The Expression Kinetics and Immunogenicity of Lipid Nanoparticles Delivering Plasmid DNA and mRNA in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of pDNA- and mRNA-Encoding Luciferase
2.2. Lipid Nanoparticle (LNP) Generation
2.3. Nanoparticle Characterization
2.4. In Vitro Transfection Assay
2.5. Animal Study
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Mathematical and Statistical Analyses
3. Results
3.1. Characterization of Nanoparticle Formulations
3.2. Potency of LNP Formulations
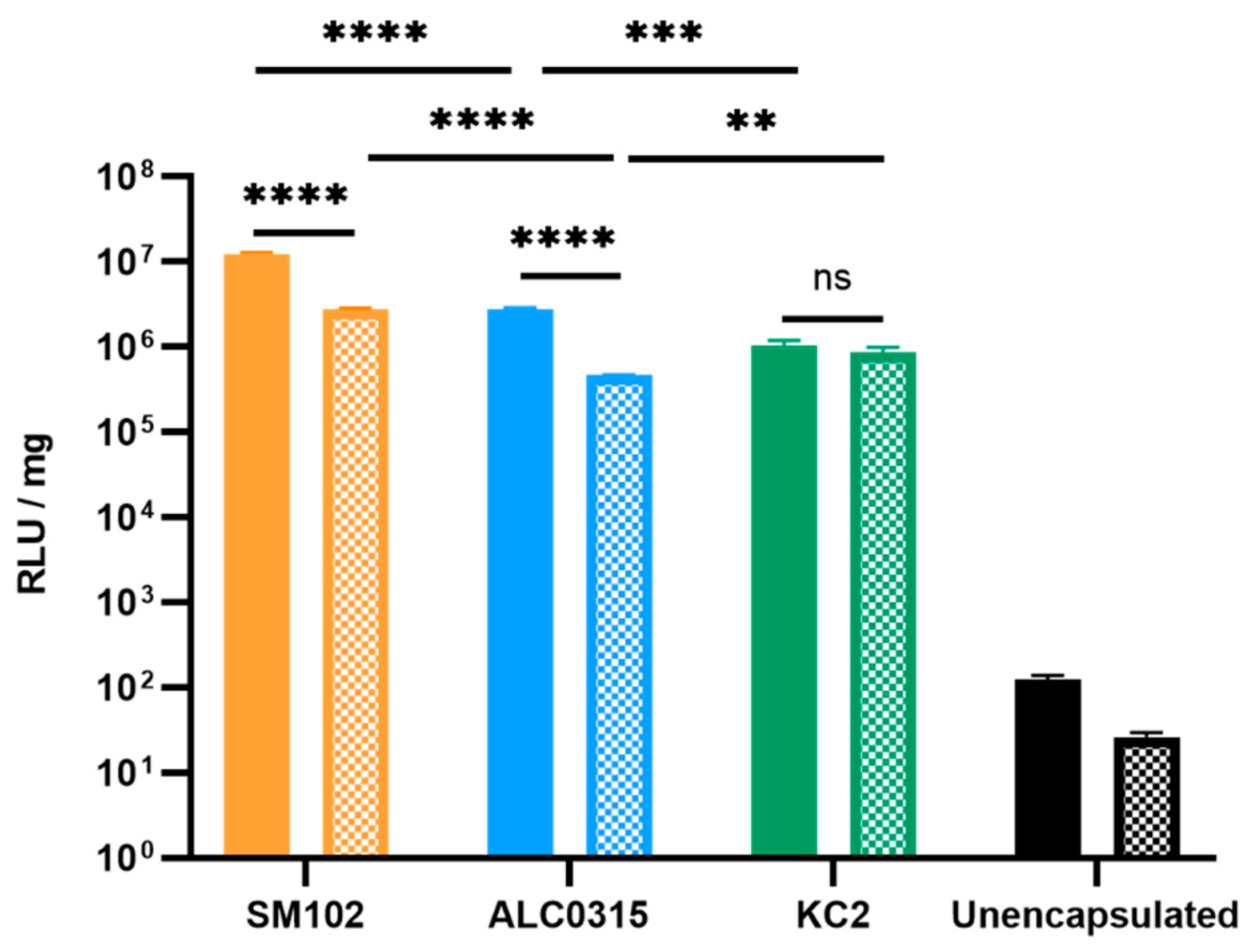
3.2.1. SM102-RNA Particles Result in the Highest Luciferase Expression In Vitro
3.2.2. SM102-RNA and ALC0315-RNA Particles Result in the Highest Luciferase Expression In Vivo
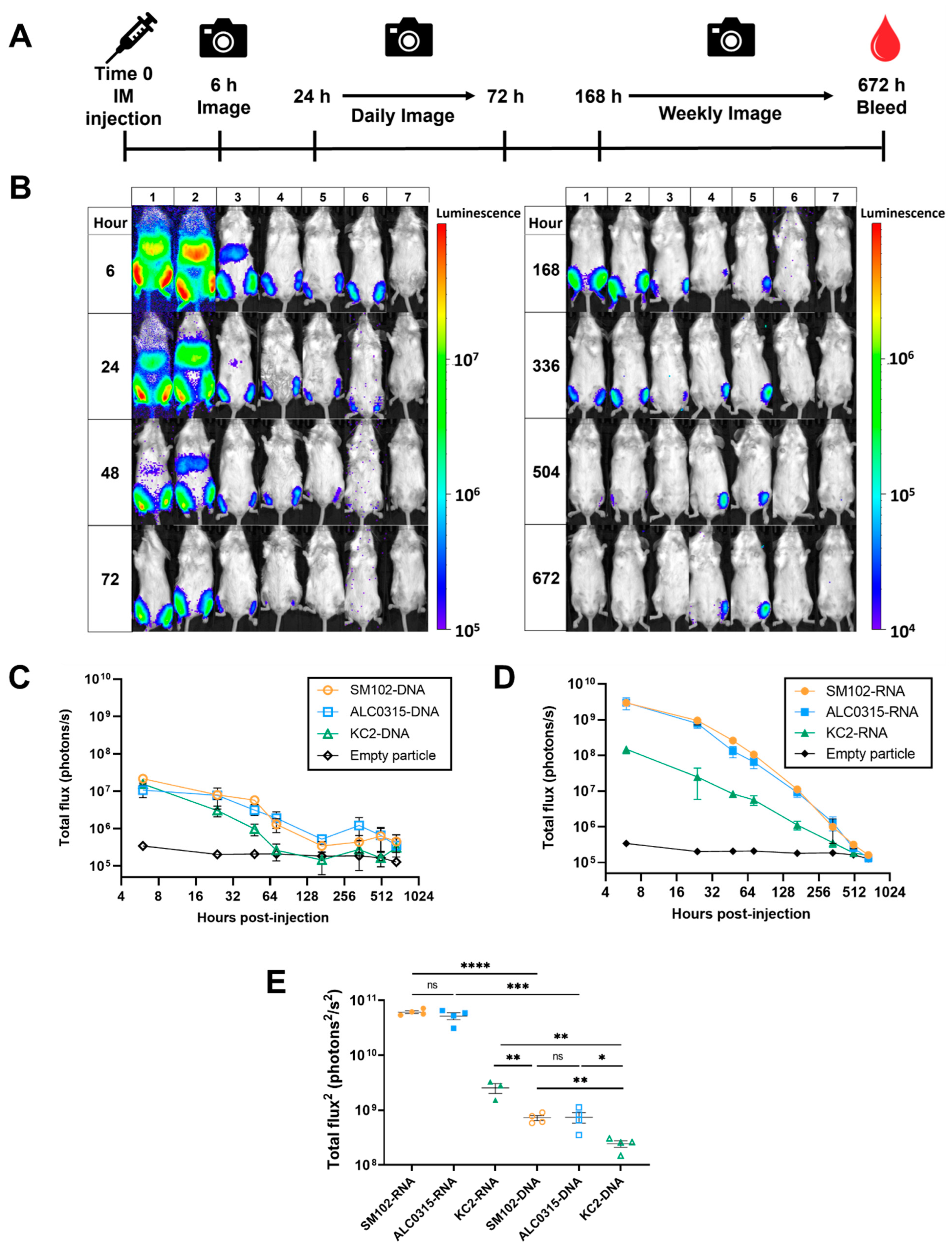
3.3. Expression Kinetics: SM102-DNA and ALC0315-DNA Particles Result in the Longest Duration of Protein Expression
