Targeted Protein-Specific Multi-Epitope-Based Vaccine Designing against Human Cytomegalovirus by Using Immunoinformatics Approaches
Abstract
1. Introduction
2. Methodology
2.1. Data Retrieval and Target Proteins Selection
2.2. Highly Immunogenic Epitopes Selection
2.3. Putative Vaccines Construction
2.4. Structural Modelling and Evaluations of MEVCs
2.5. Molecular Docking and Interaction Analysis
2.6. Cloning of MEVCs
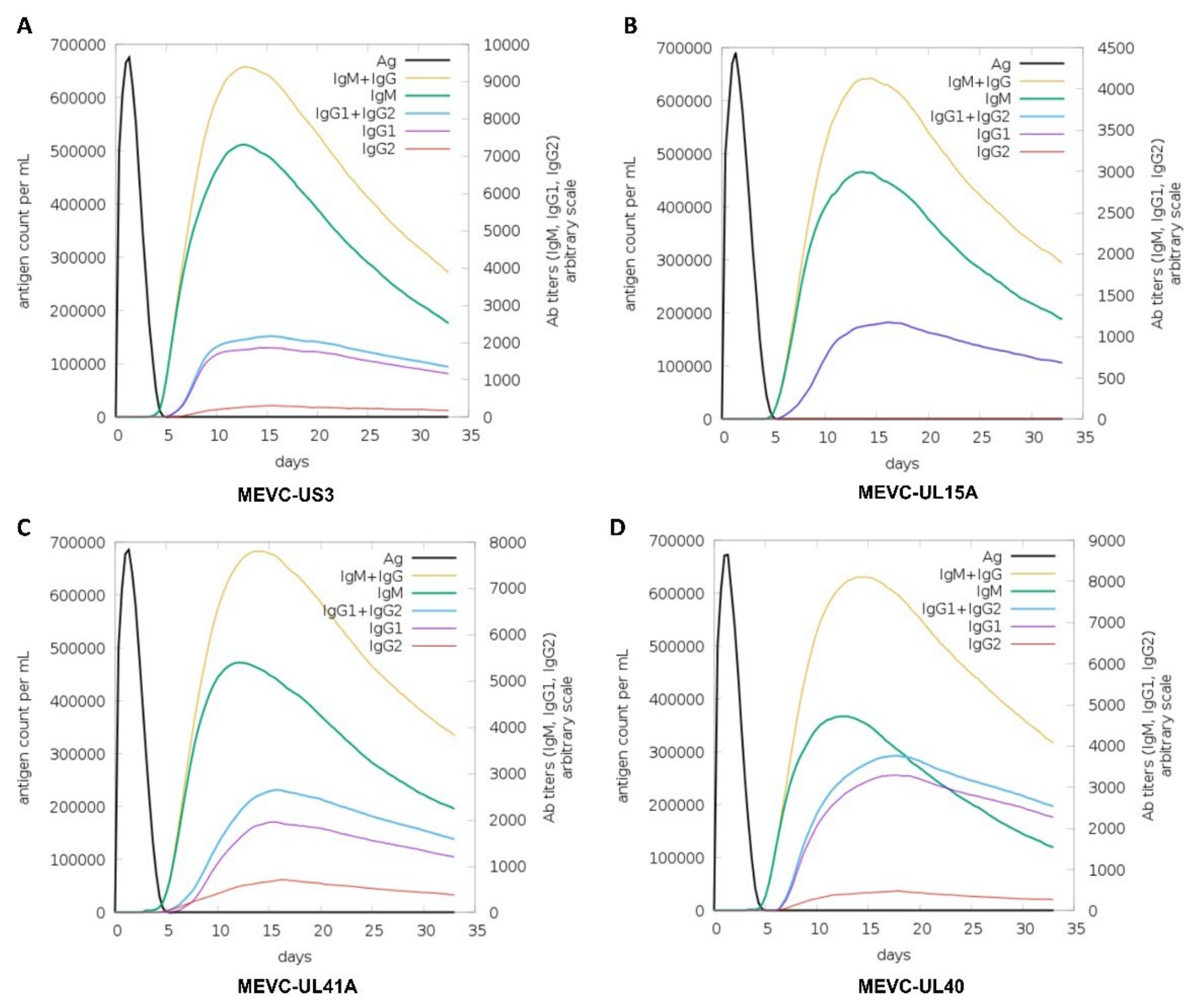
2.7. Immune Simulation of MEVCs
3. Results
3.1. Target proteins Selection for Potential Epitopes Prioritization in Vaccine Construction
3.2. Multi-Epitope-Based Vaccine Constructs
3.3. Physiochemical Properties Evaluations
3.4. Structural Modelling and Validations of the Designed MEVCs
3.5. Molecular Docking and Interactions of MEVCs with Human TLR4
3.6. Codon Optimization and In silico Cloning
3.7. Immune Simulation of the Proposed MEVCs
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Emery, V.C.; Sabin, C.A.; Cope, A.V.; Gor, D.; Hassan-Walker, A.F.; Griffiths, P.D. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 2000, 355, 2032–2036. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.A. Preventing infection by human cytomegalovirus. J. Infect. Dis. 2020, 221, S123–S127. [Google Scholar] [CrossRef]
- Modlin, J.F.; Arvin, A.M.; Fast, P.; Myers, M.; Plotkin, S.; Rabinovich, R. Vaccine development to prevent cytomegalovirus disease: Report from the National Vaccine Advisory Committee. Clin. Infect. Dis. 2004, 39, 233–239. [Google Scholar] [CrossRef]
- Cannon, M.J.; Schmid, D.S.; Hyde, T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 2010, 20, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Pembrey, L.; Raynor, P.; Griffiths, P.; Chaytor, S.; Wright, J.; Hall, A.J. Seroprevalence of cytomegalovirus, Epstein Barr virus and varicella zoster virus among pregnant women in Bradford: A cohort study. PLoS ONE 2013, 8, e81881. [Google Scholar] [CrossRef]
- Zuhair, M.; Smit, G.S.A.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef]
- Cannon, M.J.; Hyde, T.B.; Schmid, D.S. Review of cytomegalovirus shedding in bodily fluids and relevance to congenital cytomegalovirus infection. Rev. Med. Virol. 2011, 21, 240–255. [Google Scholar] [CrossRef]
- Hyde, T.B.; Schmid, D.S.; Cannon, M.J. Cytomegalovirus seroconversion rates and risk factors: Implications for congenital CMV. Rev. Med. Virol. 2010, 20, 311–326. [Google Scholar] [CrossRef]
- Manicklal, S.; Emery, V.C.; Lazzarotto, T.; Boppana, S.B.; Gupta, R.K. The “silent” global burden of congenital cytomegalovirus. Clin. Microbiol. Rev. 2013, 26, 86–102. [Google Scholar] [CrossRef]
