Extraordinary Titer and Broad Anti-SARS-CoV-2 Neutralization Induced by Stabilized RBD Nanoparticles from Strain BA.5
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal
2.2. Cell Line
2.3. Protein Production and Purification
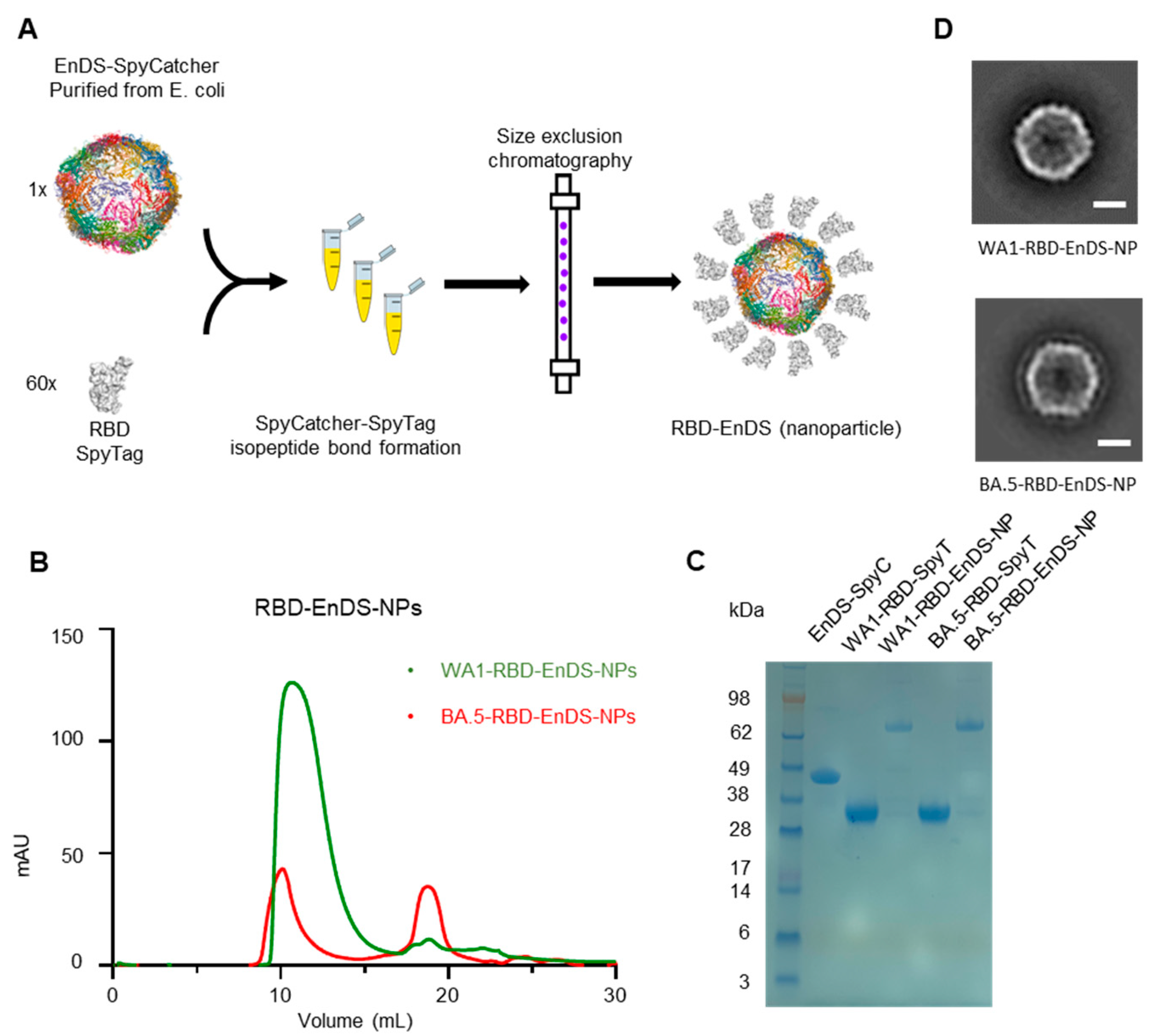
2.4. EnDS-SpyC and RBD-SpyT Conjugation
2.5. Negative-Stain Electron Microscopy (EM)
2.6. Preparation of SARS-CoV-2 Antibodies
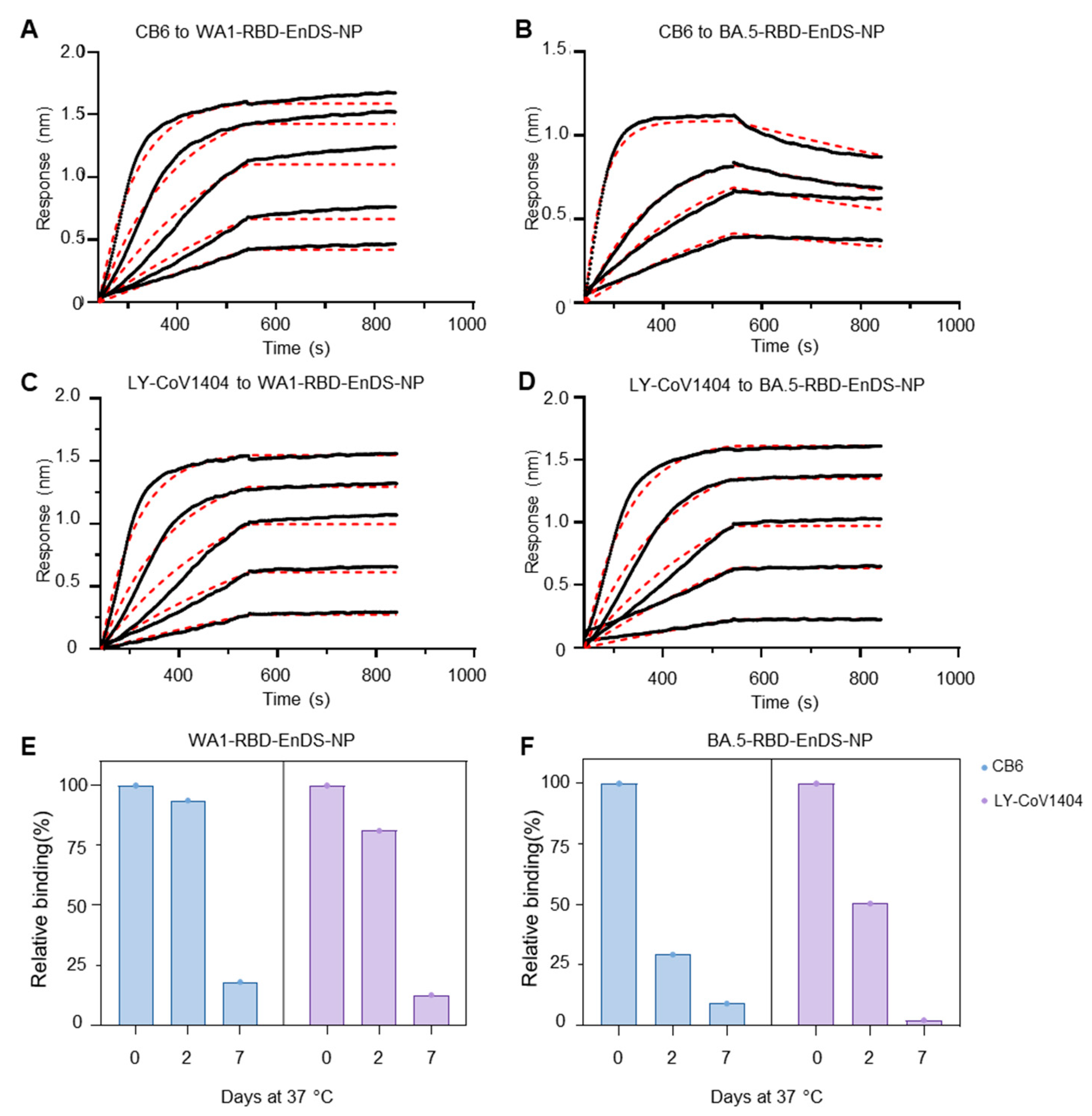
2.7. Bio-Layer Interferometry
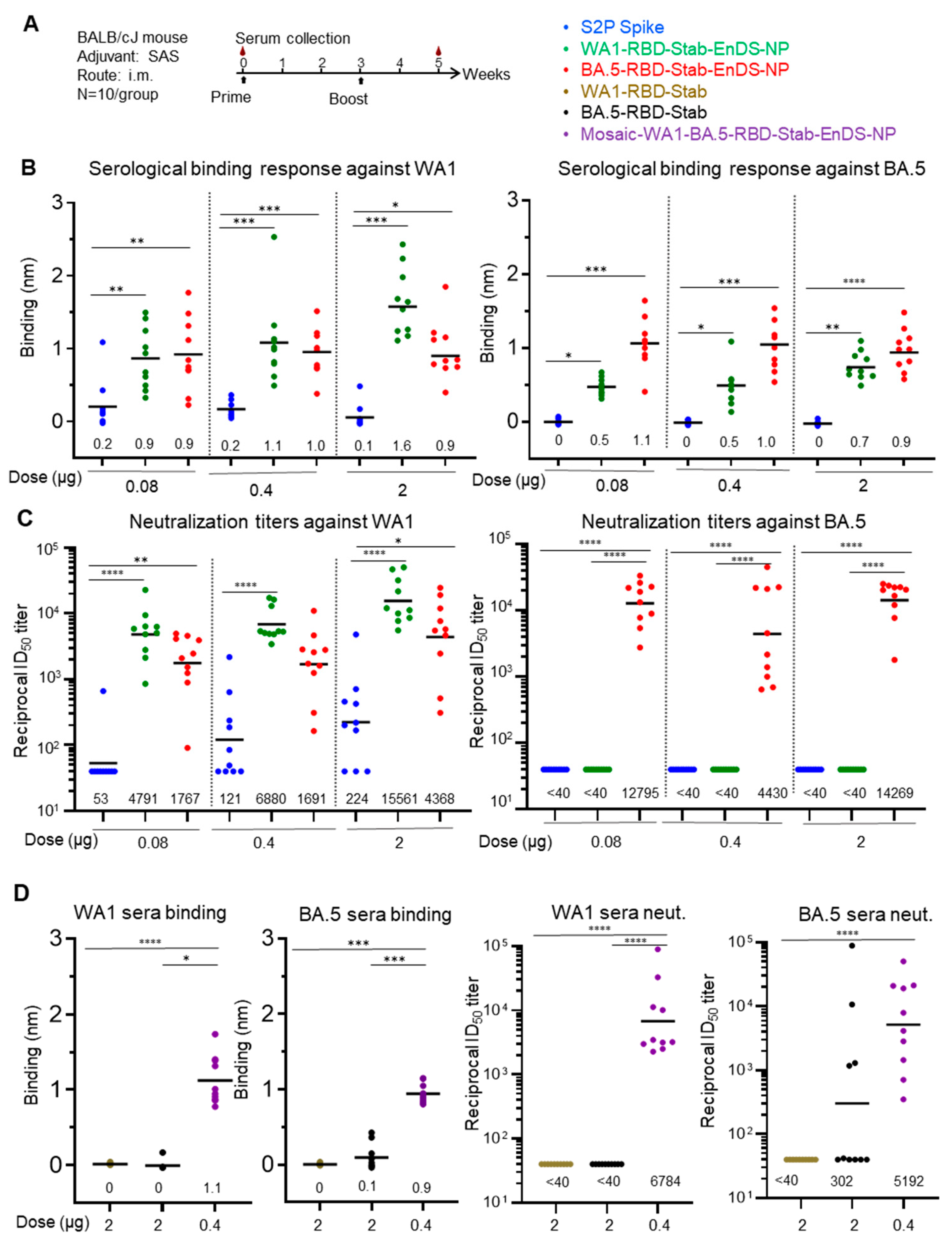
2.8. Mouse Immunization
2.9. Lentivirus-Based Pseudovirus Neutralization Assay
2.10. Statistical Analysis
3. Results
3.1. Design and Characterization of EnDS Nanoparticles Displaying WA1 or BA.5 RBDs
3.2. Antigenic Analysis and Stability Assessment for EnDS Nanoparticles Displaying WA1- and BA.5-RBDs
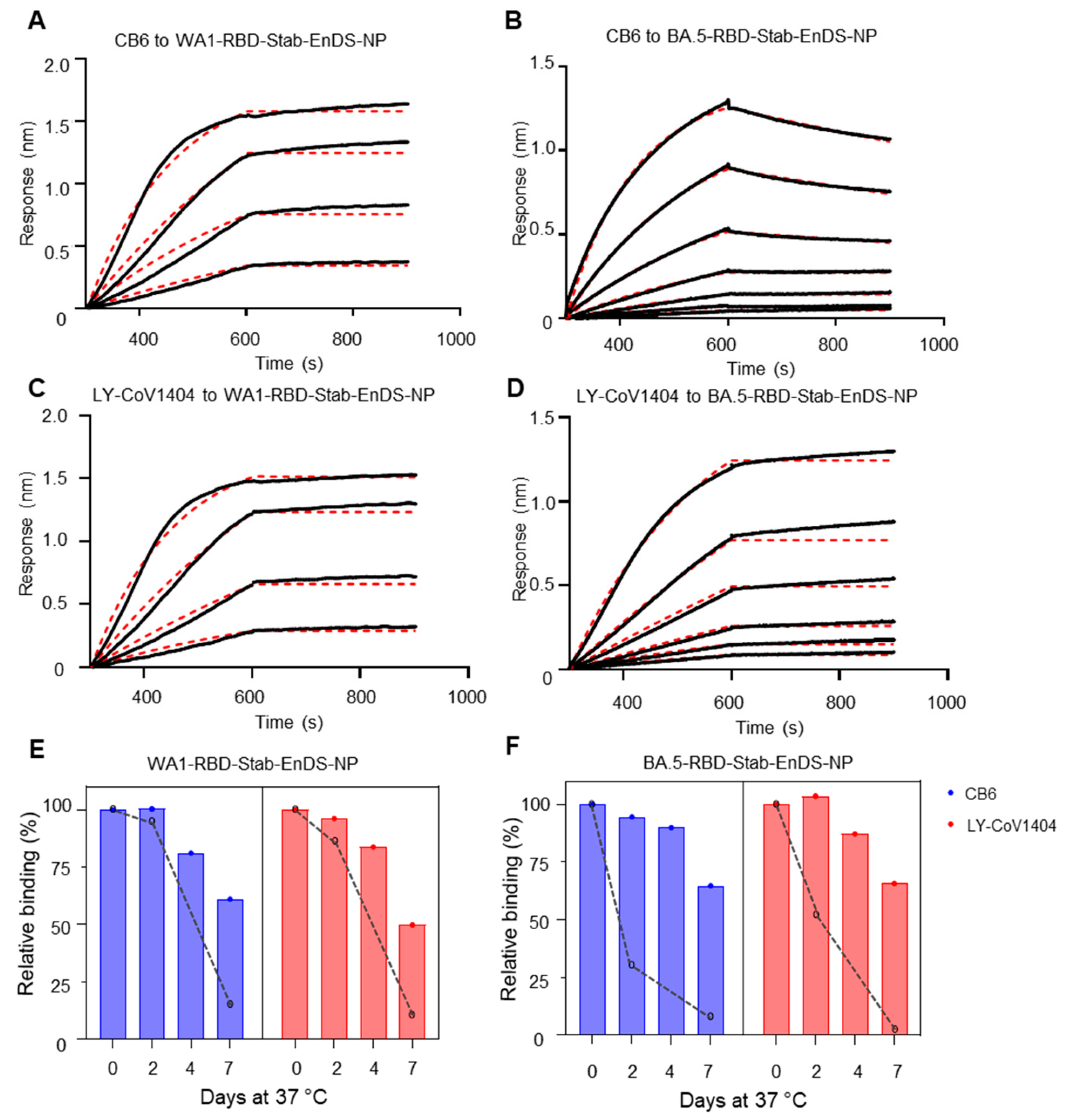
3.3. Development of Stabilized Nanoparticle Immunogens and Characterization
3.4. Stabilized WA1 and BA.5 Nanoparticles Show Superior Antigenicity and Improved Stability at 37 °C
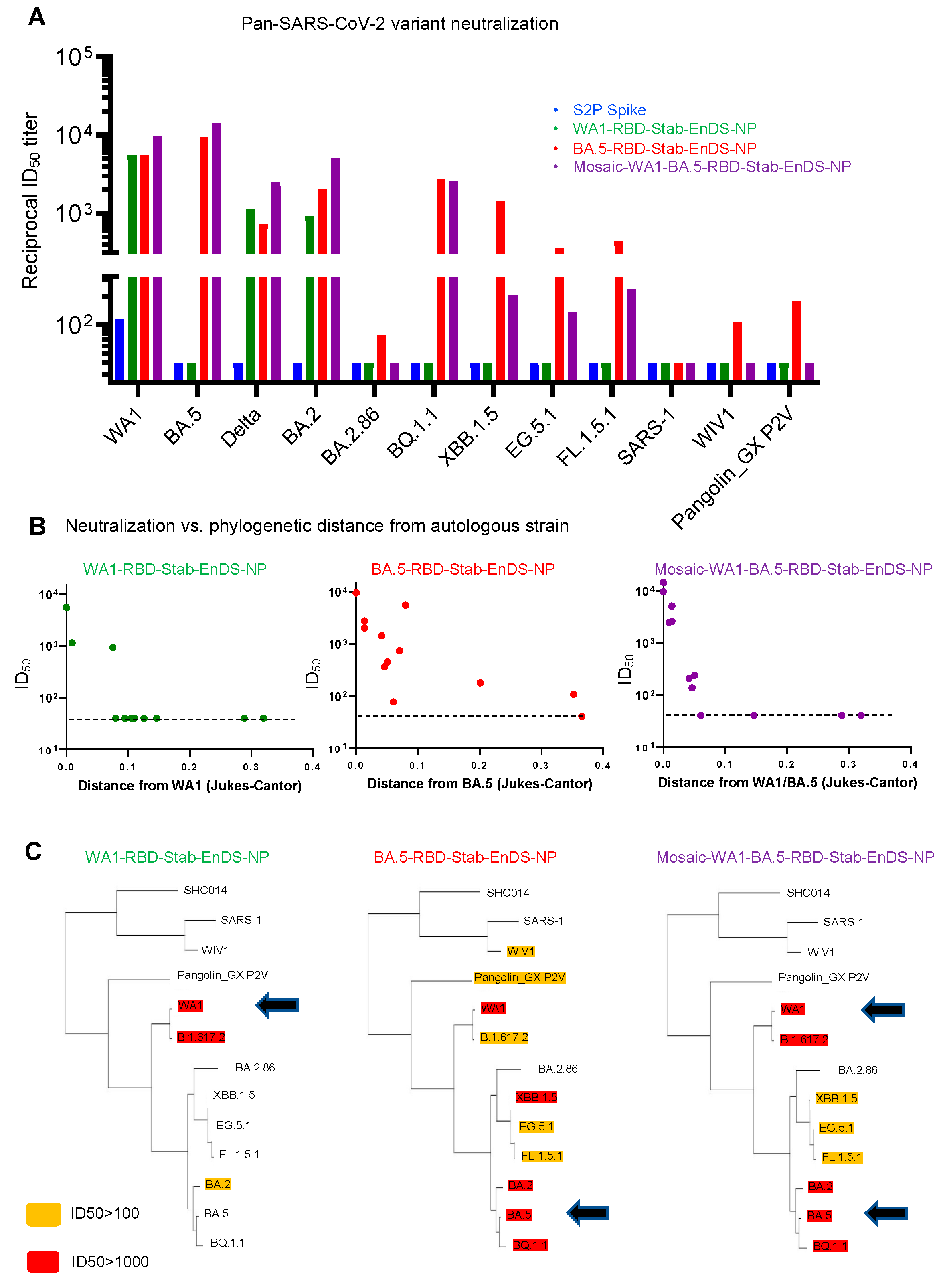
3.5. Immunization of Stabilized RBD-EnDS Nanoparticles Elicited Strong Anti-SARS-CoV-2 Pseudovirus Neutralizing Responses in Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lazarus, J.V.; Romero, D.; Kopka, C.J.; Karim, S.A.; Abu-Raddad, L.J.; Almeida, G.; Baptista-Leite, R.; Barocas, J.A.; Barreto, M.L.; Bar-Yam, Y. A multinational Delphi consensus to end the COVID-19 public health threat. Nature 2022, 611, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J. COVID-19 will continue but the end of the pandemic is near. Lancet 2022, 399, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Painter, M.M.; Johnston, T.S.; Lundgreen, K.A.; Santos, J.J.S.; Qin, J.S.; Goel, R.R.; Apostolidis, S.A.; Mathew, D.; Fulmer, B.; Williams, J.C.; et al. Prior vaccination promotes early activation of memory T cells and enhances immune responses during SARS-CoV-2 breakthrough infection. Nat. Immunol. 2023, 24, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Lipsitch, M.; Krammer, F.; Regev-Yochay, G.; Lustig, Y.; Balicer, R.D. SARS-CoV-2 breakthrough infections in vaccinated individuals: Measurement, causes and impact. Nat. Rev. Immunol. 2022, 22, 57–65. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Follmann, D.; Neuzil, K.M.; August, A.; Clouting, H.; Fortier, G.; Deng, W.; Han, S.; et al. Phase 3 Trial of mRNA-1273 during the Delta-Variant Surge. N. Engl. J. Med. 2021, 385, 2485–2487. [Google Scholar] [CrossRef]
