Design and Characterization of a New Formulation for the Delivery of COVID-19-mRNA Vaccine to the Nasal Mucosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Stock Solutions and Lipid Components
2.2. Preparation of mRNA-Based Formulations and Experimental Design
2.2.1. Preparation of Liposomes
2.2.2. Design of Experiment (DoE)
2.2.3. Preparation of Vaccine Formulations
2.3. Freeze-Drying
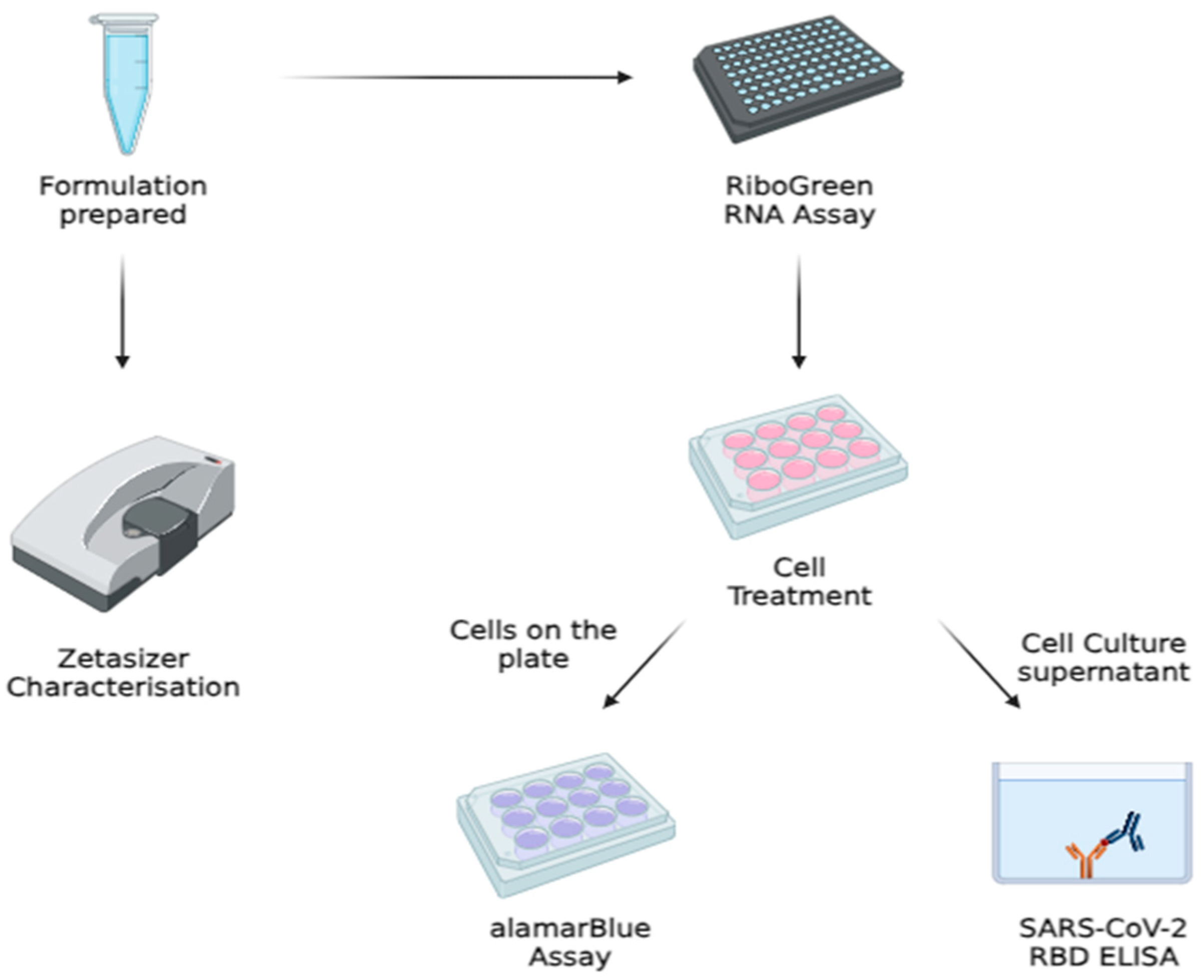
2.4. Cell Culture Work
2.5. Physicochemical Characterization of the Formulation
2.6. TEM Imaging
2.7. mRNA Encapsulation and Quantification via Quant-ItTM RiboGreen RNA Assay
2.8. alamarBlue® Cytotoxicity Assay
2.9. SARS-CoV-2 Spike RBD Quantification via Enzyme-Linked Immunosorbent Assay (ELISA)
2.10. rt-qPCR
2.11. In Vivo Animal Study
2.12. IgG and IgA Quantification via ELISA
2.13. Statistical Analysis
3. Results and Discussion
3.1. Optimization of Vaccine Formulations via DoE
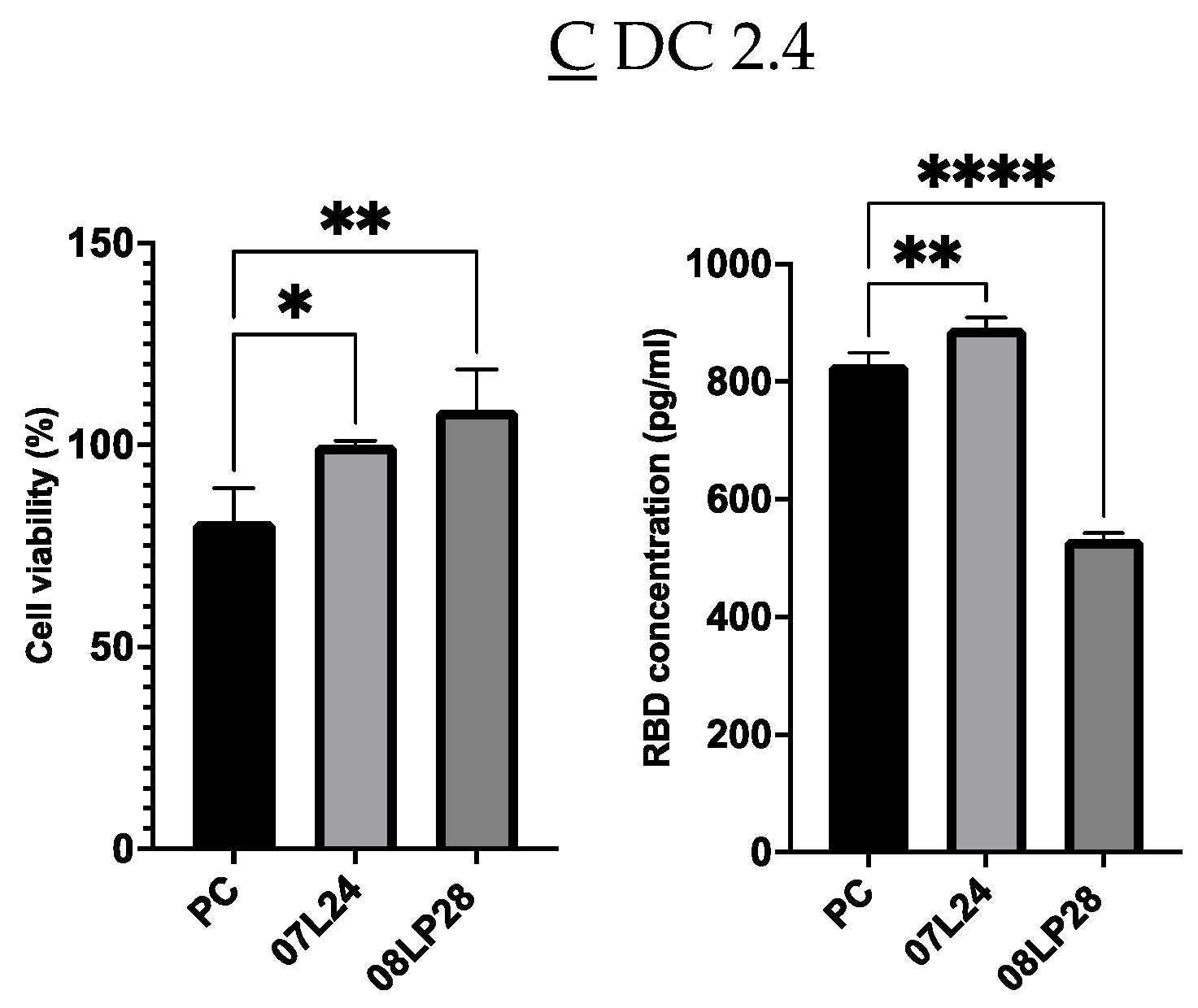
3.2. Physicochemical Characterization: Particle Size, Zeta Potential, and mRNA Loading
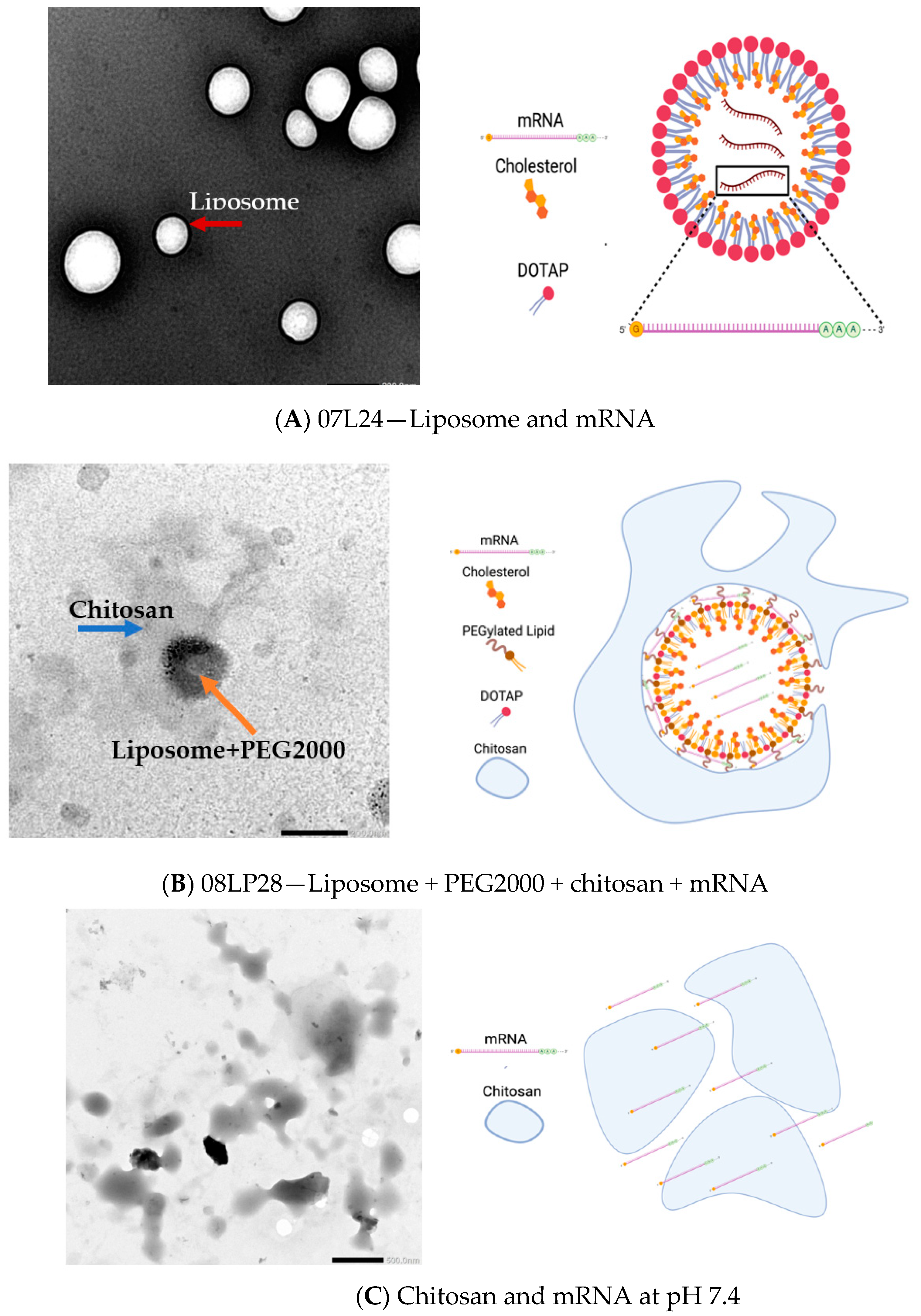
3.3. TEM Imaging of Formulations
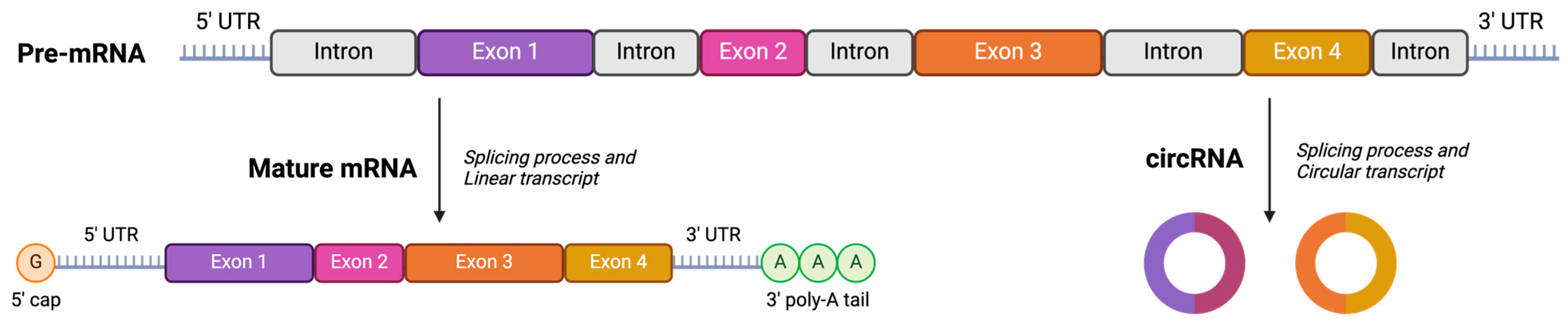
3.4. Stability Studies: Loading Circular RNA (cRNA) and Freeze-Drying Process
3.5. Inflammatory Profile from rt-qPCR
