Genomic Characterization of Dengue Virus Outbreak in 2022 from Pakistan
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sample Collection
2.3. RNA Extraction and Serotype Specific MultiPlex PCR Amplification
2.4. Next Generation Sequencing
2.5. NGS Data Analysis
2.6. Phylogenetic Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mutheneni, S.R.; Morse, A.P.; Caminade, C.; Upadhyayula, S.M. Dengue burden in India: Recent trends and importance of climatic parameters. Emerg. Microbes Infect. 2017, 6, 1–10. [Google Scholar] [CrossRef]
- Perera, R.; Kuhn, R.J. Structural proteomics of dengue virus. Curr. Opin. Microbiol. 2008, 11, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Uno, N.; Ross, T.M. Dengue virus and the host innate immune response. Emerg. Microbes Infect. 2018, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, M.S.; Rasotgi, V.; Jain, S.; Gupta, V. Discovery of fifth serotype of dengue virus (DENV-5): A new public health dilemma in dengue control. Med. J. Armed Forces India 2015, 71, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Bashyam, H.S.; Green, S.; Rothman, A.L. Dengue virus-reactive CD8+ T cells display quantitative and qualitative differences in their response to variant epitopes of heterologous viral serotypes. J. Immunol. 2006, 176, 2817–2824. [Google Scholar] [CrossRef] [PubMed]
- Simmons, C.P.; Farrar, J.J.; van Vinh Chau, N.; Wills, B. Dengue. N. Engl. J. Med. 2012, 366, 1423–1432. [Google Scholar] [CrossRef]
- Phadungsombat, J.; Lin, M.Y.; Srimark, N.; Yamanaka, A.; Nakayama, E.E.; Moolasart, V.; Suttha, P.; Shioda, T.; Uttayamakul, S. Emergence of genotype Cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand. PLoS ONE 2018, 13, e0207220. [Google Scholar] [CrossRef]
- Limkittikul, K.; Brett, J.; L’Azou, M. Epidemiological trends of dengue disease in Thailand (2000–2011): A systematic literature review. PLoS Negl. Trop. Dis. 2014, 8, e3241. [Google Scholar] [CrossRef]
- Gould, E.; Solomon, E.A. Pathogenic flaviviruses. Lancet 2008, 371, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Green, S.; Rothman, A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr. Opin. Infect. Dis. 2006, 19, 429–436. [Google Scholar] [CrossRef]
- Domingues, R.B.; Kuster, G.W.; Onuki-Castro, F.L.; Souza, V.A.; Levi, J.E.; Pannuti, C.S. Involvement of the central nervous system in patients with dengue virus infection. J. Neurol. Sci. 2008, 267, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Wiyono, L.; Rocha, I.C.N.; Cedeño, T.D.D.; Miranda, A.V.; Lucero-Prisno, D.E., III. Dengue and COVID-19 infections in the ASEAN region: A concurrent outbreak of viral diseases. Epidemiol. Health 2021, 43, e2021070. [Google Scholar] [CrossRef]
- Dengue Worldwide Overview; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2022.
- Rana, M.S.; Usman, M.; Alam, M.M.; Ikram, A.; Salman, M.; Faryal, R.; Umair, M. The outbreak of dengue during the COVID-19 pandemic in Pakistan: The emergence of overlapping crises. Asian Pac. J. Trop. Med. 2022, 15, 53–55. [Google Scholar] [CrossRef]
- Hanan, F.; Ahmad, J.; Hadi, S.; Ali, I.; Fahim, M.; Qayum, A. Analysis of Dengue Virus Genotypes and Further Investigations for Mixed Infections through RT-PCR in Classical Dengue Fever Patients in Pakistan. Int. J. Pathol. 2022, 20, 72–78. [Google Scholar]
- Rana, M.S.; Alam, M.M.; Salman, M.; Ikram, A. Prevention and control of escalating dengue epidemics in Pakistan. J. Med. Virol. 2020, 92, 927–928. [Google Scholar] [CrossRef] [PubMed]
- Pakistan: Dengue Response Emergency Plan of Action (EPoA) DREF Operation no. MDRPK022. 23 October 2021. Available online: https://reliefweb.int/report/pakistan/pakistan-dengue-response-emergency-plan-action-epoa-dref-operation-n-mdrpk022 (accessed on 2 November 2022).