- Bergwerk, M.; Gonen, T.; Lustig, Y.; Amit, S.; Lipsitch, M.; Cohen, C.; Mandelboim, M.; Levin, E.G.; Rubin, C.; Indenbaum, V.; et al. COVID-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021, 385, 1474–1484. [Google Scholar] [CrossRef]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.G.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022, 602, 654–656. [Google Scholar] [CrossRef]
- Mao, T.; Israelow, B.; Pena-Hernandez, M.A.; Suberi, A.; Zhou, L.; Luyten, S.; Reschke, M.; Dong, H.; Homer, R.J.; Saltzman, W.M.; et al. Unadjuvanted intranasal spike vaccine elicits protective mucosal immunity against sarbecoviruses. Science 2022, 378, eabo2523. [Google Scholar] [CrossRef]
- Mudgal, R.; Nehul, S.; Tomar, S. Prospects for mucosal vaccine: Shutting the door on SARS-CoV-2. Hum. Vaccines Immunother. 2020, 16, 2921–2931. [Google Scholar] [CrossRef]
- Lapuente, D.; Fuchs, J.; Willar, J.; Vieira Antao, A.; Eberlein, V.; Uhlig, N.; Issmail, L.; Schmidt, A.; Oltmanns, F.; Peter, A.S.; et al. Protective mucosal immunity against SARS-CoV-2 after heterologous systemic prime-mucosal boost immunization. Nat. Commun. 2021, 12, 6871. [Google Scholar] [CrossRef]
- Sui, Y.; Li, J.; Zhang, R.; Prabhu, S.K.; Andersen, H.; Venzon, D.; Cook, A.; Brown, R.; Teow, E.; Velasco, J.; et al. Protection against SARS-CoV-2 infection by a mucosal vaccine in rhesus macaques. JCI Insight 2021, 6, e148494. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Gnanapragasam, P.N.P.; Lee, Y.E.; Hoffman, P.R.; Ou, S.; Kakutani, L.M.; Keeffe, J.R.; Wu, H.J.; Howarth, M.; West, A.P.; et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 2021, 371, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Van de Steen, A.; Khalife, R.; Colant, N.; Khan, H.M.; Deveikis, M.; Charalambous, S.; Robinson, C.M.; Dabas, R.; Serna, S.E.; Catana, D.A. Bioengineering bacterial encapsulin nanocompartments as targeted drug delivery system. Synth. Syst. Biotechnol. 2021, 6, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Miranda, M.C.; Schäfer, A.; Pham, M.N.; Greaney, A.; Arunachalam, P.S.; Navarro, M.-J.; Tortorici, M.A.; Rogers, K.; O’Connor, M.A. