Immunological Studies to Understand Hybrid/Recombinant Variants of SARS-CoV-2
Abstract
1. Introduction
2. Variants of SARS-CoV-2
3. Hybrid Variants
4. Techniques for Identification of Hybrid Variant
5. Immunological Studies and Assays
6. Challenges in the Immunological Analysis of Hybrid Variants
7. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C.; et al. The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Lytras, S.; Xia, W.; Hughes, J.; Jiang, X.; Robertson, D.L. The animal origin of SARS-CoV-2. Science 2021, 373, 968–970. [Google Scholar] [CrossRef] [PubMed]
- Frutos, R.; Pliez, O.; Gavotte, L.; Devaux, C.A. There is no “origin” to SARS-CoV-2. Environ. Res. 2022, 207, 112173. [Google Scholar] [CrossRef] [PubMed]
- Voskarides, K. SARS-CoV-2: Tracing the origin, tracking the evolution. BMC Med. Genom. 2022, 15, 62. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard. WHO Coronavirus (COVID-19) Dashboard with Vaccination Data; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Koelle, K.; Martin, M.A.; Antia, R.; Lopman, B.; Dean, N.E. The changing epidemiology of SARS-CoV-2. Science 2022, 375, 1116. [Google Scholar] [CrossRef]
- Salzberger, B.; Buder, F.; Lampl, B.; Ehrenstein, B.; Hitzenbichler, F.; Holzmann, T.; Schmidt, B.; Hanses, F. Epidemiology of SARS-CoV-2. Infection 2021, 49, 233. [Google Scholar] [CrossRef]
- Chang, M.R.; Ke, H.; Coherd, C.D.; Wang, Y.; Mashima, K.; Kastrunes, G.M.; Huang, C.Y.; Marasco, W.A. Analysis of a SARS-CoV-2 convalescent cohort identified a common strategy for escape of vaccine-induced anti-RBD antibodies by Beta and Omicron variants. EBioMedicine 2022, 80, 104025. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Lauring, A.S.; Malani, P.N. Variants of SARS-CoV-2. JAMA 2021, 326, 880. [Google Scholar] [CrossRef]
- Hirabara, S.M.; Serdan, T.D.A.; Gorjao, R.; Masi, L.N.; Pithon-Curi, T.C.; Covas, D.T.; Curi, R.; Durigon, E.L. SARS-COV-2 Variants: Differences and Potential of Immune Evasion. Front. Cell. Infect. Microbiol. 2022, 11, 1401. [Google Scholar] [CrossRef] [PubMed]
- SARS-CoV-2 Variant Classifications and Definitions Centers for Disease Control and Prevention. 2022. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 9 November 2022).
- Chavda, V.P.; Vuppu, S.; Mishra, T.; Kamaraj, S.; Patel, A.B.; Sharma, N.; Chen, Z.S. Recent review of COVID-19 management: Diagnosis, treatment and vaccination. Pharmacol. Rep. 2022; Online first. [Google Scholar]
- Otto, S.P.; Day, T.; Arino, J.; Colijn, C.; Dushoff, J.; Li, M.; Mechai, S.; Domselaar, G.V.; Wu, J.; Earn, D.J.D.; et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021, 31, R918–R929. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.M.; Capistrano, K.; Thakkar, P.; Ranade, H.; Soni, V.; Datta, M.; Naqvi, A. Current molecular diagnostics assays for SARS-CoV-2 and emerging variants. Methods Microbiol. 2022, 50, 83–121. [Google Scholar]
- Chavda, V.P.; Apostolopoulos, V. Global impact of delta plus variant and vaccination. Expert Rev. Vaccines 2022, 21, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.; Shaik Syed Ali, P.; Sheeza, A. Omicron (B.1.1.529)—Variant of concern—Molecular profile and epidemiology: A mini review. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 8019–8022. [Google Scholar]
- Chavda, V.P.; Apostolopoulos, V. Omicron Variant (B.1.1.529) of SARS-CoV-2: Threat for the elderly? Maturitas 2022, 158, 78–81. [Google Scholar] [CrossRef]
- WHO. Tracking SARS-CoV-2 Variants. 2022. Available online: https://www.who.int/health-topics/typhoid/tracking-SARS-CoV-2-variants (accessed on 31 March 2022).