3.4. Biodistribution: LNPs Result in Protein Expression in the Liver Proportional to the Expression at the Injection Site
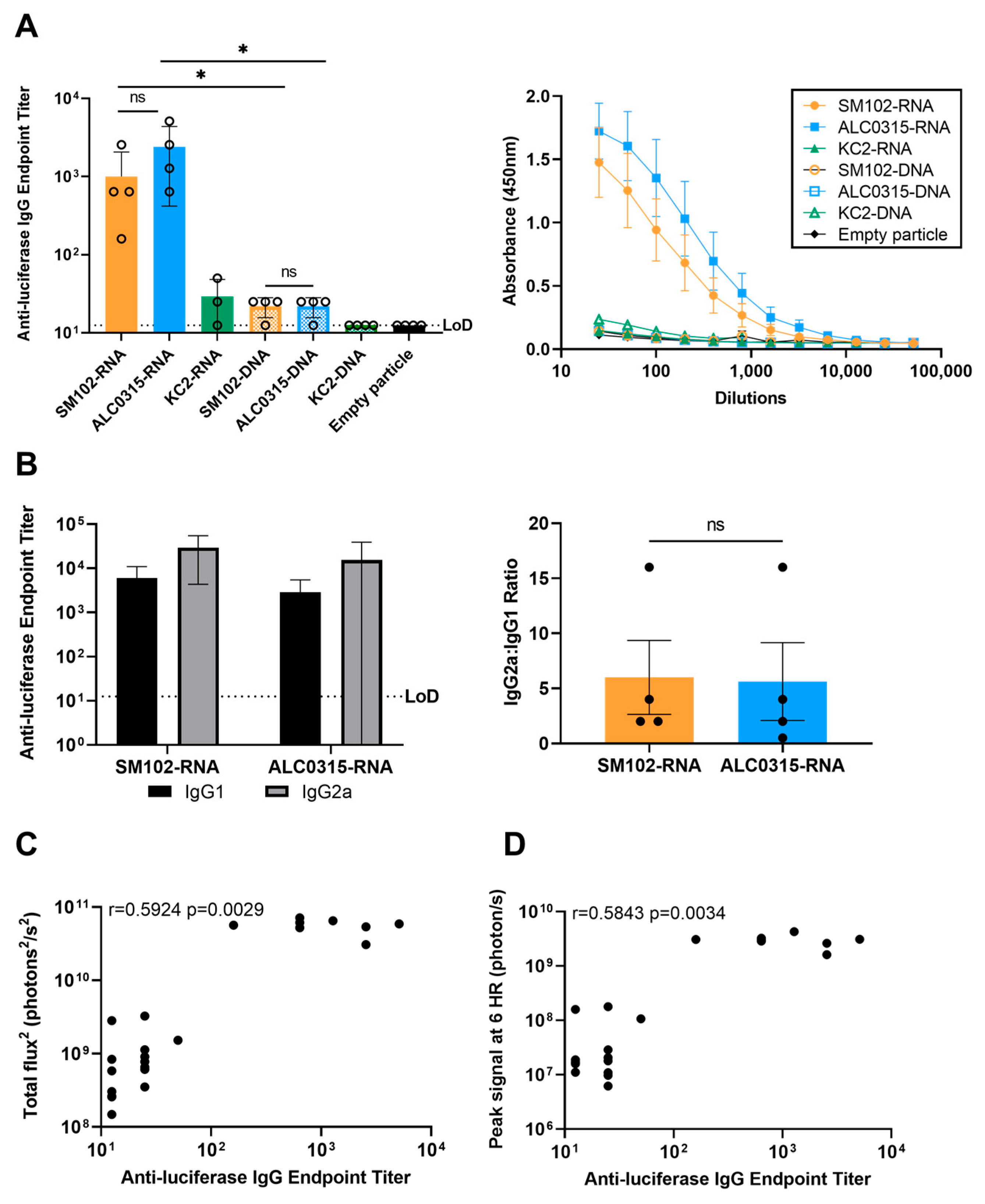
3.5. Immunogenicity: SM102-RNA and ALC0315-RNA Particles Result in the Highest Antibody Response with Th1 Bias
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- COVID-19 Vaccine Doses Administered by Manufacturer, Canada. Available online: https://ourworldindata.org/grapher/covid-vaccine-doses-by-manufacturer?country=~CAN (accessed on 22 March 2023).