- Dengue Fever; World Health Organization: Geneva, Switzerland, 2022.
- Bharaj, P.; Chahar, H.S.; Pandey, A.; Diddi, K.; Dar, L.; Guleria, R.; Kabra, S.K.; Broor, S. Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, India. Virol. J. 2008, 5, 1–5. [Google Scholar] [CrossRef]
- Dhanoa, A.; Hassan, S.S.; Ngim, C.F.; Lau, C.F.; Chan, T.S.; Adnan, N.A.A.; Eng, W.W.H.; Gan, H.M.; Rajasekaram, G. Impact of dengue virus (DENV) co-infection on clinical manifestations, disease severity and laboratory parameters. BMC Infect. Dis. 2016, 16, 1–14. [Google Scholar] [CrossRef]
- Yousaf, M.Z.; Siddique, A.; Ashfaq, U.A.; Ali, M. Scenario of dengue infection & its control in Pakistan: An up—Date and way forward. Asian Pac. J. Trop. Med. 2018, 11, 15. [Google Scholar]
- Suleman, M.; Faryal, R.; Alam, M.M.; Sharif, S.; Shaukat, S.; Khurshid, A.; Angez, M.; Umair, M.; Syed, S. Identification of concurrent infection by multiple dengue virus serotypes during an epidemic in 2011 in Pakistan. J. Microbiol. Exp. 2016, 3, 00088. [Google Scholar]
- Atif, M.; Raheel, U.; Alam, F.; Arshad, H.U.; Baloch, F.; Imran, M.; Zaidi, N.; Waqar, A. Serotyping of dengue virus from deadly outbreaks of Pakistan. J. Hum. Virol. Retro-Virol. 2016, 3, 00092. [Google Scholar]
- Khan, E.; Prakoso, D.; Imtiaz, K.; Malik, F.; Farooqi, J.Q.; Long, M.T.; Barr, K.L. The clinical features of co-circulating dengue viruses and the absence of dengue hemorrhagic fever in Pakistan. Front. Public Health 2020, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.W.; Russell, B.J.; Lanciotti, R.S. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 2005, 43, 4977–4983. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Institute, B. Picard Toolkit; Broad Institute: Cambridge, MA, USA, 2019. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. In Bioinformatics for DNA Sequence Analysis; Humana Press: Totowa, NJ, USA, 2009; pp. 39–64. [Google Scholar]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A. FigTree v1. 4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2010. [Google Scholar]
- World Health Organization. Dengue and Severe Dengue. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 10 January 2022).
- Khatri, G.; Hasan, M.M.; Shaikh, S.; Mir, S.L.; Sahito, A.M.; Rocha, I.C.N.; Elmahi, O.K.O. The simultaneous crises of dengue and COVID-19 in Pakistan: A double hazard for the country’s debilitated healthcare system. Trop. Med. Health 2022, 50, 1–5. [Google Scholar] [CrossRef]
- WHO EMRO|Dengue Fever. Available online: http://www.emro.who.int/pdf/pak/programmes/dengue-fever.pdf (accessed on 1 December 2022).
- Khan, A. Pakistan Reports over 52,000 Dengue Cases in 2021. Available online: https://pakaffairs.pk/health/15369/pakistan-reports-over-52000-dengue-cases-in-2021/ (accessed on 4 January 2022).