- Thakur, P.; Thakur, V.; Kumar, P.; Singh Patel, S.K. Emergence of novel omicron hybrid variants: BA(x), XE, XD, XF more than just alphabets. Int. J. Surg. 2022, 104, 106727. [Google Scholar] [CrossRef]
- Jung, C.; Kmiec, D.; Koepke, L.; Zech, F.; Jacob, T.; Sparrer, K.M.J.; Kirchhoff, F. Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning? J. Virol. 2022, 96, e02077-21. [Google Scholar] [CrossRef]
- Lee, I.J.; Sun, C.P.; Wu, P.Y.; Lan, Y.H.; Wang, I.H.; Liu, W.C.; Yuan, J.P.Y.; Chang, Y.W.; Tseng, S.C.; Tsung, S.I.; et al. A booster dose of Delta × Omicron hybrid mRNA vaccine produced broadly neutralizing antibody against Omicron and other SARS-CoV-2 variants. J. Biomed. Sci. 2022, 29, 49. [Google Scholar] [CrossRef]
- Rahimi, F.; Talebi Bezmin Abadi, A. Hybrid SARS-CoV-2 variants. Int. J. Surg. 2022, 102, 106656. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 2015, 30, 296. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, R.K.; Kandi, V.; Tuli, H.S.; Chakraborty, C.; Dhama, K. The recombinant variants of SARS-CoV-2: Concerns continues amid COVID-19 pandemic. J. Med. Virol. 2022, 94, 3506–3508. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Kumar, R. XE variant of the novel coronavirus. Indian J. Microbiol. Res. 2022, 9, 92–94. [Google Scholar] [CrossRef]
- Ma, K.; Chen, J. Omicron XE emerges as SARS-CoV-2 keeps evolving. Innovation 2022, 3, 100248. [Google Scholar] [CrossRef]
- Vo, V.; Harrington, A.; Afzal, S.; Papp, K.; Chang, C.L.; Baker, H.; Aguilar, P.; Buttery, E.; Picker, M.A.; Lockett, C.; et al. Identification of a rare SARS-CoV-2 XL hybrid variant in wastewater and the subsequent discovery of two infected individuals in Nevada. Sci. Total Environ. 2023, 858, 160024. [Google Scholar] [CrossRef] [PubMed]
- Berno, G.; Fabeni, L.; Matusali, G.; Gruber, C.E.M.; Rueca, M.; Giombini, E.; Garbuglia, A.R. SARS-CoV-2 Variants Identification: Overview of Molecular Existing Methods. Pathogens 2022, 11, 1058. [Google Scholar] [CrossRef] [PubMed]
- Methods for the Detection and Characterisation of SARS-CoV-2 Variants—Second Update. Available online: https://www.ecdc.europa.eu/en/publications-data/methods-detection-and-characterisation-sars-cov-2-variants-second-update (accessed on 10 November 2022).