- Blakney, A.K.; McKay, P.F.; Yus, B.I.; Aldon, Y.; Shattock, R.J. Inside out: Optimization of Lipid Nanoparticle Formulations for Exterior Complexation and in Vivo Delivery of SaRNA. Gene Ther. 2019, 26, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Geall, A.J.; Verma, A.; Otten, G.R.; Shaw, C.A.; Hekele, A.; Banerjee, K.; Cu, Y.; Beard, C.W.; Brito, L.A.; Krucker, T.; et al. Nonviral Delivery of Self-Amplifying RNA Vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 14604–14609. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Eygeris, Y.; Gupta, M.; Sahay, G. Self-Assembled MRNA Vaccines. Adv. Drug Deliv. Rev. 2021, 170, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Francis, J.E.; Skakic, I.; Dekiwadia, C.; Shukla, R.; Taki, A.C.; Walduck, A.; Smooker, P.M. Solid Lipid Nanoparticle Carrier Platform Containing Synthetic TLR4 Agonist Mediates Non-Viral DNA Vaccine Delivery. Vaccines 2020, 8, 551. [Google Scholar] [CrossRef]
- Mucker, E.M.; Karmali, P.P.; Vega, J.; Kwilas, S.A.; Wu, H.; Joselyn, M.; Ballantyne, J.; Sampey, D.; Mukthavaram, R.; Sullivan, E.; et al. Lipid Nanoparticle Formulation Increases Efficiency of DNA-Vectored Vaccines/Immunoprophylaxis in Animals Including Transchromosomic Bovines. Sci. Rep. 2020, 10, 8764. [Google Scholar] [CrossRef]
- Lim, M.; Badruddoza, A.Z.M.; Firdous, J.; Azad, M.; Mannan, A.; Al-Hilal, T.A.; Cho, C.-S.; Islam, M.A. Engineered Nanodelivery Systems to Improve DNA Vaccine Technologies. Pharmaceutics 2020, 12, 30. [Google Scholar] [CrossRef]
- Yan, M.; Wu, S.; Wang, Y.; Liang, M.; Wang, M.; Hu, W.; Yu, G.; Mao, Z.; Huang, F.; Zhou, J. Recent Progress of Supramolecular Chemotherapy Based on Host-Guest Interactions. Adv. Mater. 2023, e2304249. [Google Scholar] [CrossRef]
- Liu, L.; Xiong, M.; Rong, Q.; Zhang, M.; Zhang, X. Nucleic Acid Sensors in Vivo: Challenges and Opportunities. VIEW 2023, 4, 20220064. [Google Scholar] [CrossRef]
- Alameh, M.-G.; Tombácz, I.; Bettini, E.; Lederer, K.; Sittplangkoon, C.; Wilmore, J.R.; Gaudette, B.T.; Soliman, O.Y.; Pine, M.; Hicks, P.; et al. Lipid Nanoparticles Enhance the Efficacy of MRNA and Protein Subunit Vaccines by Inducing Robust T Follicular Helper Cell and Humoral Responses. Immunity 2021, 54, 2877–2892.e7. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Naradikian, M.S.; Parkhouse, K.; Cain, D.W.; Jones, L.; Moody, M.A.; Verkerke, H.P.; Myles, A.; Willis, E.; et al. Nucleoside-Modified MRNA Vaccines Induce Potent T Follicular Helper and Germinal Center B Cell Responses. J. Exp. Med. 2018, 215, 1571–1588. [Google Scholar] [CrossRef]
- Ulmer, J.B.; Mason, P.W.; Geall, A.; Mandl, C.W. RNA-Based Vaccines. Vaccine 2012, 30, 4414–4418. [Google Scholar] [CrossRef] [PubMed]
- Crommelin, D.J.A.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of MRNA Vaccine Stability. J. Pharm. Sci. 2021, 110, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of MRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef] [PubMed]
- Oude Blenke, E.; Örnskov, E.; Schöneich, C.; Nilsson, G.A.; Volkin, D.B.; Mastrobattista, E.; Almarsson, Ö.; Crommelin, D.J.A. The Storage and In-Use Stability of MRNA Vaccines and Therapeutics: Not A Cold Case. J. Pharm. Sci. 2023, 112, 386–403. [Google Scholar] [CrossRef]
- Rosa, S.S.; Prazeres, D.M.F.; Azevedo, A.M.; Marques, M.P.C. MRNA Vaccines Manufacturing: Challenges and Bottlenecks. Vaccine 2021, 39, 2190–2200. [Google Scholar] [CrossRef]