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell 2021, 184, 5432–5447.e16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chao, C.W.; Tsybovsky, Y.; Abiona, O.M.; Hutchinson, G.B.; Moliva, J.I.; Olia, A.S.; Pegu, A.; Phung, E.; Stewart-Jones, G.B.E.; et al. A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 2020, 10, 18149. [Google Scholar] [CrossRef] [PubMed]
- Dickey, T.H.; Ma, R.; Orr-Gonzalez, S.; Ouahes, T.; Patel, P.; McAleese, H.; Butler, B.; Eudy, E.; Eaton, B.; Murphy, M.; et al. Design of a stabilized RBD enables potently neutralizing SARS-CoV-2 single-component nanoparticle vaccines. Cell Rep. 2023, 42, 112266. [Google Scholar] [CrossRef]
- Fan, C.; Cohen, A.A.; Park, M.; Hung, A.F.; Keeffe, J.R.; Gnanapragasam, P.N.P.; Lee, Y.E.; Gao, H.; Kakutani, L.M.; Wu, Z.; et al. Neutralizing monoclonal antibodies elicited by mosaic RBD nanoparticles bind conserved sarbecovirus epitopes. Immunity 2022, 55, 2419–2435.e10. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, D.A.; Gonzalez Ruiz, G.; Escalante-Bermudez, C.; Garcia Artalejo, J.A.; Gomez Pena, T.; Gomez, J.A.; Gonzalez-Martinez, E.; Cazanas Quintana, Y.; Fundora Barrios, T.; Hernandez, T.; et al. Efficient capture of recombinant SARS-CoV-2 receptor-binding domain (RBD) with citrate-coated magnetic iron oxide nanoparticles. Nanoscale 2023, 15, 7854–7869. [Google Scholar] [CrossRef]
- Hutchinson, G.B.; Abiona, O.M.; Ziwawo, C.T.; Werner, A.P.; Ellis, D.; Tsybovsky, Y.; Leist, S.R.; Palandjian, C.; West, A.; Fritch, E.J.; et al. Nanoparticle display of prefusion coronavirus spike elicits S1-focused cross-reactive antibody response against diverse coronavirus subgenera. Nat. Commun. 2023, 14, 6195. [Google Scholar] [CrossRef]
- Joyce, M.G.; Chen, W.H.; Sankhala, R.S.; Hajduczki, A.; Thomas, P.V.; Choe, M.; Martinez, E.J.; Chang, W.C.; Peterson, C.E.; Morrison, E.B.; et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 2021, 37, 110143. [Google Scholar] [CrossRef]
- Lee, D.B.; Kim, H.; Jeong, J.H.; Jang, U.S.; Jang, Y.; Roh, S.; Jeon, H.; Kim, E.J.; Han, S.Y.; Maeng, J.Y.; et al. Mosaic RBD nanoparticles induce intergenus cross-reactive antibodies and protect against SARS-CoV-2 challenge. Proc. Natl. Acad. Sci. USA 2023, 120, e2208425120. [Google Scholar] [CrossRef]