- Nagy-Szakal, D.; Couto-Rodriguez, M.; Wells, H.L.; Barrows, J.E.; Debieu, M.; Butcher, K.; Chen, S.; Berki, A.; Hager, C.; Boorstein, R.J.; et al. Targeted Hybridization Capture of SARS-CoV-2 and Metagenomics Enables Genetic Variant Discovery and Nasal Microbiome Insights. Microbiol. Spectr. 2021, 9, e00197-21. [Google Scholar] [CrossRef]
- Bechtold, P.; Wagner, P.; Hosch, S.; Siegrist, D.; Ruiz-Serrano, A.; Gregorini, M.; Siegrist, D.; Engler, O.; Stark, W.J.; Daubenberger, C.A.; et al. Rapid Identification of SARS-CoV-2 Variants of Concern Using a Portable peakPCR Platform. Anal. Chem. 2021, 93, 16350–16359. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Zhang, L.; Sun, W.; Zhang, Z.; Chen, W.; Zhu, A.; Huang, Y.; Xiao, F.; Yao, J.; et al. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021, 13, 30. [Google Scholar] [CrossRef]
- Joloba, M.L.; Mboowa, G.; Mwesigwa, S.; Kateete, D.; Wayengera, M.; Nasinghe, E.; Katagirya, E.; Katabazi, A.F.; Kigozi, E.; Kirimunda, S.; et al. Whole-genome sequencing of SARS-CoV-2 in Uganda: Implementation of the low-cost ARTIC protocol in resource-limited settings. F1000Research 2021, 10, 598. [Google Scholar]
- Yin, R.; Kwoh, C.K.; Zheng, J. Whole Genome Sequencing Analysis. Encycl. Bioinform. Comput. Biol. ABC Bioinform. 2019, 1, 176–183. [Google Scholar]
- CDC Centre for Disease Control and Prevention. Whole Genome Sequencing. 2022. Available online: https://www.cdc.gov/pulsenet/pathogens/wgs.html (accessed on 21 November 2022).
- Ekblom, R.; Wolf, J.B.W. A field guide to whole-genome sequencing, assembly and annotation. Evol. Appl. 2014, 7, 1026. [Google Scholar] [CrossRef]
- CDC Centre for Disease Control and Prevention. Nucleic Acid Amplification Tests (NAATs). 2021. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/naats.html (accessed on 21 November 2022).
- Monis, P.T.; Giglio, S. Nucleic acid amplification-based techniques for pathogen detection and identification. Infect. Genet. Evol. 2006, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.; Lu, J.; Yu, T.; Long, Y.; Liu, G. Advances in nucleic acid amplification techniques (NAATs): COVID-19 point-of-care diagnostics as an example. Biosens. Bioelectron. 2022, 206, 114109. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Cartagena, C.; Fernández-García, L.; Blasco, L.; Pacios, O.; Bleriot, I.; López, M.; Cantón, R.; Tomás, M. Reverse Transcription-Loop-Mediated Isothermal Amplification-CRISPR-Cas13a Technology as a Promising Diagnostic Tool for SARS-CoV-2. Microbiol. Spectr. 2022, 10, e02398-22. [Google Scholar] [CrossRef]