- Kafetzis, K.N.; Papalamprou, N.; McNulty, E.; Thong, K.X.; Sato, Y.; Mironov, A.; Purohit, A.; Welsby, P.J.; Harashima, H.; Yu-Wai-Man, C.; et al. The Effect of Cryoprotectants and Storage Conditions on the Transfection Efficiency, Stability, and Safety of Lipid-Based Nanoparticles for MRNA and DNA Delivery. Adv. Healthc. Mater. 2023, 12, 2203022. [Google Scholar] [CrossRef]
- Jahanafrooz, Z.; Baradaran, B.; Mosafer, J.; Hashemzaei, M.; Rezaei, T.; Mokhtarzadeh, A.; Hamblin, M.R. Comparison of DNA and MRNA Vaccines against Cancer. Drug Discov. Today 2020, 25, 552–560. [Google Scholar] [CrossRef]
- Middaugh, C.R.; Evans, R.K.; Montgomery, D.L.; Casimiro, D.R. Analysis of Plasmid DNA from a Pharmaceutical Perspective. J. Pharm. Sci. 1998, 87, 130–146. [Google Scholar] [CrossRef]
- Khobragade, A.; Bhate, S.; Ramaiah, V.; Deshpande, S.; Giri, K.; Phophle, H.; Supe, P.; Godara, I.; Revanna, R.; Nagarkar, R.; et al. Efficacy, Safety, and Immunogenicity of the DNA SARS-CoV-2 Vaccine (ZyCoV-D): The Interim Efficacy Results of a Phase 3, Randomised, Double-Blind, Placebo-Controlled Study in India. Lancet 2022, 399, 1313–1321. [Google Scholar] [CrossRef]
- Juba, R.J.; Samawova, A.; Seals, K.; Kinney, P.; Brandreth, E.J. 2140. INO-4800, a DNA Plasmid Vaccine Encoding the Ancestral SARS-CoV-2 Spike Protein, Is Stable Over a Range of Temperatures Not Requiring Ultra-Cold Chain Storage. Open Forum Infect. Dis. 2022, 9 (Suppl. S2). [Google Scholar] [CrossRef]
- Liu, M.A. A Comparison of Plasmid DNA and MRNA as Vaccine Technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.R.d.S.; de Macêdo, L.S.; Invenção, M.d.C.V.; de Moura, I.A.; da Gama, M.A.T.M.; de Melo, C.M.L.; Silva, A.J.D.; Batista, M.V.d.A.; Freitas, A.C.D. Third-Generation Vaccines: Features of Nucleic Acid Vaccines and Strategies to Improve Their Efficiency. Genes 2022, 13, 2287. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C. First COVID-19 DNA Vaccine Approved, Others in Hot Pursuit. Available online: https://www.nature.com/articles/d41587-021-00023-5 (accessed on 22 March 2023).
- Ledesma-Feliciano, C.; Chapman, R.; Hooper, J.W.; Elma, K.; Zehrung, D.; Brennan, M.B.; Spiegel, E.K. Improved DNA Vaccine Delivery with Needle-Free Injection Systems. Vaccines 2023, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Shafaati, M.; Saidijam, M.; Soleimani, M.; Hazrati, F.; Mirzaei, R.; Amirheidari, B.; Tanzadehpanah, H.; Karampoor, S.; Kazemi, S.; Yavari, B.; et al. A Brief Review on DNA Vaccines in the Era of COVID-19. Future Virol. 2021, 17, 49–66. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Myhre, J.L.; Chen, S.; Tam, Y.Y.C.; Danescu, A.; Richman, J.M.; Cullis, P.R. Design of Lipid Nanoparticles for in Vitro and in Vivo Delivery of Plasmid DNA. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1377–1387. [Google Scholar] [CrossRef]
- Wong, J.P.; Zabielski, M.A.; Schmaltz, F.L.; Brownlee, G.G.; Bussey, L.A.; Marshall, K.; Borralho, T.; Nagata, L.P. DNA Vaccination against Respiratory Influenza Virus Infection. Vaccine 2001, 19, 2461–2467. [Google Scholar] [CrossRef]
- Algarni, A.; Pilkington, E.H.; Suys, E.J.A.; Al-Wassiti, H.; Pouton, C.W.; Truong, N.P. In Vivo Delivery of Plasmid DNA by Lipid Nanoparticles: The Influence of Ionizable Cationic Lipids on Organ-Selective Gene Expression. Biomater. Sci. 2022, 10, 2940–2952. [Google Scholar] [CrossRef]