3.6. In Vivo Animal Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ozpolat, B.; Sood, A.K.; Lopez-Berestein, G. Liposomal siRNA nanocarriers for cancer therapy. Adv. Drug Deliv. Rev. 2014, 66, 110–116. [Google Scholar] [CrossRef]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles from Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [Google Scholar] [CrossRef] [PubMed]
- Eygeris, Y.; Gupta, M.; Kim, J.; Sahay, G. Chemistry of Lipid Nanoparticles for RNA Delivery. Acc. Chem. Res. 2022, 55, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Mai, Y.; Guo, J.; Zhao, Y.; Ma, S.; Hou, Y.; Yang, J. Intranasal delivery of cationic liposome-protamine complex mRNA vaccine elicits effective anti-tumor immunity. Cell. Immunol. 2020, 354, 104143. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xiong, M.; Bi, Q.; Wang, Y.; Li, C. Self-enhanced targeted delivery of a cell wall– and membrane-active antibiotics, daptomycin, against staphylococcal pneumonia. Acta Pharm. Sin. B 2016, 6, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Sala, M.; Diab, R.; Elaissari, A.; Fessi, H. Lipid nanocarriers as skin drug delivery systems: Properties, mechanisms of skin interactions and medical applications. Int. J. Pharm. 2018, 535, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hossen, S.; Hossain, M.K.; Basher, M.K.; Mia, M.N.H.; Rahman, M.T.; Uddin, M.J. Smart nanocarrier-based drug delivery systems for cancer therapy and toxicity studies: A review. J. Adv. Res. 2019, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.H.; Radtke, M.; Wissing, S.A. Solid lipid nanoparticles (SLN) and nanostructured lipid carriers (NLC) in cosmetic and dermatological preparations. Adv. Drug Deliv. Rev. 2002, 54, 131–155. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, A.; Rezaei-sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N. Liposome: Classification, prepNew aspects of liposomesaration, and applications. Nanoscale Res. Lett. 2013, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef] [PubMed]
- Billingsley, M.M.; Singh, N.; Ravikumar, P.; Zhang, R.; June, C.H.; Mitchell, M.J. Ionizable Lipid Nanoparticle-Mediated mRNA Delivery for Human CAR T Cell Engineering. Nano Lett. 2020, 20, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Chen, Z.; Li, G.; Welte, T.; Shen, H. Nanoplatforms for mRNA Therapeutics. Adv. Ther. 2021, 4, 2000099. [Google Scholar] [CrossRef]
- Ho, W.; Gao, M.; Li, F.; Li, Z.; Zhang, X.Q.; Xu, X. Next-Generation Vaccines: Nanoparticle-Mediated DNA and mRNA Delivery. Adv. Healthc. Mater. 2021, 10, 2001812. [Google Scholar] [CrossRef] [PubMed]