- Shahid, A.; Azeem, S.; Shahzil, M.; Ghafoor, M.S.; Shah, J.; Cheema, H.A. Catastrophic floods in Pakistan: An urgent appeal for action. In Disaster Medicine and Public Health Preparedness; Cambridge University Press: Cambridge, UK, 2022; pp. 1–4. [Google Scholar]
- Umair, M.; Ikram, A.; Salman, M.; Rana, M.S.; Ashraf, A.; Ali, Q.; Alam, M.M. Serotype diversity of dengue virus reveals the predominance of type 2 in Pakistan during 2019. J. Med. Virol. 2020, 92, 2900–2902. [Google Scholar] [CrossRef]
- Khan, N.U.; Danish, L.; Khan, H.U.; Shah, M.; Ismail, M.; Ali, I.; Petruzziello, A.; Sabatino, R.; Guzzo, A.; Botti, G.; et al. Prevalence of dengue virus serotypes in the 2017 outbreak in Peshawar, KP, Pakistan. J. Clin. Lab. Anal. 2020, 34, e23371. [Google Scholar] [CrossRef]
- Ali, A.; Ahmad, H.; Idrees, M.; Zahir, F.; Ali, I. Circulating serotypes of dengue virus and their incursion into non-endemic areas of Pakistan; a serious threat. Virol. J. 2016, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bosch, I.; Reddy, A.; de Puig, H.; Ludert, J.E.; Perdomo-Celis, F.; Narváez, C.F.; Versiani, A.; Fandos, D.; Nogueira, M.L.; Singla, M.; et al. Serotype-specific detection of dengue viruses in a nonstructural protein 1-based enzyme-linked immunosorbent assay validated with a multi-national cohort. PLoS Negl. Trop. Dis. 2020, 14, e0008203. [Google Scholar] [CrossRef] [PubMed]
- Senaratne, U.; Murugananthan, K.; Sirisena, P.D.N.N.; Carr, J.M.; Noordeen, F. Dengue virus co-infections with multiple serotypes do not result in a different clinical outcome compared to mono-infections. Epidemiol. Infect. 2020, 148, e119. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Fatima, Z.; Wahid, B.; Rafique, S.; Idrees, M. Cosmopolitan A1 lineage of dengue virus serotype 2 is circulating in Pakistan: A study from 2017 dengue viral outbreak. J. Med. Virol. 2019, 91, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Yenamandra, S.P.; Koo, C.; Chiang, S.; Lim, H.S.J.; Yeo, Z.Y.; Ng, L.C.; Hapuarachchi, H.C. Evolution, heterogeneity and global dispersal of cosmopolitan genotype of Dengue virus type 2. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Akram, M.; Fatima, Z.; Purdy, M.A.; Sue, A.; Saleem, S.; Amin, I.; Shahid, M.; Idrees, M.; Nawaz, R. Introduction and evolution of dengue virus type 2 in Pakistan: A phylogeographic analysis. Virol. J. 2015, 12, 1–11. [Google Scholar] [CrossRef]
- Holmes, E.C.; Twiddy, S.S. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 2003, 3, 19–28. [Google Scholar] [CrossRef]
- Khan, M.A.; Ellis, E.M.; Tissera, H.A.; Alvi, M.Y.; Rahman, F.F.; Masud, F.; Chow, A.; Howe, S.; Dhanasekaran, V.; Ellis, B.R.; et al. Emergence and diversification of dengue 2 cosmopolitan genotype in Pakistan, 2011. PLoS ONE 2013, 8, e56391. [Google Scholar] [CrossRef]
- Ma, M.; Wu, S.; He, Z.; Yuan, L.; Bai, Z.; Jiang, L.; Marshall, J.; Lu, J.; Yang, Z.; Jing, Q. New genotype invasion of dengue virus serotype 1 drove massive outbreak in Guangzhou, China. Parasites Vectors 2021, 14, 1–12. [Google Scholar] [CrossRef]
- Teoh, B.-T.; Sam, S.S.; Tan, K.K.; Johari, J.; Shu, M.H.; Danlami, M.B.; Abd-Jamil, J.; MatRahim, N.; Mahadi, N.M.; AbuBakar, S. Dengue virus type 1 clade replacement in recurring homotypic outbreaks. BMC Evol. Biol. 2013, 13, 1–10. [Google Scholar] [CrossRef]