- Griffiths, P.; Baboonian, C. A prospective study of primary cytomegalovirus infection during pregnancy. BJOG: Int. J. Obstet. Gynaecol. 1984, 91, 307–315. [Google Scholar] [CrossRef]
- de Vries, J.J.; van Zwet, E.W.; Dekker, F.W.; Kroes, A.C.; Verkerk, P.H.; Vossen, A.C. The apparent paradox of maternal seropositivity as a risk factor for congenital cytomegalovirus infection: A population-based prediction model. Rev. Med. Virol. 2013, 23, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Kenneson, A.; Cannon, M.J. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev. Med. Virol. 2007, 17, 253–276. [Google Scholar] [CrossRef]
- Khan, M.; Khan, S.; Ali, A.; Akbar, H.; Sayaf, A.M.; Khan, A.; Wei, D.Q. Immunoinformatics approaches to explore Helicobacter Pylori proteome (Virulence Factors) to design B and T cell multi-epitope subunit vaccine. Sci. Rep. 2019, 9, 13321. [Google Scholar] [CrossRef] [PubMed]
- Sauer, A.; Wang, J.B.; Hahn, G.; McVoy, M.A. A human cytomegalovirus deleted of internal repeats replicates with near wild type efficiency but fails to undergo genome isomerization. Virology 2010, 401, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W. Structure and assembly of the virion. Intervirology 1996, 39, 389–400. [Google Scholar] [CrossRef]
- Patro, A. Subversion of immune response by human cytomegalovirus. Front. Immunol. 2019, 10, 1155. [Google Scholar] [CrossRef]
- Borst, E.-M.; Mathys, S.; Wagner, M.; Muranyi, W.; Messerle, M. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 2001, 75, 1450–1458. [Google Scholar] [CrossRef]
- Cheeran, M.C.-J.; Lokensgard, J.R.; Schleiss, M.R. Neuropathogenesis of congenital cytomegalovirus infection: Disease mechanisms and prospects for intervention. Clin. Microbiol. Rev. 2009, 22, 99–126. [Google Scholar] [CrossRef]
- Lawrence, R.S.; Durch, J.S.; Stratton, K.R. Vaccines for the 21st Century: A Tool for Decisionmaking; National Academies Press: Washington, DC, USA, 2001. [Google Scholar]
- Plotkin, S.A.; Higgins, R.; Kurtz, J.B.; Morris, P.J.; Campbell Jr, D.A.; Shope, T.C.; Spector, S.A.; Dankner, W.M. Multicenter trial of Towne strain attenuated virus vaccine in seronegative renal transplant recipients. Transplantation 1994, 58, 1176–1178. [Google Scholar]
- Pass, R.F.; Zhang, C.; Evans, A.; Simpson, T.; Andrews, W.; Huang, M.-L.; Corey, L.; Hill, J.; Davis, E.; Flanigan, C. Vaccine prevention of maternal cytomegalovirus infection. N. Engl. J. Med. 2009, 360, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, P.D.; Stanton, A.; McCarrell, E.; Smith, C.; Osman, M.; Harber, M.; Davenport, A.; Jones, G.; Wheeler, D.C.; O’Beirne, J. Cytomegalovirus glycoprotein-B vaccine with MF59 adjuvant in transplant recipients: A phase 2 randomised