- Schwendener, R.A. Liposomes as vaccine delivery systems: A review of the recent advances. Ther. Adv. Vaccines 2014, 2, 159–182. [Google Scholar] [CrossRef] [PubMed]
- Chheda, U.; Pradeepan, S.; Esposito, E.; Strezsak, S.; Fernandez-Delgado, O.; Kranz, J. Factors Affecting Stability of RNA—Temperature, Length, Concentration, pH, and Buffering Species. J. Pharm. Sci. 2024, 113, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Blenke, E.O.; Örnskov, E.; Schöneich, C.; Nilsson, G.A.; Volkin, D.B.; Mastrobattista, E.; Almarsson; Crommelin, D.J. The Storage and In-Use Stability of mRNA Vaccines and Therapeutics: Not A Cold Case. J. Pharm. Sci. 2023, 112, 386–403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jiang, Y.; He, Y.; Boucetta, H.; Wu, J.; Chen, Z.; He, W. Lipid carriers for mRNA delivery. Acta Pharm. Sin. B 2023, 13, 4105–4126. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Gao, Y.; Sun, W.; Cao, J.; Liang, Y.; Han, S.; Wang, X.; Sun, Y. Self-Assembled chitosan/phospholipid nanoparticles: From fundamentals to preparation for advanced drug delivery. Drug Deliv. 2020, 27, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Zhou, S.; Dain, L.; Mei, L.; Zhu, G. Circular RNA: An emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J. Control. Release 2022, 348, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.H.; Shunmuganathan, B.; Zhang, L.; Lim, A.; Gupta, R.; Wang, Y.; Chua, B.L.; Wang, Y.; Gu, Y.; Qian, X.; et al. Durable cross-protective neutralizing antibody responses elicited by lipid nanoparticle-formulated SARS-CoV-2 mRNA vaccines. NPJ Vaccines 2024, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y.; Kuo, H.C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 29. [Google Scholar] [CrossRef] [PubMed]
- Riccardo, L. CAT (Chemometric Agile Tool). 2017. Available online: http://www.gruppochemiometria.it/index.php/software (accessed on 23 March 2020).
- Reagent. Quant-iT RiboGreen RNA Reagent and Kit. Invitrogen. 2008. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/mp11490.pdf (accessed on 27 February 2024).
- Invitrogen. Alamarblue® Assay. U.S. Patent No 5501959, 2007. Available online: https://tools.thermofisher.com/content/sfs/manuals/PI-DAL1025-1100_TI alamarBlue Rev 1.1.pdf (accessed on 27 February 2024).
- Kit, S.E. SARS-CoV-2 (COVID-19) Spike RBD Protein Sandwich ELISA Kit. Available online: https://www.genetex.com/PDF/Download?catno=GTX536267 (accessed on 27 February 2024).
- Kit, R.M. RNAeasy Handbook. 2023. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=f646813a-efbb-4672-9ae3-e665b3045b2b&lang=en (accessed on 27 February 2024).