- Hassett, K.J.; Benenato, K.E.; Jacquinet, E.; Lee, A.; Woods, A.; Yuzhakov, O.; Himansu, S.; Deterling, J.; Geilich, B.M.; Ketova, T.; et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of MRNA Vaccines. Mol. Ther.-Nucleic Acids 2019, 15, 1–11. [Google Scholar] [CrossRef]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissman, D. Expression Kinetics of Nucleoside-Modified MRNA Delivered in Lipid Nanoparticles to Mice by Various Routes. J. Control. Release 2015, 217, 345–351. [Google Scholar] [CrossRef]
- Pfeifle, A.; Thulasi Raman, S.N.; Lansdell, C.; Zhang, W.; Tamming, L.; Cecillon, J.; Laryea, E.; Patel, D.; Wu, J.; Gravel, C.; et al. DNA Lipid Nanoparticle Vaccine Targeting Outer Surface Protein C Affords Protection against Homologous Borrelia Burgdorferi Needle Challenge in Mice. Front. Immunol. 2023, 14, 1020134. [Google Scholar] [CrossRef]
- European Medicines Agency. Comirnaty Assessment Report; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Bachmann, M.F.; Jennings, G.T. Vaccine Delivery: A Matter of Size, Geometry, Kinetics and Molecular Patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- Hoshyar, N.; Gray, S.; Han, H.; Bao, G. The Effect of Nanoparticle Size on in Vivo Pharmacokinetics and Cellular Interaction. Nanomedicine 2016, 11, 673–692. [Google Scholar] [CrossRef]
- Hassett, K.J.; Higgins, J.; Woods, A.; Levy, B.; Xia, Y.; Hsiao, C.J.; Acosta, E.; Almarsson, Ö.; Moore, M.J.; Brito, L.A. Impact of Lipid Nanoparticle Size on MRNA Vaccine Immunogenicity. J. Control. Release 2021, 335, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.H.; Choi, Y.; Kang, J.H.; Kim, C.W.; Jeong, J.M.; Lee, D.S.; Chung, J.-K. Immune Response to Firefly Luciferase as a Naked DNA. Cancer Biol. Ther. 2007, 6, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, M.J.; Alishetty, S.; Alameh, M.G.; Said, H.; Wright, L.; Paige, M.; Soliman, O.; Weissman, D.; Cleveland, T.E.; Grishaev, A.; et al. Ionization and Structural Properties of MRNA Lipid Nanoparticles Influence Expression in Intramuscular and Intravascular Administration. Commun. Biol. 2021, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nantz, M.H.; Dicus, C.W.; Hilliard, B.; Yellayi, S.; Zou, S.; Hecker, J.G. The Benefit of Hydrophobic Domain Asymmetry on the Efficacy of Transfection as Measured by in Vivo Imaging. Mol. Pharm. 2010, 7, 786–794. [Google Scholar] [CrossRef]
- Ly, H.H.; Daniel, S.; Soriano, S.K.V.; Kis, Z.; Blakney, A.K. Optimization of Lipid Nanoparticles for SaRNA Expression and Cellular Activation Using a Design-of-Experiment Approach. Mol. Pharm. 2022, 19, 1892–1905. [Google Scholar] [CrossRef]
- Goldman, R.L.; Vittala Murthy, N.T.; Northen, T.P.; Balakrishnan, A.; Chivukula, S.; Danz, H.; Tibbitts, T.; Dias, A.; Vargas, J.; Cooper, D.; et al. Understanding Structure Activity Relationships of Good HEPES Lipids for Lipid Nanoparticle MRNA Vaccine Applications. Biomaterials 2023, 301, 122243. [Google Scholar] [CrossRef]
- Escalona-Rayo, O.; Zeng, Y.; Knol, R.A.; Kock, T.J.F.; Aschmann, D.; Slütter, B.; Kros, A. In Vitro and in Vivo Evaluation of Clinically-Approved Ionizable Cationic Lipids Shows Divergent Results between MRNA Transfection and Vaccine Efficacy. Biomed. Pharmacother. 2023, 165, 115065. [Google Scholar] [CrossRef]
- Hajj, K.A.; Ball, R.L.; Deluty, S.B.; Singh, S.R.; Strelkova, D.; Knapp, C.M.; Whitehead, K.A. Branched-Tail Lipid Nanoparticles Potently Deliver MRNA In Vivo Due to Enhanced Ionization at Endosomal PH. Small 2019, 15, 1805097. [Google Scholar] [CrossRef]
