz, M.T.C.; de Freitas, A.C. Prevalence of human papillomavirus variants and genetic diversity in the L1 gene and long control region of HPV16, HPV31, and HPV58 found in North-East Brazil. Biomed. Res. Int. 2015, 2015, 130828. [Google Scholar] [CrossRef][Green Version]
- Chen, Z.; Jing, Y.; Wen, Q.; Ding, X.; Zhang, S.; Wang, T.; Zhang, Y.; Zhang, J. L1 and L2 gene polymorphisms in HPV-58 and HPV-33: Implications for vaccine design and diagnosis. Virol. J. 2016, 13, 167. [Google Scholar] [CrossRef] [PubMed]
- Oumeslakht, L.; Ababou, M.; Badaoui, B.; Qmichou, Z. Worldwide genetic variations in high-risk human papillomaviruses capsid L1 gene and their impact on vaccine efficiency. Gene 2021, 782, 145533. [Google Scholar] [CrossRef] [PubMed]
- Godi, A.; Boampong, D.; Elegunde, B.; Panwar, K.; Fleury, M.; Li, S.; Zhao, Q.; Xia, N.; Christensen, N.D.; Beddows, S. Comprehensive Assessment of the Antigenic Impact of Human Papillomavirus Lineage Variation on Recognition by Neutralizing Monoclonal Antibodies Raised against Lineage A Major Capsid Proteins of Vaccine-Related Genotypes. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Godi, A.; Bissett, S.L.; Masloh, S.; Fleury, M.; Li, S.; Zhao, Q.; Xia, N.; Cocuzza, C.E.; Beddows, S. Impact of naturally occurring variation in the human papillomavirus 52 capsid proteins on recognition by type-specific neutralising antibodies. J. Gen. Virol. 2019, 100, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Godi, A.; Kemp, T.J.; Pinto, L.A.; Beddows, S. Sensitivity of Human Papillomavirus (HPV) Lineage and Sublineage Variant Pseudoviruses to Neutralization by Nonavalent Vaccine Antibodies. J. Infect. Dis. 2019, 220, 1940–1945. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zheng, W.; Li, Y.; Pearce, R.; Zhang, C.; Bell, E.W.; Zhang, G.; Zhang, Y. I-TASSER-MTD: A deep-learning-based platform for multi-domain protein structure and function prediction. Nat. Protoc. 2022, 17, 2326–2353. [Google Scholar] [CrossRef]
- Shilling, H.; Garland, S.M.; Atchison, S.; Cornall, A.M.; Brotherton, J.M.; Bateson, D.; McNamee, K.; Kaldor, J.M.; Hocking, J.S.; Chen, M.Y.; et al. Human papillomavirus prevalence and risk factors among Australian women 9-12 years after vaccine program introduction. Vaccine 2021, 39, 4856–4863. [Google Scholar] [CrossRef] [PubMed]
- Schlecht, N.F.; Diaz, A.; Nucci-Sack, A.; Shyhalla, K.; Shankar, V.; Guillot, M.; Hollman, D.; Strickler, H.D.; Burk, R.D. Incidence and Types of Human Papillomavirus Infections in Adolescent Girls and Young Women Immunized With the Human Papillomavirus Vaccine. JAMA Netw. Open 2021, 4, e2121893. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, M.; Doorbar, J.; Wentzensen, N.; de Sanjosé, S.; Fakhry, C.; Monk, B.J.; Stanley, M.A.; Franceschi, S. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers 2016, 2, 16086. [Google Scholar] [CrossRef] [PubMed]
- Bee, K.J.; Gradissimo, A.; Chen, Z.; Harari, A.; Schiffman, M.; Raine-Bennett, T.; Castle, P.E.; Clarke, M.; Wentzensen, N.; Burk, R.D. Genetic and Epigenetic Variations of HPV52 in Cervical Precancer. Int. J. Mol. Sci. 2021, 22, 6463. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ho, W.C.S.; Boon, S.S.; Law, P.T.Y.; Chan, M.C.W.; DeSalle, R.; Burk, R.D.; Chan, P.K.S. Ancient Evolution and Dispersion of Human Papillomavirus 58 Variants. J. Virol. 2017, 91, e01285-17. [Google Scholar] [CrossRef] [PubMed]
- Tsakogiannis, D.; Nikolaidis, M.; Zagouri, F.; Zografos, E.; Kottaridi, C.; Kyriakopoulou, Z.; Tzioga, L.; Markoulatos, P.; Amoutzias, G.D.; Bletsa, G. Mutation Profile of HPV16 L1 and L2 Genes in Different Geographic Areas. Viruses 2022, 15, 141. [Google Scholar] [CrossRef] [PubMed]