- Lock, J.Y.; Carlson, T.L.; Carrier, R.L. Mucus models to evaluate the diffusion of drugs and particles. Adv. Drug Deliv. Rev. 2018, 124, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, A.; Voigt, E.; Archer, M.; Reed, S.; Larson, E.; Van Hoeven, N.; Kramer, R.; Fox, C.; Casper, C. A Thermostable, Flexible RNA Vaccine Delivery Platform for Pandemic Response. bioRxiv 2021. bioRxiv:02.01.429283. [Google Scholar]
- Zylberberg, C.; Matosevic, S. Pharmaceutical liposomal drug delivery: A review of new delivery systems and a look at the regulatory landscape. Drug Deliv. 2016, 23, 3319–3329. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.H.; Beirag, N.; Murugaiah, V.; Chou, Y.-C.; Kuo, W.-S.; Kao, H.-F.; Madan, T.; Kishore, U.; Wang, J.-Y. Human Surfactant Protein D Binds Spike Protein and Acts as an Entry Inhibitor of SARS-CoV-2 Pseudotyped Viral Particles. Front. Immunol. 2021, 12, 641360. [Google Scholar] [CrossRef] [PubMed]
- Paidi, R.K.; Jana, M.; Mishra, R.K.; Dutta, D.; Pahan, K. Selective Inhibition of the Interaction between SARS-CoV-2 Spike S1 and ACE2 by SPIDAR Peptide Induces Anti-Inflammatory Therapeutic Responses. J. Immunol. 2021, 207, 2521–2533. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; To, K.K.-W.; Wong, Y.-C.; Liu, L.; Zhou, B.; Li, X.; Huang, H.; Mo, Y.; Luk, T.-Y.; Lau, T.T.-K.; et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity 2020, 53, 864–877.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, Z.; Lihe, H.; Lu, L.; Zhang, Z.; Yang, S.; Meng, N.; Xiong, Y.; Fan, X.; Chen, Z.; et al. Intranasal G5-BGG/pDNA Vaccine Elicits Protective Systemic and Mucosal Immunity against SARS-CoV-2 by Transfecting Mucosal Dendritic Cells. Adv. Healthc. Mater. 2023, 13, 2303261. [Google Scholar] [CrossRef] [PubMed]
- Barreda, D.; Santiago, C.; Rodríguez, J.R.; Rodríguez, J.F.; Casasnovas, J.M.; Mérida, I.; Ávila-Flores, A. SARS-CoV-2 Spike Protein and Its Receptor Binding Domain Promote a Proinflammatory Activation Profile on Human Dendritic Cells. Cells 2021, 10, 3279. [Google Scholar] [CrossRef] [PubMed]
- Petrey, A.C.; Qeadan, F.; Middleton, E.A.; Pinchuk, I.V.; Campbell, R.A.; Beswick, E.J. Cytokine release syndrome in COVID-19: Innate immune, vascular, and platelet pathogenic factors differ in severity of disease and sex. J. Leukoc. Biol. 2021, 109, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Tucureanu, M.M.; Rebleanu, D.; Constantinescu, C.A.; Deleanu, M.; Voicu, G.; Butoi, E.; Calin, M.; Manduteanu, I. Lipopolysaccharide-induced inflammation in monocytes/macrophages is blocked by liposomal delivery of Gi-protein inhibitor. Int. J. Nanomed. 2018, 13, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Hou, X.; Yan, J.; Du, S.; Xue, Y.; Li, W.; Xiang, G.; Dong, Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact. Mater. 2020, 5, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; He, Z.; Chen, Q.; He, X.; Su, L.; Yu, W.; Zhang, M.; Yang, H.; Huang, X.; Li, J. Helper-Polymer Based Five-Element Nanoparticles (FNPs) for Lung-Specific mRNA Delivery with Long-Term Stability after Lyophilization. Nano Lett. 2022, 22, 6580–6589. [Google Scholar] [CrossRef] [PubMed]