- Li, D.; Martinez, D.R.; Schafer, A.; Chen, H.; Barr, M.; Sutherland, L.L.; Lee, E.; Parks, R.; Mielke, D.; Edwards, W.; et al. Breadth of SARS-CoV-2 neutralization and protection induced by a nanoparticle vaccine. Nat. Commun. 2022, 13, 6309. [Google Scholar] [CrossRef] [PubMed]
- Lovell, J.F.; Baik, Y.O.; Choi, S.K.; Lee, C.; Lee, J.Y.; Miura, K.; Huang, W.C.; Park, Y.S.; Woo, S.J.; Seo, S.H.; et al. Interim analysis from a phase 2 randomized trial of EuCorVac-19: A recombinant protein SARS-CoV-2 RBD nanoliposome vaccine. BMC Med. 2022, 20, 462. [Google Scholar] [CrossRef] [PubMed]
- Meena, J.; Singhvi, P.; Srichandan, S.; Dandotiya, J.; Verma, J.; Singh, M.; Ahuja, R.; Panwar, N.; Wani, T.Q.; Khatri, R.; et al. RBD decorated PLA nanoparticle admixture with aluminum hydroxide elicit robust and long lasting immune response against SARS-CoV-2. Eur. J. Pharm. Biopharm. 2022, 176, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Royal, J.M.; Simpson, C.A.; McCormick, A.A.; Phillips, A.; Hume, S.; Morton, J.; Shepherd, J.; Oh, Y.; Swope, K.; DeBeauchamp, J.L.; et al. Development of a SARS-CoV-2 Vaccine Candidate Using Plant-Based Manufacturing and a Tobacco Mosaic Virus-like Nano-Particle. Vaccines 2021, 9, 1347. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Kong, D.; Guo, E.; Zhao, J.; Jia, J.; Wang, R.; Ma, J.; Chen, M.; Lu, J.; Yu, C.; et al. A Polysaccharide-RBD-Fc-Conjugated COVID-19 Vaccine, SCTV01A, Showed High Immunogenicity and Low Toxicity in Animal Models. Vaccines 2023, 11, 526. [Google Scholar] [CrossRef]
- Tabynov, K.; Solomadin, M.; Turebekov, N.; Babayeva, M.; Fomin, G.; Yadagiri, G.; Renu, S.; Yerubayev, T.; Petrovsky, N.; Renukaradhya, G.J.; et al. An intranasal vaccine comprising SARS-CoV-2 spike receptor-binding domain protein entrapped in mannose-conjugated chitosan nanoparticle provides protection in hamsters. Sci. Rep. 2023, 13, 12115. [Google Scholar] [CrossRef]
- Xia, M.; Lopez, K.; Vago, F.S.; Huang, P.; Auguste, D.I.; Jiang, W.; Auguste, A.J.; Tan, M. A bioengineered pseudovirus nanoparticle displaying SARS-CoV 2 RBD fully protects mice from mortality and weight loss caused by SARS-CoV 2 challenge. Biotechnol. J. 2023, 18, e2300130. [Google Scholar] [CrossRef]
- Xiao, L.; Yu, W.; Shen, L.; Yan, W.; Qi, J.; Hu, T. Mucosal SARS-CoV-2 Nanoparticle Vaccine Based on Mucosal Adjuvants and Its Immune Effectiveness by Intranasal Administration. ACS Appl. Mater. Interfaces 2023, 15, 35895–35905. [Google Scholar] [CrossRef]
