Nanoparticle- and Microparticle-Based Vaccines against Orbiviruses of Veterinary Importance
Abstract
1. Introduction
2. Nanoparticle- and Microparticle-Based Vaccine Platforms
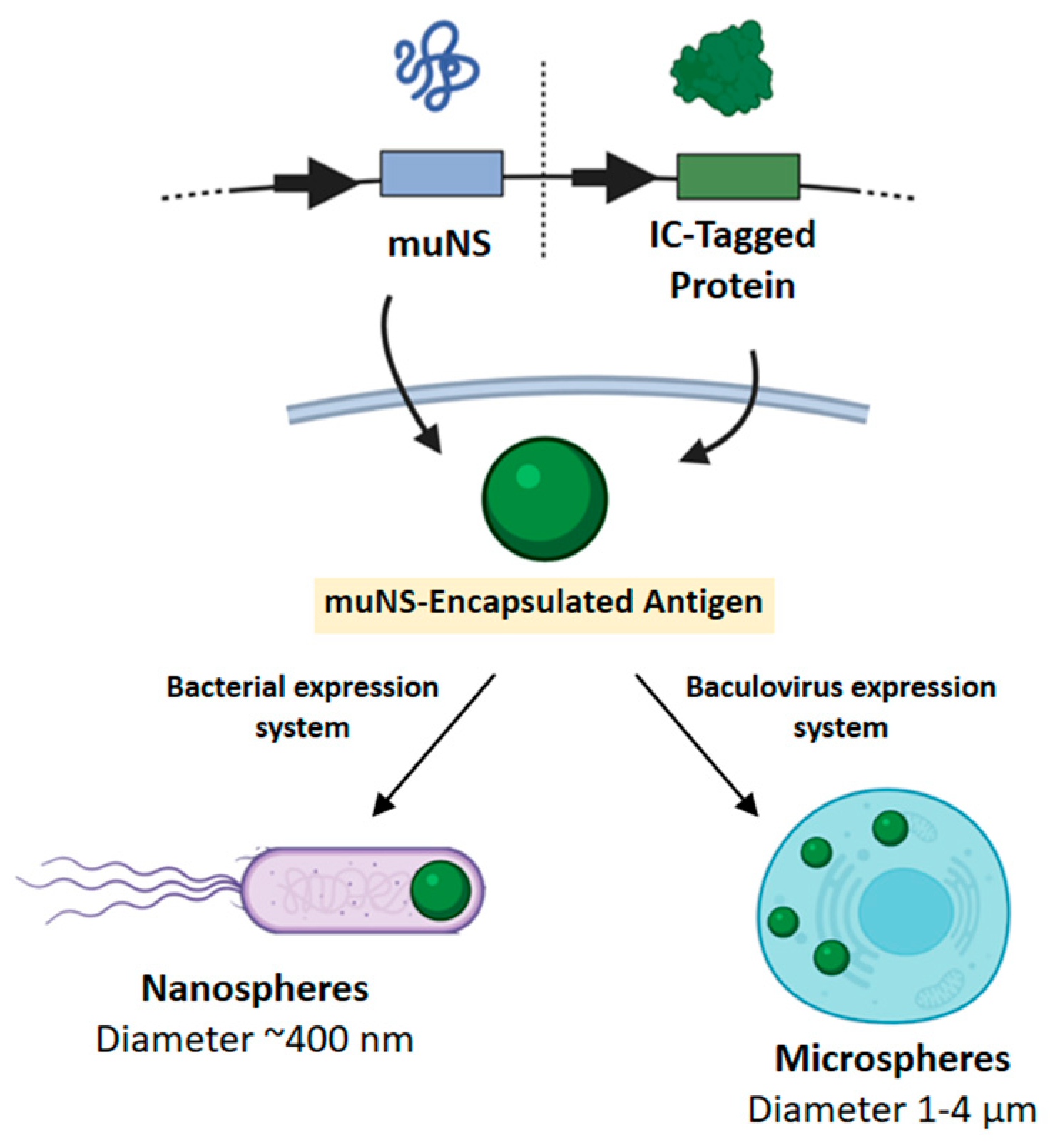
µNS Avian Reovirus Protein as an Antigen Carrier System
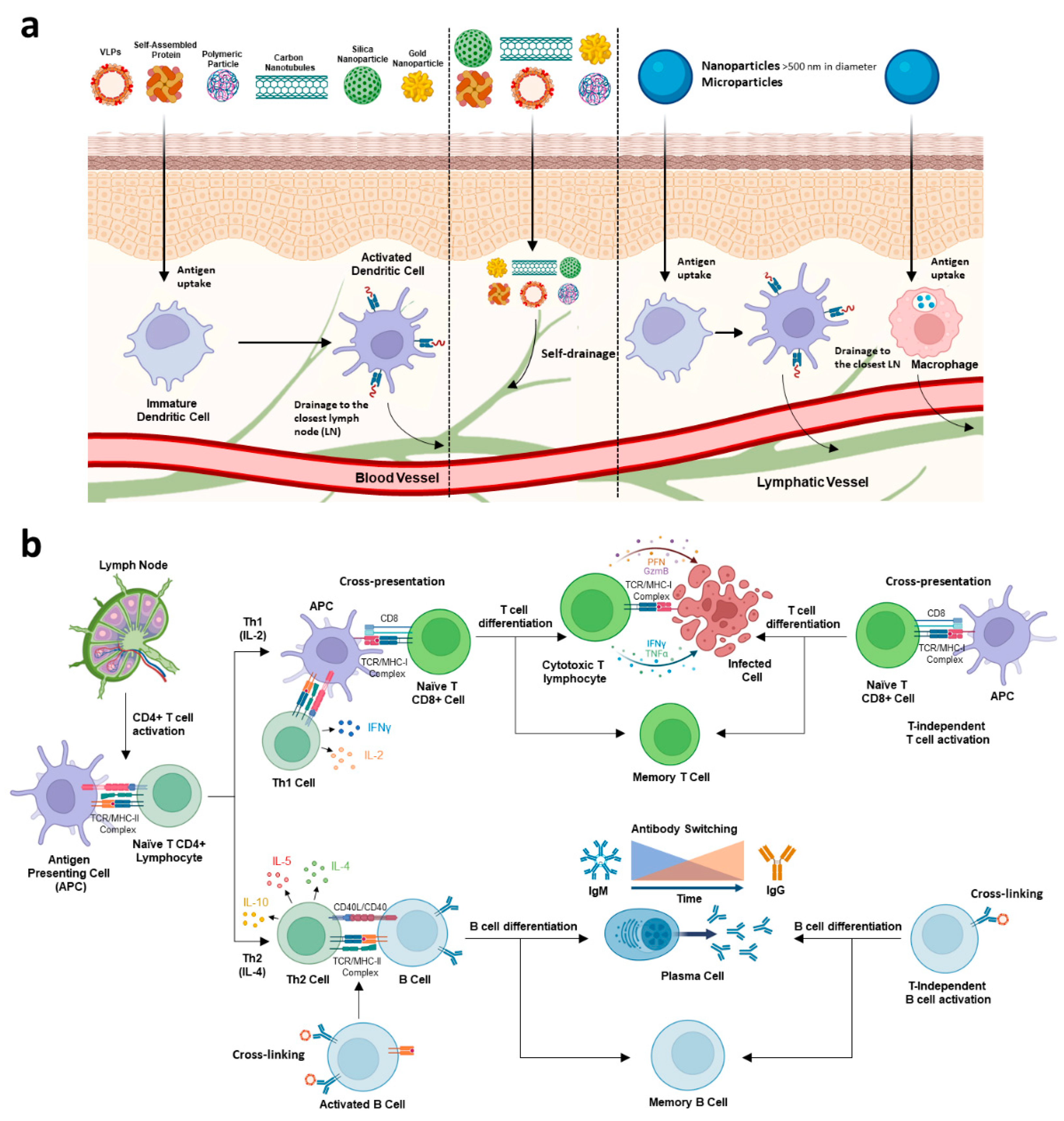
3. Interaction of Nano- or Microcarriers with the Immune System
4. Bluetongue Virus (BTV)
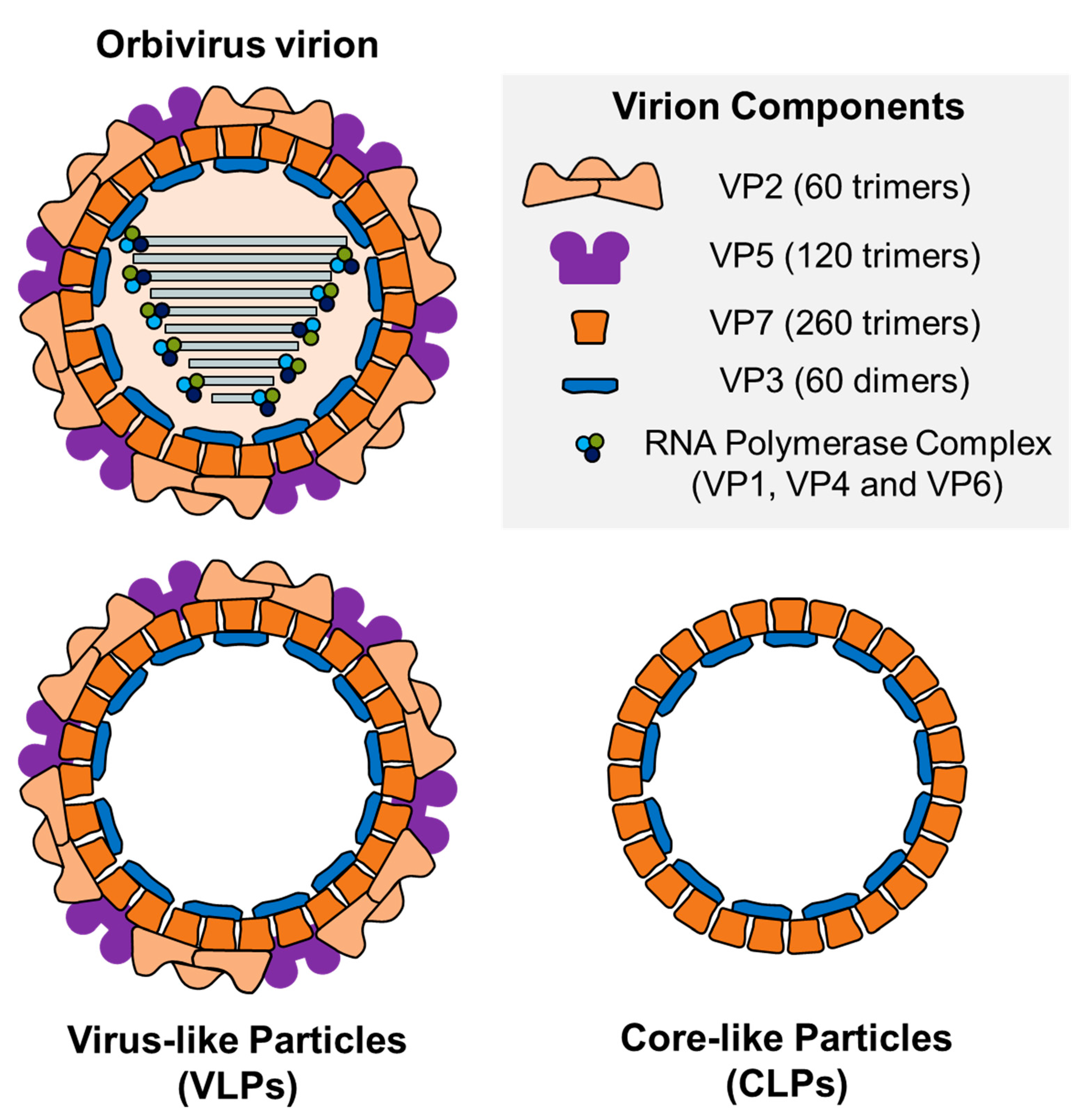
4.1. Virus-Like Particles (VLPs)
4.2. Avian Reovirus µNS- Microspheres Carrying BTV Antigens
| Vaccine Type | Antigen Included | Dose | Animal Model | Challenge | Adjuvant | Immunogenicity and/or Protection | Ref. |
|---|---|---|---|---|---|---|---|
| CLP a | VP7, VP3 | Not evaluated as vaccine candidate | - | - | - | - | [162] |
| CLP a | VP7, VP3 | Four doses (75 µg per dose) | Guinea pig | Not challenged | Adjuvant-free | Absence of Nabs. Protection not Assessed | [138] |
| VLP a | VP2 (BTV-10), VP5, VP7, VP3 | Neutralization against BTV-10. Protection not Assessed | |||||
| VLP a | VP2 (BTV-1 or BTV-17), VP5, VP7, VP3 | Four doses (100 µg per dose) | Guinea pig | Not challenged | Incomplete Freunds’ adjuvant | Neutralization against BTV-1 or BTV-17. Protection not Assessed | [163] |
| VLP a | VP2 (BTV-10), VP5, VP7, VP3 | Two doses (10, 50, 100 or 200 µg per dose) | 1-year-old Merino sheep | BTV-10 | Adjuvant-free | Neutralization against BTV-10. Partial Homologous protection c,d. | [172] |
| IFA | |||||||
| AI(OH)3 | |||||||
| ISA-50 | Neutralization against BTV-10. Homologous protection c,d. | ||||||