- Heljo, P. Comparison of Disaccharides and Polyalcohols as Stabilizers in Freeze-Dried Protein Formulations. Ph.D. Thesis, Faculty of Pharmacy, University of Helsinki, Helsinki, Finland, 2013; pp. 23–50. [Google Scholar]
- Hao, L.T.; Lee, M.; Jeon, H.; Koo, J.M.; Hwang, S.Y.; Oh, D.X.; Park, J. Tamper-Proof Time-Temperature Indicator for Inspecting Ultracold Supply Chain. ACS Omega 2021, 6, 8598–8604. [Google Scholar] [CrossRef] [PubMed]
- Schultheiß, C.; Willscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.-S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T.; et al. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep. Med. 2022, 3, 100663. [Google Scholar] [CrossRef] [PubMed]
- Jonny, J.; Putranto, T.A.; Sitepu, E.C.; Irfon, R. Dendritic cell vaccine as a potential strategy to end the COVID-19 pandemic. Why should it be Ex Vivo? Expert Rev. Vaccines 2022, 21, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodríguez, L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Biscari, L.; Kaufman, C.D.; Farré, C.; Huhn, V.; Pacini, M.F.; Balbi, C.B.; Gómez, K.A.; Pérez, A.R.; Alloatti, A. Immunization With Lipopolysaccharide-Activated Dendritic Cells Generates a Specific CD8+ T Cell Response That Confers Partial Protection Against Infection With Trypanosoma cruzi. Front. Cell. Infect. Microbiol. 2022, 12, 897133. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Cai, L.; Xu, C.; Sun, S.; Liu, Y.; Rosenecker, J.; Guan, S. Nanotechnologies in Delivery of DNA and mRNA Vaccines to the Nasal and Pulmonary Mucosa. Nanomaterials 2022, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Suberi, A.; Suberi, A.; Grun, M.K.; Grun, M.K.; Mao, T.; Mao, T.; Israelow, B.; Israelow, B.; Reschke, M.; Reschke, M.; et al. Polymer nanoparticles deliver mRNA to the lung for mucosal vaccination. Sci. Transl. Med. 2023, 15, eabq0603. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Roni, M.A. Challenges of storage and stability of mRNA-based covid-19 vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef] [PubMed]









| (A) | |||||
| Selected Variables | Number of Levels | Levels | |||
| DOPC neutral lipid | 3 | 0–2.5 mg/mL | |||
| Chitosan | 2 | 0–0.1 mg/mL | |||
| pH of Chitosan | 2 | 5.5–7.4 pH | |||
| DOTAP cationic lipid | 3 | 0–2.5 mg/mL | |||
| PEG 2000 | 3 | 0–0.5 mg/mL | |||
| (B) | |||||
| Code | DOPC | Chitosan | pH | DOTAP | PEG 2000 |
| F1 | −1 | 1 | −1 | −1 | −1 |
| F2 | 1 | 1 | −1 | −1 | −1 |
| F3 | 1 | 1 | −1 | −1 | −1 |
| F4 | 1 | 1 | −1 | −1 | 1 |
| F5 | 1 | −1 | 1 | −1 | 1 |
| F6 | 1 | −1 | 1 | 1 | 1 |
| F7 | −1 | −1 | 1 | 0.64 | −1 |
| F8 | 1 | 1 | −1 | 1 | −1 |
| F9 | −1 | 1 | −1 | 0.64 | 0.16 |
| F10 | 0.64 | −1 | 1 | 1 | 1 |
| F11 | 0.64 | −1 | −1 | 0.64 | 0.16 |
| F12 | −1 | −1 | 1 | 1 | 1 |
| F13 | 0.64 | 1 | 1 | 0.64 | 0.16 |
| F14 | −1 | −1 | 1 | 0.64 | −1 |
| F15 | −1 | −1 | 1 | 1 | 0.16 |
| F16 | 0.64 | −1 | 1 | 0.64 | 0.16 |