- Yan, W.; Yu, W.; Shen, L.; Xiao, L.; Qi, J.; Hu, T. A SARS-CoV-2 nanoparticle vaccine based on chemical conjugation of loxoribine and SpyCatcher/SpyTag. Int. J. Biol. Macromol. 2023, 253, 127159. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, S.; Liu, J.; Chen, R.; Zhang, Y.; Lin, Y.; Xi, Z.; Deng, J.; Pu, Z.; Liang, C.; et al. A Mosaic Nanoparticle Vaccine Elicits Potent Mucosal Immune Response with Significant Cross-Protection Activity against Multiple SARS-CoV-2 Sublineages. Adv. Sci. 2023, 10, e2301034. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; van Doremalen, N.; Greaney, A.J.; Andersen, H.; Sharma, A.; Starr, T.N.; Keeffe, J.R.; Fan, C.; Schulz, J.E.; Gnanapragasam, P.N.P.; et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science 2022, 377, eabq0839. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.F.; Sun, C.; Zhuang, Z.; Yuan, R.Y.; Zheng, Q.; Li, J.P.; Zhou, P.P.; Chen, X.C.; Liu, Z.; Zhang, X.; et al. Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano 2021, 15, 2738–2752. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.S.; Walls, A.C.; Golden, N.; Atyeo, C.; Fischinger, S.; Li, C.; Aye, P.; Navarro, M.J.; Lai, L.; Edara, V.V.; et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 2021, 594, 253–258. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lin, X.; Wang, Y.; Abraham, C.; Sou, C.; Ngo, T.; Zhang, Y.; Wilson, I.A.; Zhu, J. Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci. Adv. 2021, 7, eabf1591. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.I.; Kim, D.; Yu, K.M.; Seo, H.D.; Lee, S.A.; Casel, M.A.B.; Jang, S.G.; Kim, S.; Jung, W.; Lai, C.J.; et al. Development of Spike Receptor-Binding Domain Nanoparticles as a Vaccine Candidate against SARS-CoV-2 Infection in Ferrets. MBio 2021, 12, e00230-21. [Google Scholar] [CrossRef]
- King, H.A.D.; Joyce, M.G.; Lakhal-Naouar, I.; Ahmed, A.; Cincotta, C.M.; Subra, C.; Peachman, K.K.; Hack, H.R.; Chen, R.E.; Thomas, P.V.; et al. Efficacy and breadth of adjuvanted SARS-CoV-2 receptor-binding domain nanoparticle vaccine in macaques. Proc. Natl. Acad. Sci. USA 2021, 118, e2106433118. [Google Scholar] [CrossRef]