| VLP a | VP2 (BTV-10 or BTV-17), VP5, VP7, VP3 | Two doses (10 or 50 µg per dose) | 1-year-old Merino sheep | Homologous (BTV-10 or BTV-17) | ISA-50 | Neutralization against BTV-10 and BTV-17. Partial cross-neutralization against BTV-4. Homologous protection c,d. | [169] |
| VP2 (BTV-10 and BTV-17), VP5, VP7, VP3 | Heterologous (BTV-4 or BTV-11) | Neutralization against BTV-10 and BTV-17. Partial cross-neutralization against BTV-4. Partial heterologous protection c,d. | |||||
| VP2 (BTV-1, BTV-2, BTV-10, BTV-13 and BTV-17), VP5, VP7, VP3 | Two doses (50 µg per dose) | Homologous (BTV-13) and Heterologous (BTV-16) | Neutralization against BTV-10, BTV-13 and BTV-17. Homologous protection c,d. Mild heterologous protection c,d. | ||||
| VLP a | VP2 (BTV-2), VP5, VP7, VP3 | Two doses (10 or 20 µg per dose) | 10-month-old cross-bred Pre-Alpes sheep | Homologous (BTV-2) | SEPPIC | Neutralization against BTV-2. Homologous protection c,e,f. | [170] |
| VLP a | VP2 (BTV-1), VP5, VP7, VP3 | Two doses (unspecified dose) | 7–8-month-old male Merino sheep | Homologous (BTV-1) | SEPPIC | Neutralization against BTV-1. Homologous protection c,e,f. | [171] |
| VP2 (BTV-1 and BTV-4), VP5, VP7, VP3 | Homologous (BTV-1 and BTV-4) | Neutralization against BTV-1 and BTV-4. Protection against BTV-1 c,e,f. Partial Protection against BTV-4 c,e,f. | |||||
| CLP a | VP7, VP3 | Two doses (~50–100 µg per dose) | 18-month-old female Karagouniko cross-bred sheep | Homologous (BTV-1) | SEPPIC | Low protection c,d,e,f. | [166] |
| VLP a | VP2 (BTV-1), VP5, VP7, VP3 | Neutralization against BTV-1. Homologous protection c,d,e,f. | |||||