- Cheng, S.M.S.; Mok, C.K.P.; Leung, Y.W.Y.; Ng, S.S.; Chan, K.C.K.; Ko, F.W.; Chen, C.; Yiu, K.; Lam, B.H.S.; Lau, E.H.Y.; et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat. Med. 2022, 28, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Roy, V.; Fischinger, S.; Atyeo, C.; Slein, M.; Loos, C.; Balazs, A.; Luedemann, C.; Astudillo, M.G.; Yang, D.; Wesemann, D.R.; et al. SARS-CoV-2-specific ELISA development. J. Immunol. Methods 2020, 484, 112832. [Google Scholar] [CrossRef]
- Sidiq, Z.; Hanif, M.; Dwivedi, K.K.; Chopra, K.K. Benefits and limitations of serological assays in COVID-19 infection. Indian J. Tuberc. 2020, 67, S163. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Wei, H.X.; Li, Q.; Liu, L.; Li, B. Evaluation and Comparison of Serological Methods for COVID-19 Diagnosis. Front. Mol. Biosci. 2021, 8, 683. [Google Scholar] [CrossRef] [PubMed]
- Li, F.F.; Liu, A.; Gibbs, E.; Tanunliong, G.; Marquez, A.C.; Gantt, S.; Frykman, H.; Krajden, M.; Morshed, M.; Prystajecky, N.A.; et al. A novel multiplex electrochemiluminescent immunoassay for detection and quantification of anti-SARS-CoV-2 IgG and anti-seasonal endemic human coronavirus IgG. J. Clin. Virol. 2022, 146, 105050. [Google Scholar] [CrossRef]
- Liu, H.; Varvel, S.; Chen, G.; McConnell, J.; Caffrey, R.; Galdzicka, M.; Shabahang, S. Simultaneous measurement of multiple variant-specific SARS-CoV-2 neutralizing antibodies with a multiplexed flow cytometric assay. Front. Immunol. 2022, 13, 6978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, C.; Das, T.; Luo, S.; Tang, H.; Yao, X.; Cho, C.Y.; Lv, J.; Maravillas, K.; Jones, V.; et al. The spike-ACE2 binding assay: An in vitro platform for evaluating vaccination efficacy and for screening SARS-CoV-2 inhibitors and neutralizing antibodies. J. Immunol. Methods 2022, 503, 113244. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, F.; Chattopadhyay, S.; Thiruvengadam, R.; Yadav, S.; Kumar, M.; Sinha, S.K.; Goswami, S.; Kshetrapal, P.; Wadhwa, N.; Natchu, U.C.; et al. Development of a Fast SARS-CoV-2 IgG ELISA, Based on Receptor-Binding Domain, and Its Comparative Evaluation Using Temporally Segregated Samples From RT-PCR Positive Individuals. Front. Microbiol. 2021, 11, 3597. [Google Scholar] [CrossRef] [PubMed]