- Hashiba, K.; Sato, Y.; Taguchi, M.; Sakamoto, S.; Otsu, A.; Maeda, Y.; Shishido, T.; Murakawa, M.; Okazaki, A.; Harashima, H. Branching Ionizable Lipids Can Enhance the Stability, Fusogenicity, and Functional Delivery of MRNA. Small Sci. 2023, 3, 2200071. [Google Scholar] [CrossRef]
- Paunovska, K.; Sago, C.D.; Monaco, C.M.; Hudson, W.H.; Castro, M.G.; Rudoltz, T.G.; Kalathoor, S.; Vanover, D.A.; Santangelo, P.J.; Ahmed, R.; et al. A Direct Comparison of in Vitro and in Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation. Nano Lett. 2018, 18, 2148–2157. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.A.; Matthews, J.; Chang, P.H.; Niroui, F.; Dorkin, J.R.; Severgnini, M.; Anderson, D.G. In Vitro-in Vivo Translation of Lipid Nanoparticles for Hepatocellular SiRNA Delivery. ACS Nano 2012, 6, 6922–6929. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Kuipers, F.; Havekes, L.M.; Havinga, R.; Dontje, B.; Poelstra, K.; Scherphof, G.L.; Kamps, J.A.A.M. The Role of Apolipoprotein E in the Elimination of Liposomes from Blood by Hepatocytes in the Mouse. Biochem. Biophys. Res. Commun. 2005, 328, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Querbes, W.; De, S.; Qin, J.; Frank-Kamenetsky, M.; Jayaprakash, K.N.; Jayaraman, M.; Rajeev, K.G.; Cantley, W.L.; Dorkin, J.R.; et al. Targeted Delivery of RNAi Therapeutics with Endogenous and Exogenous Ligand-Based Mechanisms. Mol. Ther. 2010, 18, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jeong, M.; Hur, S.; Cho, Y.; Park, J.; Jung, H.; Seo, Y.; Woo, H.A.; Nam, K.T.; Lee, K.; et al. Engineered Ionizable Lipid Nanoparticles for Targeted Delivery of RNA Therapeutics into Different Types of Cells in the Liver. Sci. Adv. 2021, 7, eabf4398. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.A. Correlates of Protection Induced by Vaccination. Clin. Vaccine Immunol. 2010, 17, 1055–1065. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing Antibody Levels Are Highly Predictive of Immune Protection from Symptomatic SARS-CoV-2 Infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Taylor, J.M.; Ziman, M.E.; Canfield, D.R.; Vajdy, M.; Solnick, J. V Effects of a Th1- versus a Th2-Biased Immune Response in Protection against Helicobacter Pylori Challenge in Mice. Microb. Pathog. 2008, 44, 20–27. [Google Scholar] [CrossRef]
- Rivera-Hernandez, T.; Rhyme, M.S.; Cork, A.J.; Jones, S.; Segui-Perez, C.; Brunner, L.; Richter, J.; Petrovsky, N.; Lawrenz, M.; Goldblatt, D.; et al. Vaccine-Induced Th1-Type Response Protects against Invasive Group A Streptococcus Infection in the Absence of Opsonizing Antibodies. MBio 2020, 11, 10–128. [Google Scholar] [CrossRef]
- Muralidharan, A.; Li, C.; Wang, L.; Li, X. Immunopathogenesis Associated with Formaldehyde-Inactivated RSV Vaccine in Preclinical and Clinical Studies. Expert Rev. Vaccines 2017, 16, 351–360. [Google Scholar] [CrossRef]
- Hobernik, D.; Bros, M. DNA Vaccines-How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef]
- Suschak, J.J.; Williams, J.A.; Schmaljohn, C.S. Advancements in DNA Vaccine Vectors, Non-Mechanical Delivery Methods, and Molecular Adjuvants to Increase Immunogenicity. Hum. Vaccin. Immunother. 2017, 13, 2837–2848. [Google Scholar] [CrossRef] [PubMed]
- Shirai, S.; Kawai, A.; Shibuya, M.; Munakata, L.; Omata, D.; Suzuki, R.; Yoshioka, Y. Lipid Nanoparticle Acts as a Potential Adjuvant for Influenza Split Vaccine without Inducing Inflammatory Responses. Vaccines 2020, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.A. Regulatory Considerations for Nucleic Acid Vaccines. Vaccine 1994, 12, 1515–1519. [Google Scholar] [CrossRef]