| VLP a | VP2 (BTV-8), VP5, VP7, VP3 | Two doses (20 µg per dose) | 10-month-old cross-bred Pre-Alpes sheep | Homologous (BTV-8) | SEPPIC | Neutralization against BTV-8. Homologous protection c,e,f. | [72] |
| VP2 (BTV-8, BTV-1 and BTV-2), VP5, VP7, VP3 | Two doses (60 µg per dose) | Neutralization against BTV-1, BTV-2 and BTV-8. Homologous protection c,e,f. | |||||
| CLP b | VP7, VP3 | Two doses (200 µg per dose) | 1-year-old Merino sheep | Homologous (BTV-8) | SEPPIC | No protection c,e. | [164] |
| VLP b | VP2 (BTV-8), VP5, VP7, VP3 | Two doses (50 µg per dose) | Neutralization against BTV-8. Homologous protection c,e | ||||
| VLP b | VP2 (BTV-2 or BTV-4), VP5, VP7, VP3 | Two doses (~15–30 µg per dose | 6–12-month-old Merino sheep | Not challenged | SEPPIC/Alhydrogel | Neutralization against BTV-3 and BTV-4. Protection not assessed | [165] |
| Lumazine synthase complexes a | VP2 | Not evaluated as vaccine candidate | - | - | - | - | [176] |
| Carrier RHDV VLP a | Six residue epitope of VP7 (BTag) | Not evaluated as vaccine candidate | - | - | - | - | [183] |
| muNS-Mi a | NS1, VP2 (BTV-4), VP7 | Two doses (150 µg per dose) | Adult IFNAR(-/-) mice | Homologous (BTV-4) and heterologous (BTV-1) | Self-Adjuvant | Neutralization against BTV-4. Cell immune responses specific of VP7 or NS1. Homologous protection g,c,d. Partial heterologous protection g,c,d. | [191] |
| muNS-Mi/MVA a | NS1, VP2 (BTV-4), VP7 | Two doses (prime: 150 µg; boost: 107 PFU) | Adult IFNAR(-/-) mice | Homologous (BTV-4) and heterologous (BTV-1) | Self-Adjuvant | Neutralization against BTV-4. NS1-spcecific CD8+ T-cell response. Homologous and heterologous protection g,c,d. | [192] |