placebo-controlled trial. Lancet 2011, 377, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Kharfan-Dabaja, M.A.; Boeckh, M.; Wilck, M.B.; Langston, A.A.; Chu, A.H.; Wloch, M.K.; Guterwill, D.F.; Smith, L.R.; Rolland, A.P.; Kenney, R.T. A novel therapeutic cytomegalovirus DNA vaccine in allogeneic haemopoietic stem-cell transplantation: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Infect. Dis. 2012, 12, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Althurwi, H.N.; Alharthy, K.M.; Albaqami, F.F.; Altharawi, A.; Javed, M.R.; Muhseen, Z.T.; Tahir ul Qamar, M. mRNA-Based Vaccine Designing against Epstein-Barr Virus to Induce an Immune Response Using Immunoinformatic and Molecular Modelling Approaches. Int. J. Environ. Res. Public Health 2022, 19, 13054. [Google Scholar] [CrossRef]
- Ismail, S.; Alsowayeh, N.; Abbasi, H.W.; Albutti, A.; Tahir ul Qamar, M.; Ahmad, S.; Raza, R.Z.; Sadia, K.; Abbasi, S.W. Pan-genome-assisted computational design of a multi-epitopes-based vaccine candidate against Helicobacter cinaedi. Int. J. Environ. Res. Public Health 2022, 19, 11579. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Khan, A.; Rehman, A.U.; Ahmad, I.; Ullah, S.; Khan, A.A.; Ali, S.S.; Afridi, S.G.; Wei, D.Q. Immunoinformatics and structural vaccinology driven prediction of multi-epitope vaccine against Mayaro virus and validation through in-silico expression. Infect. Genet. Evol. 2019, 73, 390–400. [Google Scholar] [CrossRef]
- Rida, T.; Ahmad, S.; Ullah, A.; Ismail, S.; Tahir ul Qamar, M.; Afsheen, Z.; Khurram, M.; Saqib Ishaq, M.; Alkhathami, A.G.; Alatawi, E.A. Pan-genome analysis of oral bacterial pathogens to predict a potential novel multi-epitopes vaccine candidate. Int. J. Environ. Res. Public Health 2022, 19, 8408. [Google Scholar] [CrossRef]
- Ullah, A.; Shahid, F.A.; Haq, M.U.; Tahir ul Qamar, M.; Irfan, M.; Shaker, B.; Ahmad, S.; Alrumaihi, F.; Allemailem, K.S.; Almatroudi, A. An integrative reverse vaccinology, immunoinformatic, docking and simulation approaches towards designing of multi-epitopes based vaccine against monkeypox virus. J. Biomol. Struct. Dyn. 2022, 1–14. [Google Scholar]
- Khan, T.; Khan, A.; Wei, D.-Q. MMV-db: Vaccinomics and RNA-based therapeutics database for infectious hemorrhagic fever-causing mammarenaviruses. Database 2021, 2021, baab063. [Google Scholar] [CrossRef]
- Khan, T.; Khan, A.; Nasir, S.N.; Ahmad, S.; Ali, S.S.; Wei, D.-Q. CytomegaloVirusDb: Multi-Omics knowledge database for Cytomegaloviruses. Comput. Biol. Med. 2021, 135, 104563. [Google Scholar] [CrossRef]