- Ma, X.; Zou, F.; Yu, F.; Li, R.; Yuan, Y.; Zhang, Y.; Zhang, X.; Deng, J.; Chen, T.; Song, Z.; et al. Nanoparticle Vaccines Based on the Receptor Binding Domain (RBD) and Heptad Repeat (HR) of SARS-CoV-2 Elicit Robust Protective Immune Responses. Immunity 2020, 53, 1315–1330.e9. [Google Scholar] [CrossRef]
- Saunders, K.O.; Lee, E.; Parks, R.; Martinez, D.R.; Li, D.; Chen, H.; Edwards, R.J.; Gobeil, S.; Barr, M.; Mansouri, K.; et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature 2021, 594, 553–559. [Google Scholar] [CrossRef]
- Anselmo, A.C.; Mitragotri, S. Nanoparticles in the clinic: An update post COVID-19 vaccines. Bioeng. Transl. Med. 2021, 6, e10246. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, L.; Yu, H.; Lv, P.; Lei, Z.; Zeng, Y.; Liu, G.; Cheng, T. Ferritin nanocage-based antigen delivery nanoplatforms: Epitope engineering for peptide vaccine design. Biomater. Sci. 2019, 7, 1794–1800. [Google Scholar] [CrossRef] [PubMed]
- Kheirollahpour, M.; Mehrabi, M.; Dounighi, N.M.; Mohammadi, M.; Masoudi, A. Nanoparticles and vaccine development. Pharm. Nanotechnol. 2020, 8, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Joyce, M.G.; Gillespie, R.A.; Gallagher, J.R.; Andrews, S.F.; Yassine, H.M.; Wheatley, A.K.; Fisher, B.E.; Ambrozak, D.R.; Creanga, A.; et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019, 20, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Rivera, O.A.; Shukla, S.; Shin, M.D.; Chen, A.; Beiss, V.; Moreno-Gonzalez, M.A.; Zheng, Y.; Clark, A.E.; Carlin, A.F.; Pokorski, J.K. Cowpea mosaic virus nanoparticle vaccine candidates displaying peptide epitopes can neutralize the severe acute respiratory syndrome coronavirus. ACS Infect. Dis. 2021, 7, 3096–3110. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Eom, S.; Kim, H.-u.; Bae, Y.; Jung, H.S.; Kang, S. Load and display: Engineering encapsulin as a modular nanoplatform for protein-cargo encapsulation and protein-ligand decoration using split intein and SpyTag/SpyCatcher. Biomacromolecules 2021, 22, 3028–3039. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Giessen, T.W. Advances in encapsulin nanocompartment biology and engineering. Biotechnol. Bioeng. 2021, 118, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Gabashvili, A.N.; Vodopyanov, S.S.; Chmelyuk, N.S.; Sarkisova, V.A.; Fedotov, K.A.; Efremova, M.V.; Abakumov, M.A. Encapsulin based self-assembling iron-containing protein nanoparticles for stem cells MRI visualization. Int. J. Mol. Sci. 2021, 22, 12275. [Google Scholar] [CrossRef]