4.3. A Self-Assembled Nanoparticle Vaccine Platform Based on BTV-NS1 Tubules
5. African Horse Sickness Virus (AHSV)
6. Epizootic Hemorrhagic Disease Virus (EHDV)
| Species | Vaccine Type | Antigen Included | Dose | Animal Model | Challenge | Adjuvant | Immunogenicity and/or Protection | Ref. |
|---|---|---|---|---|---|---|---|---|
| African horse sickness virus (AHSV) | CLP a | VP7,VP3 | Not evaluated in animal model | - | - | - | - | [224] |
| CLP a | VP7,VP3 | Not evaluated in animal model | - | - | - | - | [225] | |
| Partial VLP a | VP2 (AHSV-9),VP7,VP3 | |||||||
| VP5,VP7,VP3 | ||||||||
| VLP a | VP2 (AHSV-9),VP5,VP7,VP3 | |||||||
| VLP b | VP2 (AHSV-5),VP5,VP7,VP3 | Two doses (Prime: 16.5 µg; Boost: 50 µg) | Guinea pig | Not challenged | 5% Montanide PET Gel A adjuvant | Neutralization against AHSV-5. Partial cross-neutralization against AHSV-8. | [227] | |
| Two doses (100 or 200 µg per dose) | Horse | Not challenged | Neutralization against AHSV-5. Cross-neutralization against AHSV-8. | [228] | ||||
| VLP b | VP2 (AHSV-6), VP5,VP7,VP3 | Two doses (~200 µg per dose) | 6–12 months foals | Not challenged | 5% Montanide PET Gel A adjuvant | Neutralization against AHSV-6. | [70] | |
| muNS-Mi a | NS1 (AHSV-4) | Two doses (50 µg per dose) | Adult IFNAR(-/-) mice | Homologous (AHSV-4) | Self-adjuvant | NS1-specific CD8+ T-cell response. Partial homologous protection c,d,e. | [128] | |
| muNS-Mi/MVA a | Two doses (prime: 50 µg; boost: 107 PFU) | Homologous (AHSV-4) and heterologous (AHSV-9) | NS1-specific CD8+ T-cell response. Sterile homologous and heterologous protection c,d,e. | |||||