- Khan, T.; Muzaffar, A.; Shoaib, R.M.; Khan, A.; Waheed, Y.; Wei, D.-Q. Towards specie-specific ensemble vaccine candidates against mammarenaviruses using optimized structural vaccinology pipeline and molecular modelling approaches. Microb. Pathog. 2022, 172, 105793. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Khan, A.; Ansari, J.K.; Najmi, M.H.; Wei, D.-Q.; Muhammad, K.; Waheed, Y. Potential Immunogenic Activity of Computationally Designed mRNA-and Peptide-Based Prophylactic Vaccines against MERS, SARS-CoV, and SARS-CoV-2: A Reverse Vaccinology Approach. Molecules 2022, 27, 2375. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Stranzl, T.; Larsen, M.V.; Lundegaard, C.; Nielsen, M. NetCTLpan: Pan-specific MHC class I pathway epitope predictions. Immunogenetics 2010, 62, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Kaliamurthi, S.; Selvaraj, G.; Junaid, M.; Khan, A.; Gu, K.; Wei, D.Q. Cancer immunoinformatics: A promising era in the development of peptide vaccines for human papillomavirus-induced cervical cancer. Curr. Pharm. Des. 2018, 24, 3791–3817. [Google Scholar]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef]
- Calis, J.J.; Maybeno, M.; Greenbaum, J.A.; Weiskopf, D.; De Silva, A.D.; Sette, A.; Keşmir, C.; Peters, B. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013, 9, e1003266. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins: Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Negahdaripour, M.; Nezafat, N.; Eslami, M.; Ghoshoon, M.B.; Shoolian, E.; Najafipour, S.; Morowvat, M.H.; Dehshahri, A.; Erfani, N.; Ghasemi, Y. Structural vaccinology considerations for in silico designing of a multi-epitope vaccine. Infect. Genet. Evol. 2018, 58, 96–109. [Google Scholar] [CrossRef]
- Parvizpour, S.; Pourseif, M.M.; Razmara, J.; Rafi, M.A.; Omidi, Y. Epitope-based vaccine design: A comprehensive overview of bioinformatics approaches. Drug Discov. Today 2020, 25, 1034–1042. [Google Scholar] [CrossRef]
- Alshammari, A.; Alharbi, M.; Alghamdi, A.; Alharbi, S.A.; Ashfaq, U.A.; Tahir ul Qamar, M.; Ullah, A.; Irfan, M.; Khan, A.; Ahmad, S. Computer-aided multi-epitope vaccine design against Enterobacter xiangfangensis. Int. J. Environ. Res. Public Health 2022, 19, 7723. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006, 34, W202–W209. [Google Scholar] [CrossRef]
- Chivian, D.; Kim, D.E.; Malmström, L.; Bradley, P.; Robertson, T.; Murphy, P.; Strauss, C.E.; Bonneau, R.; Rohl, C.A.; Baker, D. Automated prediction of CASP-5 structures using the Robetta server. Proteins Struct. Funct. Bioinform. 2003, 53, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.; Kayastha, A.M.; Singh, V.K. MFPPI–multi FASTA ProtParam interface. Bioinformation 2016, 12, 74. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handb. 2005, 571–607. [Google Scholar]