- Cornut, G.; Gagnon, S.; Hankins, C.; Money, D.; Pourreaux, K.; Franco, E.L.; Coutlée, F. Polymorphism of the capsid L1 gene of human papillomavirus types 31, 33, and 35. J. Med. Virol. 2010, 82, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-H.; Lu, Z.-T.; Wang, G.-L.; Zhou, W.-Q.; Liu, C.; Yang, L.-X.; Sun, Z.-R.; Ruan, Q. Variations of human papillomavirus type 58 E6, E7, L1 genes and long control region in strains from women with cervical lesions in Liaoning province, China. Infect Genet. Evol. 2012, 12, 1466–1472. [Google Scholar] [CrossRef]
- McClements, W.L.; Wang, X.-M.; Ling, J.C.; Skulsky, D.M.; Christensen, N.D.; Jansen, K.U.; Ludmerer, S.W. A novel human papillomavirus type 6 neutralizing domain comprising two discrete regions of the major capsid protein L1. Virology 2001, 289, 262–268. [Google Scholar] [CrossRef] [PubMed][Green Version]

| HPV Type | Gene | Primer Sequences |
|---|---|---|
| HPV31 | L1 | 5′→3′ gacgtaaacgtgtatcatatttttttacag 3′ →5′ atacagacacatgtattacatacacatc |
| HPV33 | L1 | 5′→3′ ggcgtaaacgttttccatatttt 3′ →5′ aacaacaacataacacaattacacaa |
| HPV52 | L1 | 5′→3′ ctgacattccattaccttcgttac 3′ →5′ caatggttaccttttaacctgtgtt |
| HPV58 | L1 | 5′→3′ gaacctggtccagacattgca 3′ →5′ catacaacatatacacaaacataaacaa |
| HPV Types | Lineage A | Lineage B | Lineage C | Lineage D |
|---|---|---|---|---|
| HPV31 | A1: J04353 A2: HQ537675 | B1: HQ537676 B2: HQ537680 | C1: HQ537682 C2: HQ537684 C3: HQ537685 | |
| HPV33 | A1: M12732 A2: HQ537698 A3: EU918766 | B: HQ537705 | C: KF436865 | |
| HPV52 | A1: X74481 A2: HQ537739 | B1: HQ537740 B2: HQ537743 | C1: HQ537744 C2: HQ537746 | D: HQ537748 |
| HPV58 | A1: D90400 A2: HQ537752 A3: HQ537758 | B1: HQ537762 B2: HQ537764 | C: HQ537774 | D1: HQ537768 D2: HQ537770 |
| Characteristics | Group | No. | % |
|---|---|---|---|
| HPV Genotype (n = 601) | HPV31 | 61 | 10.1 |
| HPV33 | 92 | 15.3 | |
| HPV52 | 281 | 46.8 | |
| HPV58 | 167 | 27.8 | |
| TCT Result (n = 558) | Normal | 300 | 53.8 |
| ASC-US | 125 | 22.4 | |
| LSIL | 92 | 16.5 | |
| HSIL | 23 | 4.1 | |
| ASC-H | 10 | 1.8 | |
| SCC | 1 | 0.2 | |
| LSIL-HSIL | 5 | 0.9 | |
| HSIL-AGC | 2 | 0.4 | |
| Vaccination (n = 565) | No | 478 | 84.6 |
| Yes | 87 | 15.4 | |
| Vaccination Type (n = 87) | Bivalent | 9 | 10.3 |
| Quadrivalent | 36 | 41.4 | |
| Nonvalent | 40 | 46.0 | |
| Undefined | 2 | 2.3 | |
| Multiple Infection (n = 501) | No | 272 | 54.3 |
| Yes | 229 | 45.7 | |
| Persistent Infection > 24 months (n = 515) | No | 309 | 60.0 |
| Yes | 206 | 40.0 | |
| Pathological Diagnosis of Biopsy (n = 544) | Normal | 13 | 2.4 |
| Inflammation | 203 | 37.3 | |
| LSIL | 261 | 48.0 | |
| HSIL | 64 | 11.8 | |
| SCC | 1 | 0.2 | |
| AIS | 1 | 0.2 | |
| AC | 1 | 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, W.; Hua, C.; Wang, Y.; Wang, Y.; Zhang, L.; Wang, Z.; Shi, W.; Chen, F.; Wu, Z.; Wang, Q.; et al. Lineage Replacement and Genetic Changes of Four HR-HPV Types during the Period of Vaccine Coverage: A Six-Year Retrospective Study in Eastern China. Vaccines 2024, 12, 411. https://doi.org/10.3390/vaccines12040411
Qu W, Hua C, Wang Y, Wang Y, Zhang L, Wang Z, Shi W, Chen F, Wu Z, Wang Q, et al. Lineage Replacement and Genetic Changes of Four HR-HPV Types during the Period of Vaccine Coverage: A Six-Year Retrospective Study in Eastern China. Vaccines. 2024; 12(4):411. https://doi.org/10.3390/vaccines12040411
Chicago/Turabian StyleQu, Wenjie, Chen Hua, Yaping Wang, Yan Wang, Lu Zhang, Zhiheng Wang, Wenqian Shi, Fang Chen, Zhiyong Wu, Qian Wang, and et al. 2024. "Lineage Replacement and Genetic Changes of Four HR-HPV Types during the Period of Vaccine Coverage: A Six-Year Retrospective Study in Eastern China" Vaccines 12, no. 4: 411. https://doi.org/10.3390/vaccines12040411
APA StyleQu, W., Hua, C., Wang, Y., Wang, Y., Zhang, L., Wang, Z., Shi, W., Chen, F., Wu, Z., Wang, Q., Lu, L., Jiang, S., Sui, L., & Li, Y. (2024). Lineage Replacement and Genetic Changes of Four HR-HPV Types during the Period of Vaccine Coverage: A Six-Year Retrospective Study in Eastern China. Vaccines, 12(4), 411. https://doi.org/10.3390/vaccines12040411