| F17 | −1 | 1 | −1 | 0.64 | −1 |
| F18 | −1 | 1 | 1 | 0.64 | 0.16 |
| (A) | ||
| Selected Variables | Number of Levels | Levels |
| Chitosan | 3 | 0.1–0.4 nM |
| PEG 2000 | 2 | 0–100 nM |
| (B) | ||
| PEG 2000 | Chitosan | |
| F19 | −1 | −1 |
| F20 | −1 | 1 |
| F21 | 1 | 1 |
| F22 | 1 | −1 |
| F23 | −1 | 0 |
| F24 | 1 | 0 |
| Formulation | Particle Size (nm) | Zeta Potential (mV) | mRNA Loading (%) |
|---|---|---|---|
| Pfizer vaccine (PC) | 89.4 ± 1.5 | −1.2 ± 4.0 | 97.0 ± 0.1 |
| 08LP28 BLANK | 139.6 ± 3.3 | +17.8 ± 1.1 | - |
| 08LP28 + mRNA | 587.7 ± 14.7 | +16.5 ± 1.4 | 84.6 ± 5.4 |
| 07L24 BLANK | 133.6 ± 0.3 | +23.8 ± 2.7 | - |
| 07L24 + mRNA | 359.9 ± 7.2 | +22.2 ± 0.6 | 90.3 ± 5.3 |
| Formulation | Particle Size (nm) | Zeta Potential (mV) | mRNA Loading (%) | RBD Expression HEK 293 (pg/mL) |
|---|---|---|---|---|
| 08LP28 + 4% mRNA | 570.5 ± 6.2 | +16.9 ± 3.9 | 90.9 ± 10.1 | 177.9 ± 20.1 |
| 08LP28 + 4% cRNA | 370.3 ± 16.4 | +22.4 ± 0.6 | 111.0 ± 2.7 | 170.1 ± 22.7 |
| 08LP28 + 6% mRNA (before lyophilization) | 619.0 ± 26.3 | +22.5 ± 0.9 | 88.2 ± 4.3 | 1985 ± 101.9 |
| 08LP28 + 6% mRNA (after lyophilization) | 1061 ± 196.1 | +19.9 ± 1.2 | 51.2 ± 5.1 | 729.7 ± 28.7 |
| 07L24 + 4% mRNA | 309.2 ± 7.2 | +30.7 ± 0.6 | 103.4 ± 1.3 | 9776.6 ± 49.8 |
| 07L24 + 4% cRNA | 301.5 ± 5.7 | +25.0 ± 0.6 | 98.7 ± 19.0 | 8661.5 ± 765.6 |
| 07L24 + 6% mRNA (before lyophilization) | 619.4 ± 35.1 | +32.6 ± 0.6 | 85.7 ± 9.0 | 9994.0 ± 153.6 |
| 07L24 + 6% mRNA (after lyophilization) | 1432 ± 155.7 | +32.6 ± 2.4 | 53.7 ± 3.6 | 914.8 ± 124.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altay Benetti, A.; Tan, E.Y.Z.; Chang, Z.W.; Bae, K.H.; Thwin, M.T.; Muthuramalingam, R.P.K.; Liao, K.-C.; Wan, Y.; Ng, L.F.P.; Renia, L.; et al. Design and Characterization of a New Formulation for the Delivery of COVID-19-mRNA Vaccine to the Nasal Mucosa. Vaccines 2024, 12, 409. https://doi.org/10.3390/vaccines12040409
Altay Benetti A, Tan EYZ, Chang ZW, Bae KH, Thwin MT, Muthuramalingam RPK, Liao K-C, Wan Y, Ng LFP, Renia L, et al. Design and Characterization of a New Formulation for the Delivery of COVID-19-mRNA Vaccine to the Nasal Mucosa. Vaccines. 2024; 12(4):409. https://doi.org/10.3390/vaccines12040409
Chicago/Turabian StyleAltay Benetti, Ayça, Eugene Yang Zhi Tan, Zi Wei Chang, Ki Hyun Bae, Ma Thinzar Thwin, Ram Pravin Kumar Muthuramalingam, Kuo-Chieh Liao, Yue Wan, Lisa F. P. Ng, Laurent Renia, and et al. 2024. "Design and Characterization of a New Formulation for the Delivery of COVID-19-mRNA Vaccine to the Nasal Mucosa" Vaccines 12, no. 4: 409. https://doi.org/10.3390/vaccines12040409
APA StyleAltay Benetti, A., Tan, E. Y. Z., Chang, Z. W., Bae, K. H., Thwin, M. T., Muthuramalingam, R. P. K., Liao, K.-C., Wan, Y., Ng, L. F. P., Renia, L., Liu, J., Chen, X., Yang, Y. Y., White, K. P., & Pastorin, G. (2024). Design and Characterization of a New Formulation for the Delivery of COVID-19-mRNA Vaccine to the Nasal Mucosa. Vaccines, 12(4), 409. https://doi.org/10.3390/vaccines12040409