- Zhang, X.; Ge, P.; Yu, X.; Brannan, J.M.; Bi, G.; Zhang, Q.; Schein, S.; Zhou, Z.H. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 2013, 20, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Cardosa, J.; Hanley, K.A.; Holmes, E.C.; Weaver, S.C. Fever from the forest: Prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat. Rev. Microbiol. 2011, 9, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Petit, M.J.; Kenaston, M.W.; Pham, O.H.; Nagainis, A.A.; Fishburn, A.T.; Shah, P.S. Nuclear dengue virus NS5 antagonizes expression of PAF1-dependent immune response genes. PLoS Pathog. 2021, 17, e1010100. [Google Scholar] [CrossRef]
- Lim, S.P.; Noble, C.; Shi, P.-Y. The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 2015, 119, 57–67. [Google Scholar] [CrossRef]
- Koo, C.; Nasir, A.; Hapuarachchi, H.C.; Lee, K.S.; Hasan, Z.; Ng, L.C.; Khan, E. Evolution and heterogeneity of multiple serotypes of Dengue virus in Pakistan, 2006–2011. Virol. J. 2013, 10, 1–10. [Google Scholar] [CrossRef]
- Torres-Flores, J.M.; Reyes-Sandoval, A.; Salazar, M. Dengue Vaccines: An Update. BioDrugs 2022, 36, 325–336. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Dengue vaccine: WHO position paper, September 2018—Recommendations. Vaccine 2019, 37, 4848–4849. [Google Scholar] [CrossRef]
- Turner, M.; Papadimitriou, A.; Winkle, P.; Segall, N.; Levin, M.; Doust, M.; Johnson, C.; Lucksinger, G.; Fierro, C.; Pickrell, P.; et al. Immunogenicity and safety of lyophilized and liquid dengue tetravalent vaccine candidate formulations in healthy adults: A randomized, phase 2 clinical trial. Hum. Vaccines Immunother. 2020, 16, 2456–2464. [Google Scholar] [CrossRef] [PubMed]
- Tricou, V.; Sáez-Llorens, X.; Yu, D.; Rivera, L.; Jimeno, J.; Villarreal, A.C.; Dato, E.; de Suman, O.S.; Montenegro, N.; DeAntonio, R.; et al. Safety and immunogenicity of a tetravalent dengue vaccine in children aged 2–17 years: A randomised, placebo-controlled, phase 2 trial. Lancet 2020, 395, 1434–1443. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Llorens, X.; Tricou, V.; Yu, D.; Rivera, L.; Jimeno, J.; Villarreal, A.C.; Dato, E.; Mazara, S.; Vargas, M.; Brose, M.; et al. Immunogenicity and safety of one versus two doses of tetravalent dengue vaccine in healthy children aged 2–17 years in Asia and Latin America: 18-month interim data from a phase 2, randomised, placebo-controlled study. Lancet Infect. Dis. 2018, 18, 162–170. [Google Scholar] [CrossRef]
- Jackson, L.A.; Rupp, R.; Papadimitriou, A.; Wallace, D.; Raanan, M.; Moss, K.J. A phase 1 study of safety and immunogenicity following intradermal administration of a tetravalent dengue vaccine candidate. Vaccine 2018, 36, 3976–3983. [Google Scholar] [CrossRef] [PubMed]
- Myshko, D. FDA Grants Priority Review of BLA for Dengue Vaccine. Available online: https://www.formularywatch.com/view/fda-grants-priority-review-of-bla-for-dengue-vaccine (accessed on 23 November 2022).
- Takeda Eyes Further Filings For Dengue Vaccine But Timings Unclear. Available online: https://scrip.pharmaintelligence.informa.com/SC147441/Takeda-Eyes-Further-Filings-For-Dengue-Vaccine-But-Timings-Unclear (accessed on 28 November 2022).
- Takeda’s QDENGA®▼ (Dengue Tetravalent Vaccine [Live, Attenuated]) Approved in Indonesia for Use Regardless of Prior Dengue Exposure. Available online: https://www.takeda.com/newsroom/newsreleases/2022/takedas-qdenga-dengue-tetravalent-vaccine-live-attenuated-approved-in-indonesia-for-use-regardless-of-prior-dengue-exposure/ (accessed on 22 August 2022).