- Cinquanta, L.; Fontana, D.E.; Bizzaro, N. Chemiluminescent immunoassay technology: What does it change in autoantibody detection? Auto-Immun. Highlights 2017, 8, 9. [Google Scholar] [CrossRef]
- Liang, W.; Fan, C.; Zhuo, Y.; Zheng, Y.; Xiong, C.; Chai, Y.; Yuan, R. Multiparameter Analysis-Based Electrochemiluminescent Assay for Simultaneous Detection of Multiple Biomarker Proteins on a Single Interface. Anal. Chem. 2016, 88, 4940–4948. [Google Scholar] [CrossRef] [PubMed]
- Kulmala, S.; Kankare, J. CHEMILUMINESCENCE | Electrogenerated. Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, Elsevier. 2013. Available online: https://www.sciencedirect.com/science/article/pii/B9780124095472000640 (accessed on 18 December 2022).
- Chen, M.; Ning, Z.; Chen, K.; Zhang, Y.; Shen, Y. Recent Advances of Electrochemiluminescent System in Bioassay. J. Anal. Test. 2020, 4, 57–75. [Google Scholar] [CrossRef]
- Goh, Y.S.; Ng, L.F.P.; Renia, L. A flow cytometry-based assay for serological detection of anti-spike antibodies in COVID-19 patients. STAR Protoc. 2021, 2, 100671. [Google Scholar] [CrossRef] [PubMed]
- Gil-Manso, S.; Miguens Blanco, I.; López-Esteban, R.; Carbonell, D.; López-Fernández, L.A.; West, L.; Correa-Rocha, R.; Pion, M. Comprehensive Flow Cytometry Profiling of the Immune System in COVID-19 Convalescent Individuals. Front. Immunol. 2022, 12, 5734. [Google Scholar] [CrossRef]
- Xu, X.; Nie, S.; Wang, Y.; Long, Q.; Zhu, H.; Zhang, X.; Sun, J.; Zeng, Q.; Zhao, J.; Liu, L.; et al. Dynamics of neutralizing antibody responses to SARS-CoV-2 in patients with COVID-19: An observational study. Signal Transduct. Target. Ther. 2021, 6, 1–7. [Google Scholar] [CrossRef]
- Muruato, A.E.; Fontes-Garfias, C.R.; Ren, P.; Garcia-Blanco, M.A.; Menachery, V.D.; Xie, X.; Shi, P. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020, 11, 4056. [Google Scholar] [CrossRef]
- Matusali, G.; Colavita, F.; Lapa, D.; Meschi, S.; Bordi, L.; Piselli, P.; Gagliardini, R.; Corpolongo, A.; Nicastri, E.; Antinori, A.; et al. SARS-CoV-2 Serum Neutralization Assay: A Traditional Tool for a Brand-New Virus. Viruses 2021, 13, 655. [Google Scholar] [CrossRef] [PubMed]