- Cserzö, M.; Wallin, E.; Simon, I.; von Heijne, G.; Elofsson, A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: The dense alignment surface method. Protein Eng. 1997, 10, 673–676. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Weng, G.; Wang, E.; Wang, Z.; Liu, H.; Zhu, F.; Li, D.; Hou, T. HawkDock: A web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019, 47, W322–W330. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- Rose, P.W.; Beran, B.; Bi, C.; Bluhm, W.F.; Dimitropoulos, D.; Goodsell, D.S.; Prlić, A.; Quesada, M.; Quinn, G.B.; Westbrook, J.D. The RCSB Protein Data Bank: Redesigned web site and web services. Nucleic Acids Res. 2010, 39, D392–D401. [Google Scholar] [CrossRef]
- Grote, A.; Hiller, K.; Scheer, M.; Münch, R.; Nörtemann, B.; Hempel, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005, 33 (Suppl. S2), W526–W531. [Google Scholar] [CrossRef]
- Khatoon, N.; Pandey, R.K.; Prajapati, V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 2017, 7, 8285. [Google Scholar] [CrossRef]
- Zheng, B.; Suleman, M.; Zafar, Z.; Ali, S.S.; Nasir, S.N.; Hussain, Z.; Waseem, M.; Khan, A.; Hassan, F.; Wang, Y. Towards an ensemble vaccine against the pegivirus using computational modelling approaches and its validation through in silico cloning and immune simulation. Vaccines 2021, 9, 818. [Google Scholar] [CrossRef]
- Castiglione, F.; Bernaschi, M. C-immsim: Playing with the immune response. In Proceedings of the Sixteenth International Symposium on Mathematical Theory of Networks and Systems (MTNS2004), Leuven, Belgium, 5–9 July 2004. [Google Scholar]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Oli, A.N.; Obialor, W.O.; Ifeanyichukwu, M.O.; Odimegwu, D.C.; Okoyeh, J.N.; Emechebe, G.O.; Adejumo, S.A.; Ibeanu, G.C. Immunoinformatics and vaccine development: An overview. ImmunoTargets Ther. 2020, 9, 13. [Google Scholar] [CrossRef]
- Khan, A.; Khan, S.; Ahmad, S.; Anwar, Z.; Hussain, Z.; Safdar, M.; Rizwan, M.; Waseem, M.; Hussain, A.; Akhlaq, M. HantavirusesDB: Vaccinomics and RNA-based therapeutics database for the potentially emerging human respiratory pandemic agents. Microb. Pathog. 2021, 160, 105161. [Google Scholar] [CrossRef] [PubMed]
- Noriega, V.M.; Hesse, J.; Gardner, T.J.; Besold, K.; Plachter, B.; Tortorella, D. Human cytomegalovirus US3 modulates destruction of MHC class I molecules. Mol. Immunol. 2012, 51, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Tomasec, P.; Braud, V.M.; Rickards, C.; Powell, M.B.; McSharry, B.P.; Gadola, S.; Cerundolo, V.; Borysiewicz, L.K.; McMichael, A.J.; Wilkinson, G.W. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000, 287, 1031–1033. [Google Scholar] [CrossRef]