- Robertson, J.S. Safety Considerations for Nucleic Acid Vaccines. Vaccine 1994, 12, 1526–1528. [Google Scholar] [CrossRef]
- Ledwith, B.J.; Manam, S.; Troilo, P.J.; Barnum, A.B.; Pauley, C.J.; Griffiths II, T.G.; Harper, L.B.; Beare, C.M.; Bagdon, W.J.; Nichols, W.W. Plasmid DNA Vaccines: Investigation of Integration into Host Cellular DNA Following Intramuscular Injection in Mice. Intervirology 2000, 43, 258–272. [Google Scholar] [CrossRef]
- Sheets, R.L.; Stein, J.; Manetz, T.S.; Duffy, C.; Nason, M.; Andrews, C.; Kong, W.-P.; Nabel, G.J.; Gomez, P.L. Biodistribution of DNA Plasmid Vaccines against HIV-1, Ebola, Severe Acute Respiratory Syndrome, or West Nile Virus Is Similar, without Integration, despite Differing Plasmid Backbones or Gene Inserts. Toxicol. Sci. 2006, 91, 610–619. [Google Scholar] [CrossRef]
- Petkov, S.P.; Heuts, F.; Krotova, O.A.; Kilpelainen, A.; Engström, G.; Starodubova, E.S.; Isaguliants, M.G. Evaluation of Immunogen Delivery by DNA Immunization Using Non-Invasive Bioluminescence Imaging. Hum. Vaccin. Immunother. 2013, 9, 2228–2236. [Google Scholar] [CrossRef]


| Particle Name | Encapsulated Nucleic Acid | Components | Molar Ratio | LNP Diameter * (Mean ± SD, nm) | Encapsulation Efficiency * |
|---|---|---|---|---|---|
| SM102-DNA | DNA | SM-102:DSPC:Chol:DMG-PEG 2000 | 50:10:38.5:1.5 [30,31] | 78 ± 24 | 97% |
| ALC0315-DNA | DNA | ALC-0315:DSPC:Chol:ALC-0159 | 46.3:9.4:42.7:1.6 [33] | 67 ± 19 | 97% |
| KC2-DNA | DNA | KC2:DSPC:Chol:DMG-PEG 2000 | 50:10:38.5:1.5 [30,31] | 82 ± 36 | 99% |
| SM102-RNA | RNA | SM-102:DSPC:Chol:DMG-PEG 2000 | 50:10:38.5:1.5 [30,31] | 74 ± 20 | 88% |
| ALC0315-RNA | RNA | ALC-0315:DSPC:Chol:ALC-0159 | 46.3:9.4:42.7:1.6 [33] | 71 ± 21 | 92% |
| KC2-RNA | RNA | KC2:DSPC:Chol:DMG-PEG 2000 | 50:10:38.5:1.5 [30,31] | 83 ± 31 | 97% |
| Particle Name | Signal Duration (Inj. Site) | Signal Duration (Liver) |
|---|---|---|
| SM102-DNA | >28 days | 48 h |
| ALC0315-DNA | >28 days | 72 h |
| KC2-DNA | 48 h | N/A |
| SM102-RNA | 21 days | 7 days |
| ALC0315-RNA | 21 days | 7 days |
| KC2-RNA | 14 days | 72 h |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Pfeifle, A.; Lansdell, C.; Frahm, G.; Cecillon, J.; Tamming, L.; Gravel, C.; Gao, J.; Thulasi Raman, S.N.; Wang, L.; et al. The Expression Kinetics and Immunogenicity of Lipid Nanoparticles Delivering Plasmid DNA and mRNA in Mice. Vaccines 2023, 11, 1580. https://doi.org/10.3390/vaccines11101580
Zhang W, Pfeifle A, Lansdell C, Frahm G, Cecillon J, Tamming L, Gravel C, Gao J, Thulasi Raman SN, Wang L, et al. The Expression Kinetics and Immunogenicity of Lipid Nanoparticles Delivering Plasmid DNA and mRNA in Mice. Vaccines. 2023; 11(10):1580. https://doi.org/10.3390/vaccines11101580
Chicago/Turabian StyleZhang, Wanyue, Annabelle Pfeifle, Casey Lansdell, Grant Frahm, Jonathon Cecillon, Levi Tamming, Caroline Gravel, Jun Gao, Sathya N. Thulasi Raman, Lisheng Wang, and et al. 2023. "The Expression Kinetics and Immunogenicity of Lipid Nanoparticles Delivering Plasmid DNA and mRNA in Mice" Vaccines 11, no. 10: 1580. https://doi.org/10.3390/vaccines11101580
APA StyleZhang, W., Pfeifle, A., Lansdell, C., Frahm, G., Cecillon, J., Tamming, L., Gravel, C., Gao, J., Thulasi Raman, S. N., Wang, L., Sauve, S., Rosu-Myles, M., Li, X., & Johnston, M. J. W. (2023). The Expression Kinetics and Immunogenicity of Lipid Nanoparticles Delivering Plasmid DNA and mRNA in Mice. Vaccines, 11(10), 1580. https://doi.org/10.3390/vaccines11101580