- Misra, A.; Theel, E.S. Immunity to SARS-CoV-2: What Do We Know and Should We Be Testing for It? J. Clin. Microbiol. 2022, 60, e00482-21. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, Y.; Greig, M.; Liu, G.; Driedger, M.; Langlois, M.A. Humoral Responses and Serological Assays in SARS-CoV-2 Infections. Front. Immunol. 2020, 11, 3382. [Google Scholar] [CrossRef] [PubMed]
- Sen Tan, A.; Nerurkar, S.N.; Tan, W.C.C.; Goh, D.; Lai, C.P.T.; Poh Sheng Yeong, J. The Virological, Immunological, and Imaging Approaches for COVID-19 Diagnosis and Research. SLAS Technol. 2020, 25, 522. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cui, X.; Cai, X.; An, T. Recombination in Positive-Strand RNA Viruses. Front. Microbiol. 2022, 13, 1704. [Google Scholar] [CrossRef]
| Sl. No. | Variants of Concern (VOCs) | Mutations | Property of the Variant | References |
|---|---|---|---|---|
| 1. | Alpha (B.1.1.7) | A total of 23 mutations (17 amino acid changes) from the first strain that was discovered in Wuhan. Notable mutations: spike D614G, spike N501Y, and spike HV-69–70 deletions. |
| [16] |
| 2. | Beta (B.1.351) | Notable mutations: multiple mutations in the S protein, three in the RBD (N501Y, E484K, and K417N). |
| [16] |
| 3. | Gamma (P1) | Seventeen mutations (eleven amino acid changes). Notable mutations: N501Y, E484K, and K417T. |
| [16] |
| 4. | Delta (B.1.617.2) | Notable mutations: 12 mutations, 10 of which in the S-protein (T19R, G142D, 156del, 157del, R158G, L452R, T478K, D614G, P681R, and D950N). Contains major mutations in RBD and NBD-containing S1 subunits. |
| [16,17] |
| 5. | Omicron (B.1.1.529) | The spike protein contains 32 amino acid mutations. Notable mutations: K417N, E484K, N501Y, D614G, and T478K. |
| [18,19] |
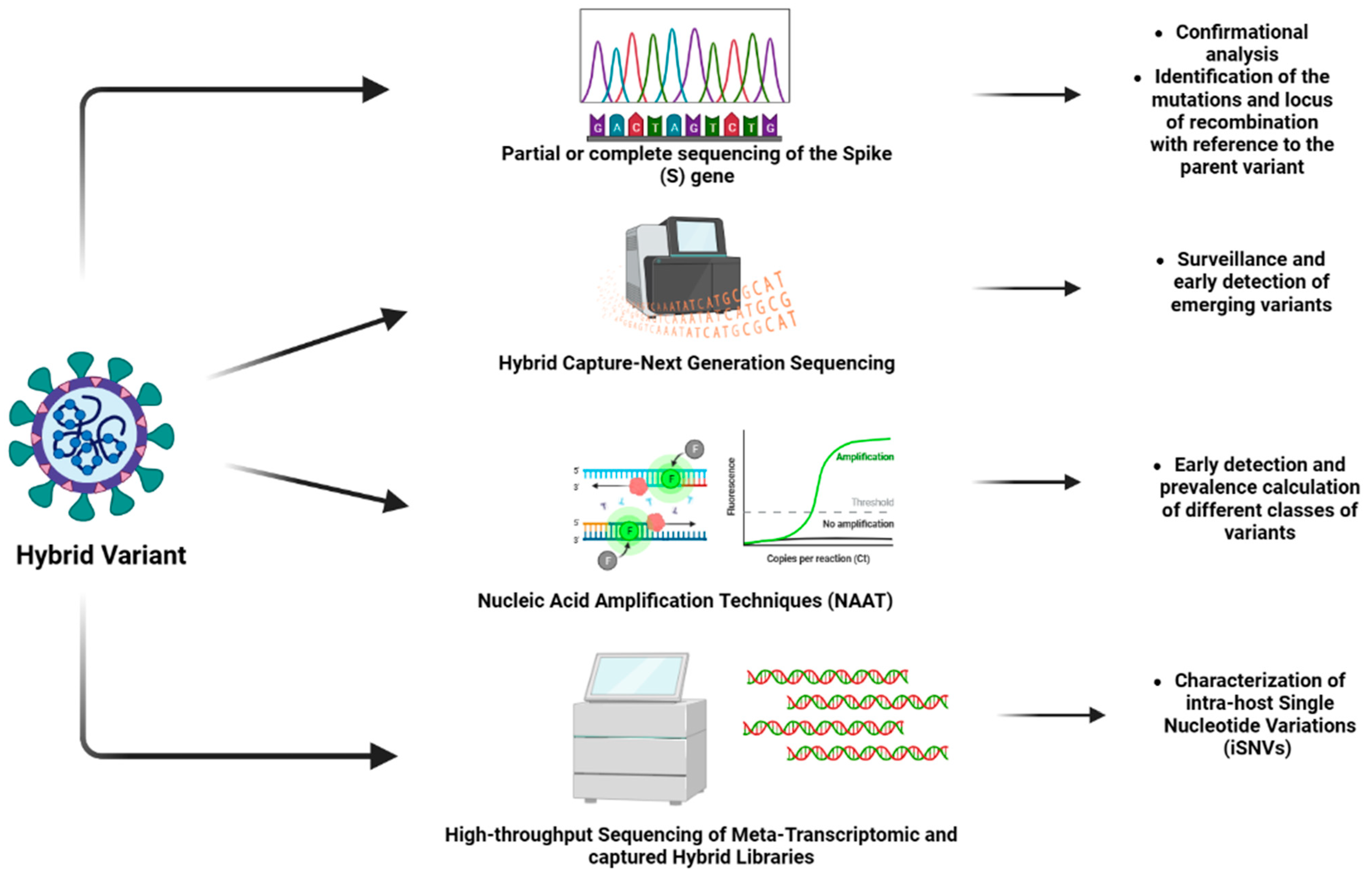
| Technique | Description | Comments | Output | Advantages | Limitations | Reference |
|---|---|---|---|---|---|---|
| Whole genome sequencing or partial or complete sequencing of the spike protein | The nucleic acid samples are fragmented into smaller segments. These sequences are independently decoded followed by the alignment of all the sequences using computer algorithms. | This technique is facilitated by high-throughput sequencing approaches. The sequencer identifies the nucleotide bases that make up the sequence of the nucleic acid chain. Computer-based tools are used for comparison as well as identification of the variations. | The result obtained is nucleotide sequence on a computer system. |