- Mallapaty, S. Dengue vaccine poised for roll-out but safety concerns linger. Nature 2022, 611, 434–435. [Google Scholar] [CrossRef] [PubMed]
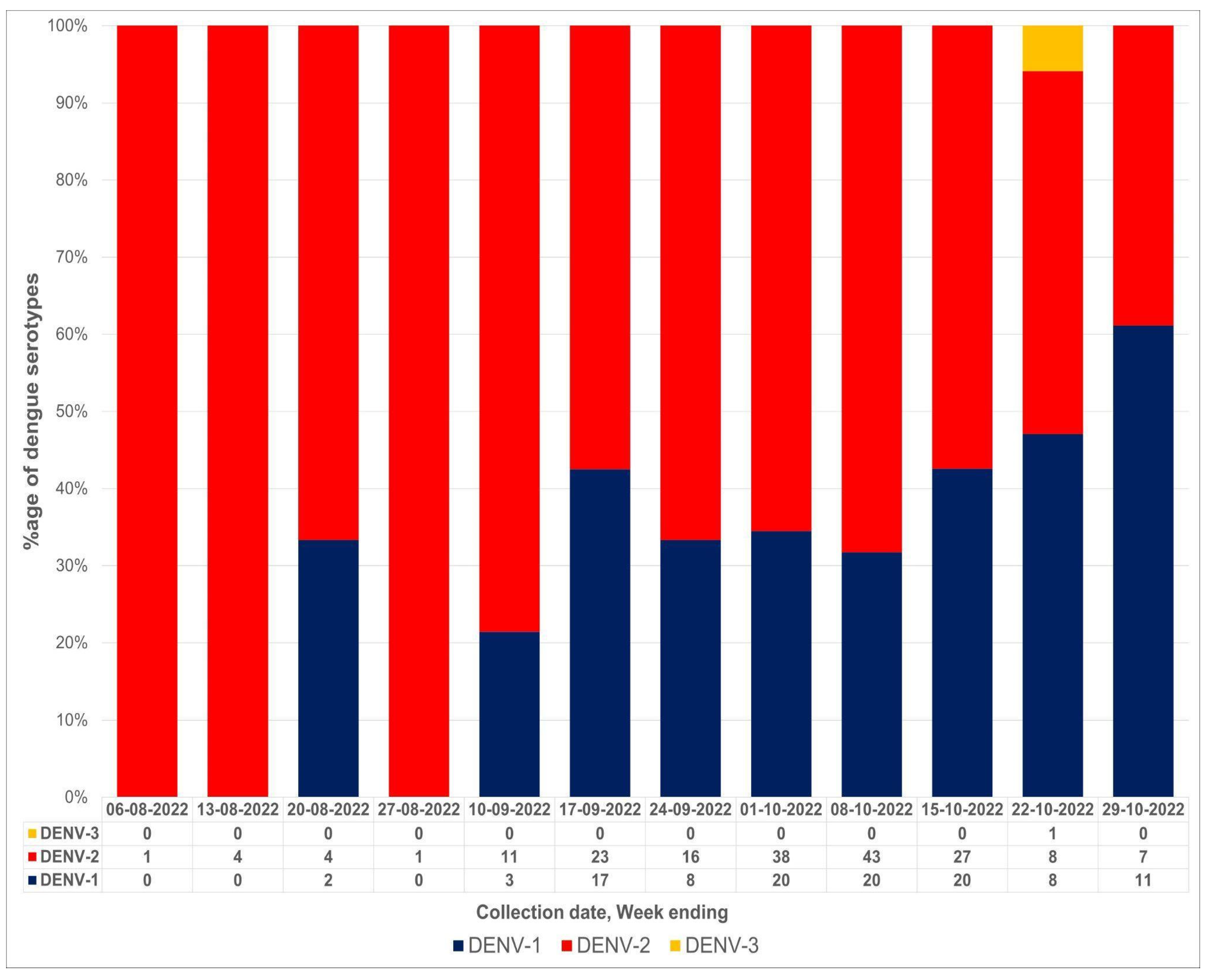
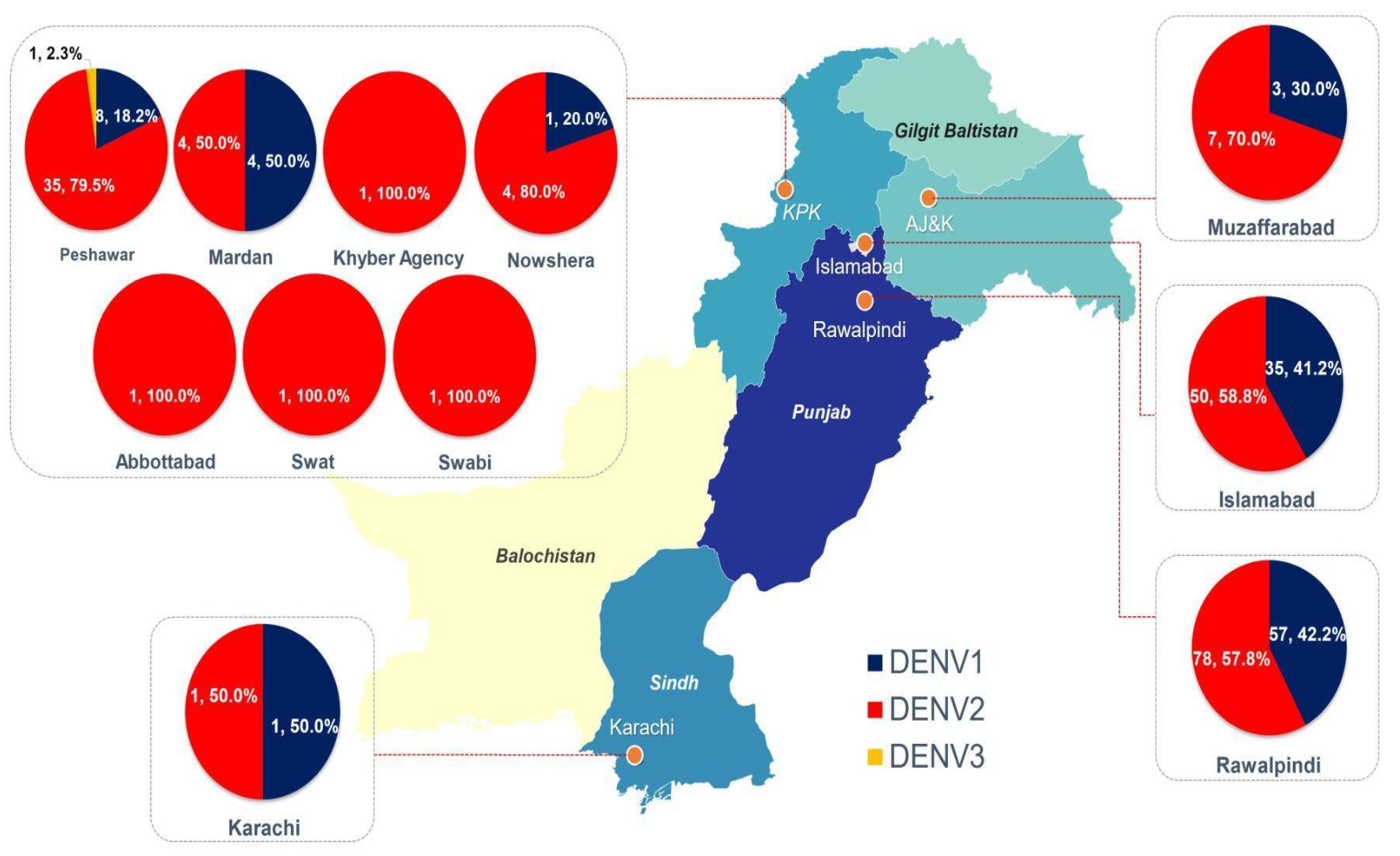
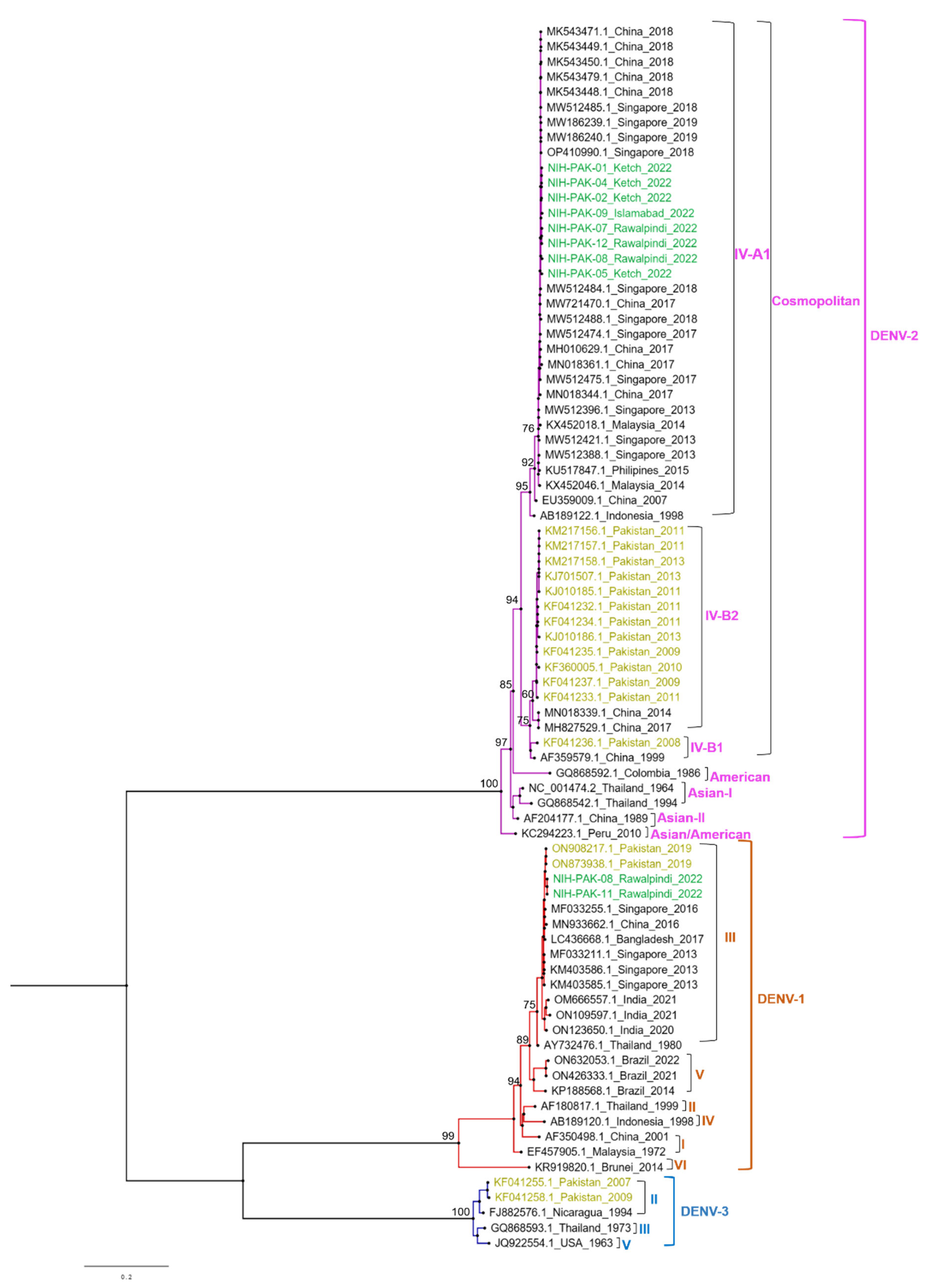
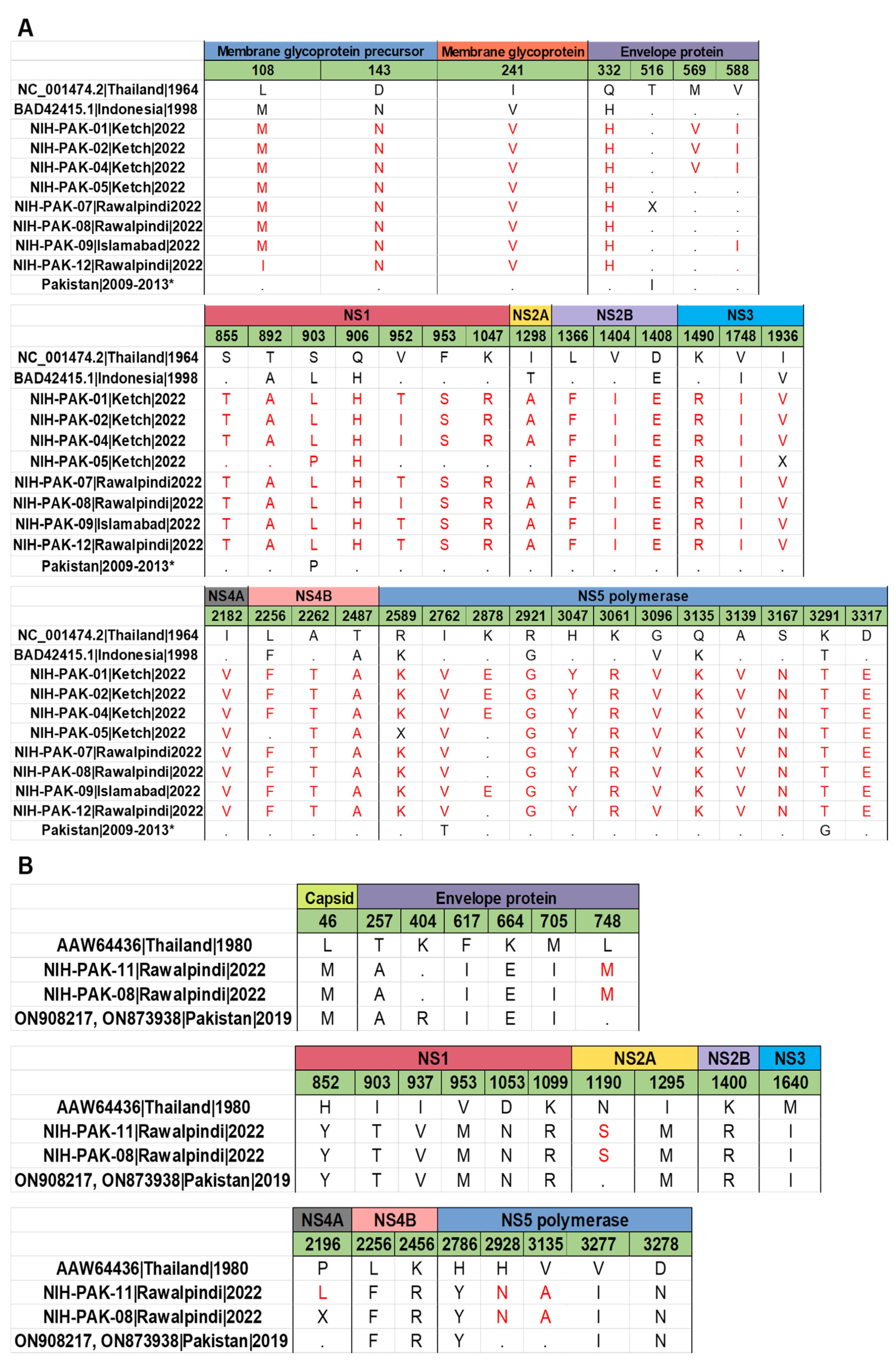
| Total No. of Cases (NS1+) | PCR Positive | PCR Negative |
|---|---|---|
| 343 | 293 (85%) | 50 (15%) |
| Gender | No. (%) | No. (%) |
| Male (213) | 184 (63%) | 29 (58%) |
| Female (130) | 109 (37%) | 21 (42%) |
| Age groups (Years) | ||