- Jiang, P.; Cai, Y.; Chen, J.; Ye, X.; Mao, S.; Zhu, S.; Xue, X.; Chen, S.; Zhang, L. Evaluation of tandem Chlamydia trachomatis MOMP multi-epitopes vaccine in BALB/c mice model. Vaccine 2017, 35, 3096–3103. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Du, W.; Xiong, Y.; Lv, Y.; Feng, J.; Zhu, S.; Xue, X.; Chen, S.; Zhang, L. Hepatitis B virus core antigen as a carrier for Chlamydia trachomatis MOMP multi-epitope peptide enhances protection against genital chlamydial infection. Oncotarget 2015, 6, 43281. [Google Scholar] [CrossRef]
- Talesh, G.A.; Ebrahimi, Z.; Badiee, A.; Mansourian, M.; Attar, H.; Arabi, L.; Jalali, S.A.; Jaafari, M.R. Poly (I: C)-DOTAP cationic nanoliposome containing multi-epitope HER2-derived peptide promotes vaccine-elicited anti-tumor immunity in a murine model. Immunol. Lett. 2016, 176, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Kuai, R.; Ochyl, L.J.; Bahjat, K.S.; Schwendeman, A.; Moon, J.J. Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat. Mater. 2017, 16, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Black, M.; Trent, A.; Tirrell, M.; Olive, C. Advances in the design and delivery of peptide subunit vaccines with a focus on toll-like receptor agonists. Expert Rev. Vaccines 2010, 9, 157–173. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Lu, C.; Lei, L.; Yu, P.; Zhong, G. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 2010, 185, 1670–1680. [Google Scholar] [CrossRef] [PubMed]
- Malonis, R.J.; Lai, J.R.; Vergnolle, O. Peptide-based vaccines: Current progress and future challenges. Chem. Rev. 2019, 120, 3210–3229. [Google Scholar] [CrossRef] [PubMed]

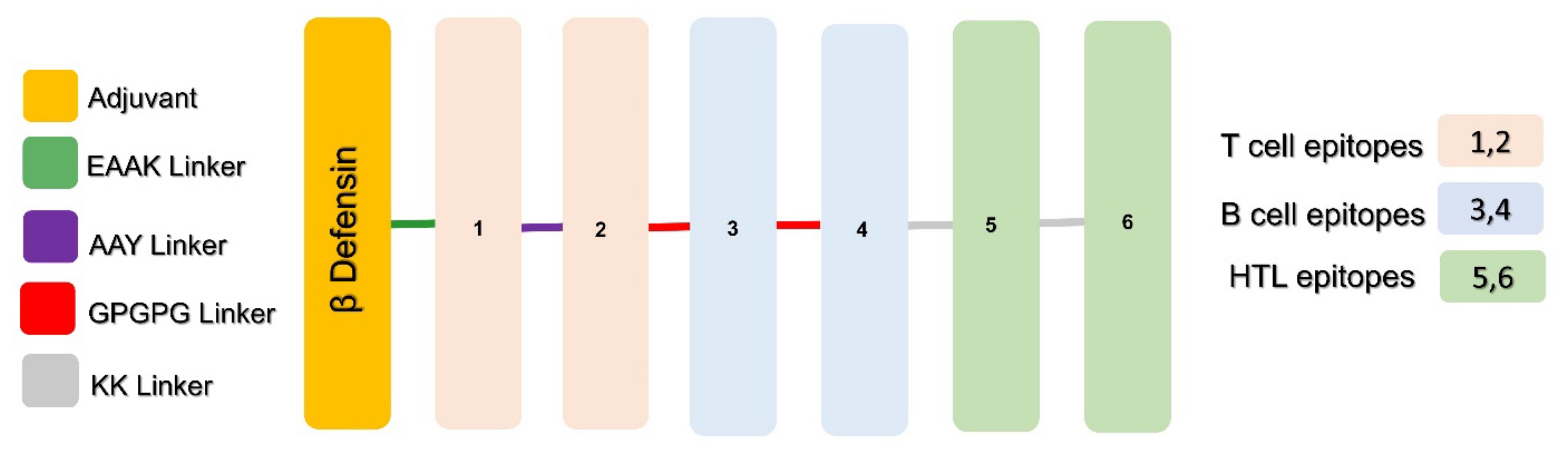



| Index | Target Protein | Description/ Function | Antigenicity Score | CTL Epitopes | B Cell Epitopes | HTL Epitopes |
|---|---|---|---|---|---|---|
| 1 | membrane glycoprotein US3 | glycoprotein/ hinders viral clearance by cytotoxic T lymphocytes (CTLs) | 0.83 | YSSQTINWY, SSQTINWYL | SGEQYHHDERGAYFEW, SQTINWYLQRSMRDDNST | FRTLLVYLFSLVVLV, GEQYHHDERGAYFEW |
| 2 | protein UL15A | uncharacterized | 0.71 | LCSWLAMRY, RTDHQKADI | GALICGSGTRRGSGAN, ACTRTDHQKADIGLWF | MFLVFGLCSWLAMRY, TRTDHQKADIGLWFM |
| 3 | protein UL41A | uncharacterized | 0.71 | RTANSTAGY, FCSTLKAFY | ETTVWEKRRMESDTDF, LFCRTANSTAGYVDMN, | ITKIMLARRKARAMV, WLVGVGIFMAGGFIA |
| 4 | membrane glycoprotein UL40 | glycoprotein/ promotes efficient cell surface expression of the non-classical MHC-I molecule | 0.7 | ALGSFSSFY, TSSNTVVAF | HQDCPAQTVHVRGVNE, VDGISCQDHFRAQHQD, | TVGILALGSFSSFYS, CMRIRSLLSSPVETT |
| Index | Vaccine-Target Protein | Protein-Specific MEVC Constructs | Antigenicity Scores |
|---|---|---|---|