|
| [35,36,37,38] |
| Hybrid capture SARS-CoV-2 NGS (Next Generation Sequencing) assay | It utilizes double-stranded probes that are labeled with biotin for the purpose of panel design integrated with software for the detection and mapping of the hybrid variants. Additionally, the microbiomes in the nasopharyngeal tract. |
| The result obtained is nucleotide sequence on a computer system. |
|
| [39] |
| Nucleic acid amplification techniques (NAAT) | The procedure is a sensitive diagnostic test that is based on the amplification of the viral genome which facilitates the detection of RNA of the virus. | This technique is a rapid, industry-standard approach. It basically involves real-time polymerase chain reactions (RT-PCR), CRISPR- related amplification, loop-mediated isothermal amplification, strand displacement amplification, as well as a ligase chain reaction. | The result of the amplification is studied using fluorescent probes computer system linked to the PCR machine. |
|
| [40,41,42] |
| SNP Assays | The technique enables rapid estimation of prevalent variants with specific mutations by RT-PCR assays that target single polymorphism. | The effectiveness of the technique can be enhanced by integrating it with whole genome sequencing. The melting curve analysis of RT-PCR has been used commercially for the detection of Variants of Concern. | The result of the amplification is studied using fluorescent probes computer system linked to the PCR machine. |
|
| [31] |
| Reverse transcription loop-mediated isothermal amplification | Alternative molecular method for identification of variants of SARS-CoV-2. | This technique when integrated with CRISPR-Cas13 can efficiently detect the hybrid variants of SARS-CoV-2 with 100% specificity, as well as 83% sensitivity. | The result is observed by visual observation of turbidity or fluorescence. |
|
| [31,43] |
| High-throughput sequencing of meta-transcriptomic and captured hybrid libraries | The sequences of the sample are mapped on a pre-determined database with genomes of coronaviridae that are used as references to eliminate low-quality data. Further, intra-host variants are identified. |