| 0–10 | 4 (1.3%) | 1 (2%) |
| 11–20 | 62 (21%) | 8 (16%) |
| 21–30 | 94 (32%) | 17 (34%) |
| 31–40 | 54 (18%) | 13 (26%) |
| 41–50 | 53 (18%) | 8 (16%) |
| >50 | 26 (9%) | 3 (6%) |
| Clinical Symptoms | ||
| Total available data = 223 patients | ||
| Fever | 218 (98%) | |
| Myalgia | 210 (94%) | |
| Backache | 156 (70%) | |
| Hematological Markers | ||
| Total available data = 144 patients | ||
| Platelet Count | ≤150,000 = 129 (89%) ≥150,000 = 15 (11%) | |
| Hematocrit Level | <40 = 96 (67%) 41–45 = 32 (22%) >45 = 15 (10%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umair, M.; Haider, S.A.; Rehman, Z.; Jamal, Z.; Ali, Q.; Hakim, R.; Bibi, S.; Ikram, A.; Salman, M. Genomic Characterization of Dengue Virus Outbreak in 2022 from Pakistan. Vaccines 2023, 11, 163. https://doi.org/10.3390/vaccines11010163
Umair M, Haider SA, Rehman Z, Jamal Z, Ali Q, Hakim R, Bibi S, Ikram A, Salman M. Genomic Characterization of Dengue Virus Outbreak in 2022 from Pakistan. Vaccines. 2023; 11(1):163. https://doi.org/10.3390/vaccines11010163
Chicago/Turabian StyleUmair, Massab, Syed Adnan Haider, Zaira Rehman, Zunera Jamal, Qasim Ali, Rabia Hakim, Shaheen Bibi, Aamer Ikram, and Muhammad Salman. 2023. "Genomic Characterization of Dengue Virus Outbreak in 2022 from Pakistan" Vaccines 11, no. 1: 163. https://doi.org/10.3390/vaccines11010163
APA StyleUmair, M., Haider, S. A., Rehman, Z., Jamal, Z., Ali, Q., Hakim, R., Bibi, S., Ikram, A., & Salman, M. (2023). Genomic Characterization of Dengue Virus Outbreak in 2022 from Pakistan. Vaccines, 11(1), 163. https://doi.org/10.3390/vaccines11010163