- Corrigan, A.R.; Duan, H.; Cheng, C.; Gonelli, C.A.; Ou, L.; Xu, K.; DeMouth, M.E.; Geng, H.; Narpala, S.; O’Connell, S.; et al. Fusion peptide priming reduces immune responses to HIV-1 envelope trimer base. Cell Rep. 2021, 35, 108937. [Google Scholar] [CrossRef]
- Dickey, T.H.; Tang, W.K.; Butler, B.; Ouahes, T.; Orr-Gonzalez, S.; Salinas, N.D.; Lambert, L.E.; Tolia, N.H. Design of the SARS-CoV-2 RBD vaccine antigen improves neutralizing antibody response. Sci. Adv. 2022, 8, eabq8276. [Google Scholar] [CrossRef]
- Aida, Y.; Pabst, M.J. Removal of endotoxin from protein solutions by phase separation using Triton X-114. J. Immunol. Methods 1990, 132, 191–195. [Google Scholar] [CrossRef]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.; Radermacher, M.; Penczek, P.; Zhu, J.; Li, Y.; Ladjadj, M.; Leith, A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996, 116, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schafer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef]
- Bottcher, E.; Matrosovich, T.; Beyerle, M.; Klenk, H.D.; Garten, W.; Matrosovich, M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006, 80, 9896–9898. [Google Scholar] [CrossRef]
- Wang, L.; Shi, W.; Joyce, M.G.; Modjarrad, K.; Zhang, Y.; Leung, K.; Lees, C.R.; Zhou, T.; Yassine, H.M.; Kanekiyo, M.; et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015, 6, 7712. [Google Scholar] [CrossRef]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Zhou, T.; Wang, L.; Misasi, J.; Pegu, A.; Zhang, Y.; Harris, D.R.; Olia, A.S.; Talana, C.A.; Yang, E.S.; Chen, M.; et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science 2022, 376, eabn8897. [Google Scholar] [CrossRef]
- Dickey, T.H.; Gupta, R.; McAleese, H.; Ouahes, T.; Orr-Gonzalez, S.; Ma, R.; Muratova, O.; Salinas, N.D.; Hume, J.C.C.; Lambert, L.E.; et al. Design of a stabilized non-glycosylated Pfs48/45 antigen enables a potent malaria transmission-blocking nanoparticle vaccine. NPJ Vaccines 2023, 8, 20. [Google Scholar] [CrossRef]
- Newsroom. UW Medicine COVID-19 Vaccine Wins South Korea Approval. 2023. Available online: https://newsroom.uw.edu/news/uw-medicine-covid-19-vaccine-wins-south-korea-approval (accessed on 14 December 2023).