| 1 | MEVC-US3 | MRVLYLLFSFLFIFLMPLPGVFG GIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCK KPEAAKYSSQTINWYAAYSSQTINWYLGPGPGSGEQYHHDE RGAYFEWGPGPGSQTIN WYLQRSMRDDNKKFRTLLVYLFSLVVLVKKGEQYHHDERG AYFEW | 0.7 |
| 2 | MEVC-UL15A | MRVLYLLFSFLFIFLMPLPGVFG GIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCK KPEAAK LCSWLAMRYAAYRTDHQKADIGPGPGGA LICGSGTRRGSGANGPGPGACTRTDHQKADIGLWFKK MFLVFGLCSWLAMRYKKTRTDHQKADIGLWFM | 0.59 |
| 3 | MEVC-UL41A | MRVLYLLFSFLFIFLMPLPGVFG GIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCK KPEAAK RTANSTAGYAAYFCSTLKAFYGPGPGETTVWE KRRMESDTDFGPGPGLFCRTANSTAGYVDMNKK ITKIM LARRKARAMVKK WLVGVGIFMAGGFIA | 0.52 |
| 4 | MEVC-UL40 | MRVLYLLFSFLFIFLMPLPGVFG GIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCK KPEAAK ALGSFSSFYAAYTSSNTVVAF GPGPGHQDCPAQTVHVRGVNEGPGPGVDGISCQDHFRAQ HQDKK TVGILALGSFSSFYSKKCMRIRSLLSSPVETT | 0.58 |
| Index | Vaccine Name | Molecular Weight (Kilo Daltons) | Theoretical pI | Negatively Charged Residues (Asp + Glu): | Positively Charged Residues (Arg + Lys) | Total Atoms | Aliphatic Index | Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|---|
| 1 | MEVC-US3 | 18 kds | 9 | 12 | 18 | 2627 | 70.30 | −0.278 |
| 2 | MEVC-UL15A | 18 kds | 9.67 | 8 | 23 | 2542 | 73.39 | 0.024 |
| 3 | MEVC-UL41A | 17.9 kds | 9.82 | 8 | 24 | 2542 | 72.18 | 0.122 |
| 4 | MEVC-UL40 | 17.6 kds | 9.25 | 8 | 17 | 2475 | 74.42 | 0.093 |
| Index | Vaccine Name | TLR Name | Salt Bridges | Hydrogen Bonds | Non Bonded Contacts | Docking Score |
|---|---|---|---|---|---|---|
| 1 | MEVC-US3 | TLR4 | 0 | 5 | 151 | −302.71 |
| 2 | MEVC-UL15A | TLR4 | 1 | 6 | 200 | −346.35 |
| 3 | MEVC-UL41A | TLR4 | 2 | 7 | 225 | −336.44 |
| 4 | MEVC-UL40 | TLR4 | 1 | 8 | 234 | −340.42 |
| 5 | MD-2 | TLR4 | 1 | 5 | 158 | −247.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakkari, M.A. Targeted Protein-Specific Multi-Epitope-Based Vaccine Designing against Human Cytomegalovirus by Using Immunoinformatics Approaches. Vaccines 2023, 11, 203. https://doi.org/10.3390/vaccines11020203
Bakkari MA. Targeted Protein-Specific Multi-Epitope-Based Vaccine Designing against Human Cytomegalovirus by Using Immunoinformatics Approaches. Vaccines. 2023; 11(2):203. https://doi.org/10.3390/vaccines11020203
Chicago/Turabian StyleBakkari, Mohammed Ali. 2023. "Targeted Protein-Specific Multi-Epitope-Based Vaccine Designing against Human Cytomegalovirus by Using Immunoinformatics Approaches" Vaccines 11, no. 2: 203. https://doi.org/10.3390/vaccines11020203
APA StyleBakkari, M. A. (2023). Targeted Protein-Specific Multi-Epitope-Based Vaccine Designing against Human Cytomegalovirus by Using Immunoinformatics Approaches. Vaccines, 11(2), 203. https://doi.org/10.3390/vaccines11020203