| The result obtained is nucleotide sequence on a computer system. |
|
| [34] |
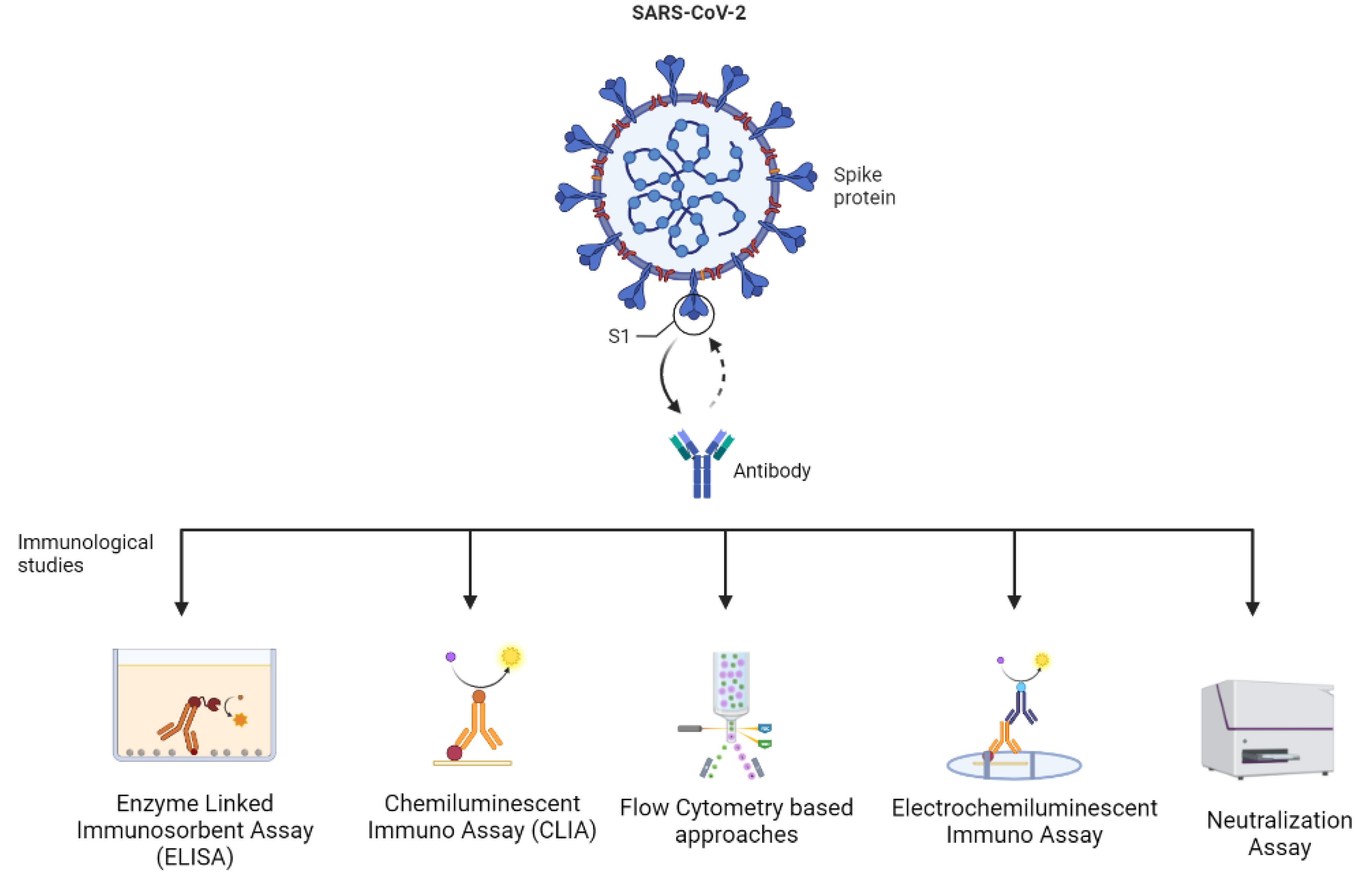
| Technique | Description | Advantages | Limitations | References |
|---|---|---|---|---|
| Enzyme-Linked Immunosorbent Assay (ELISA) | It facilitates rapid detection and quantification of IgA, IgM, and IgG against coat proteins, spike proteins, and the receptor-binding domain (RBD). |
|
| [45,46,51] |
| Chemiluminescent Immuno Assay (CLIA) | It is based on the high binding affinity of the antigen of a viral variant with specific antibodies of the host. The technique requires a chemical probe to exhibit a positive reaction. |
|
| [47,52] |
| Electro-Chemiluminescent Assay | The viral antigens are used to detect IgG corresponding to it. |
|
| [48,53,54,55] |
| Flow Cytometer-based Approaches | A novel high-throughput approach using a multiplexed flow cytometer-based assay. It can be used for a deep analysis of the immune system of people in different stages of infection. |
|
| [49,56,57] |
| Neutralization Assay | It involves techniques such as live virus plaque reduction (titer of neutralizing antibodies) and ACE-2 binding inhibition assays (binding of spike protein to the host ACE-2 receptor. It is used for validation of novel vaccine formulations, potential therapeutics, and screening of inhibitors. |
|
| [49,50,58,59] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chavda, V.P.; Mishra, T.; Vuppu, S. Immunological Studies to Understand Hybrid/Recombinant Variants of SARS-CoV-2. Vaccines 2023, 11, 45. https://doi.org/10.3390/vaccines11010045
Chavda VP, Mishra T, Vuppu S. Immunological Studies to Understand Hybrid/Recombinant Variants of SARS-CoV-2. Vaccines. 2023; 11(1):45. https://doi.org/10.3390/vaccines11010045
Chicago/Turabian StyleChavda, Vivek P., Toshika Mishra, and Suneetha Vuppu. 2023. "Immunological Studies to Understand Hybrid/Recombinant Variants of SARS-CoV-2" Vaccines 11, no. 1: 45. https://doi.org/10.3390/vaccines11010045
APA StyleChavda, V. P., Mishra, T., & Vuppu, S. (2023). Immunological Studies to Understand Hybrid/Recombinant Variants of SARS-CoV-2. Vaccines, 11(1), 45. https://doi.org/10.3390/vaccines11010045