- Wikipedia. Skycovione. 2023. Available online: https://en.wikipedia.org/wiki/Skycovione (accessed on 14 December 2023).
- Song, J.Y.; Choi, W.S.; Heo, J.Y.; Lee, J.S.; Jung, D.S.; Kim, S.W.; Park, K.H.; Eom, J.S.; Jeong, S.J.; Lee, J.; et al. Safety and immunogenicity of a SARS-CoV-2 recombinant protein nanoparticle vaccine (GBP510) adjuvanted with AS03: A randomised, placebo-controlled, observer-blinded phase 1/2 trial. EClinicalMedicine 2022, 51, 101569. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Gao, L.; Tao, L.; Hadinegoro, S.R.; Erkin, M.; Ying, Z.; He, P.; Girsang, R.T.; Vergara, H.; Akram, J.; et al. Efficacy and Safety of the RBD-Dimer-Based COVID-19 Vaccine ZF2001 in Adults. N. Engl. J. Med. 2022, 386, 2097–2111. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Bernal, F.; Ricardo-Cobas, M.C.; Martin-Bauta, Y.; Rodriguez-Martinez, E.; Urrutia-Perez, K.; Urrutia-Perez, K.; Quintana-Guerra, J.; Navarro-Rodriguez, Z.; Pinera-Martinez, M.; Rodriguez-Reinoso, J.L.; et al. A phase 3, randomised, double-blind, placebo-controlled clinical trial evaluation of the efficacy and safety of a SARS-CoV-2 recombinant spike RBD protein vaccine in adults (ABDALA-3 study). Lancet Reg. Health Am. 2023, 21, 100497. [Google Scholar] [CrossRef] [PubMed]
- Thuluva, S.; Paradkar, V.; Gunneri, S.; Yerroju, V.; Mogulla, R.; Suneetha, P.V.; Turaga, K.; Kyasani, M.; Manoharan, S.K.; Adabala, S.; et al. Immunogenicity and safety of Biological E’s CORBEVAX vaccine compared to COVISHIELD (ChAdOx1 nCoV-19) vaccine studied in a phase-3, single blind, multicentre, randomized clinical trial. Hum. Vaccin. Immunother. 2023, 19, 2203632. [Google Scholar] [CrossRef]
- Yassini, P.; Hutchens, M.; Paila, Y.D.; Schoch, L.; Aunins, A.; Siangphoe, U.; Paris, R. Interim analysis of a phase 1 randomized clinical trial on the safety and immunogenicity of the mRNA-1283 SARS-CoV-2 vaccine in adults. Hum. Vaccin. Immunother. 2023, 19, 2190690. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zhang, B.; Ou, L.; Qiu, Q.; Wang, L.; Bylund, T.; Kong, W.-P.; Shi, W.; Tsybovsky, Y.; Wu, L.; et al. Extraordinary Titer and Broad Anti-SARS-CoV-2 Neutralization Induced by Stabilized RBD Nanoparticles from Strain BA.5. Vaccines 2024, 12, 37. https://doi.org/10.3390/vaccines12010037
Wang Z, Zhang B, Ou L, Qiu Q, Wang L, Bylund T, Kong W-P, Shi W, Tsybovsky Y, Wu L, et al. Extraordinary Titer and Broad Anti-SARS-CoV-2 Neutralization Induced by Stabilized RBD Nanoparticles from Strain BA.5. Vaccines. 2024; 12(1):37. https://doi.org/10.3390/vaccines12010037
Chicago/Turabian StyleWang, Zhantong, Baoshan Zhang, Li Ou, Qi Qiu, Lingshu Wang, Tatsiana Bylund, Wing-Pui Kong, Wei Shi, Yaroslav Tsybovsky, Lingyuan Wu, and et al. 2024. "Extraordinary Titer and Broad Anti-SARS-CoV-2 Neutralization Induced by Stabilized RBD Nanoparticles from Strain BA.5" Vaccines 12, no. 1: 37. https://doi.org/10.3390/vaccines12010037
APA StyleWang, Z., Zhang, B., Ou, L., Qiu, Q., Wang, L., Bylund, T., Kong, W.-P., Shi, W., Tsybovsky, Y., Wu, L., Zhou, Q., Chaudhary, R., Choe, M., Dickey, T. H., El Anbari, M., Olia, A. S., Rawi, R., Teng, I.-T., Wang, D., ... Kwong, P. D. (2024). Extraordinary Titer and Broad Anti-SARS-CoV-2 Neutralization Induced by Stabilized RBD Nanoparticles from Strain BA.5. Vaccines, 12(1), 37. https://doi.org/10.3390/vaccines12010037