| Epizootic hemorrhagic disease virus (EHDV) | CLP a | VP7, VP3 | Two doses (prime: 500 µg; boost: 250 µg) | Rabbit | Not challenged | Incomplete Fruend’s adjuvant | Induction of VP3- and VP7-specific antibodies. | [246] |
| VLP a | VP2 (EHDV-1),VP5,VP7,VP3 | Two doses (prime: 500 µg; boost: 250 µg) | Rabbit | Not challenged | Incomplete Fruend’s adjuvant | Neutralization against EHDV-1. Mild neutralization against EHDV-2 and EHDV-6. | ||
| VLP a | VP2 (EHDV-6),VP5,VP7,VP3 | Not evaluated in animal model | - | - | - | - | [248] |
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Metcalf, P.; Cyrklaff, M.; Adrian, M. The Three-Dimensional Structure of Reovirus Obtained by Cryo-Electron Microscopy. EMBO J. 1991, 10, 3129–3136. [Google Scholar] [CrossRef] [PubMed]
- Attoui, H.; Jaafar, F.M.; de Micco, P.; de Lamballerie, X. Coltiviruses and Seadornaviruses in North America, Europe, and Asia. Emerg. Infect. Dis. 2005, 11, 1673–1679. [Google Scholar] [CrossRef] [PubMed]
- Schwartz-Cornil, I.; Mertens, P.P.C.; Contreras, V.; Hemati, B.; Pascale, F.; Bréard, E.; Mellor, P.S.; MacLachlan, N.J.; Zientara, S. Bluetongue Virus: Virology, Pathogenesis and Immunity. Vet. Res. 2008, 39, 46. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Mellor, P.S.; Fall, A.G.; Garros, C.; Venter, G.J. African Horse Sickness Virus: History, Transmission, and Current Status. Annu. Rev. Entomol. 2017, 62, 343–358. [Google Scholar] [CrossRef]
- Maclachlan, N.J. Bluetongue: History, Global Epidemiology, and Pathogenesis. Prev. Vet. Med. 2011, 102, 107–111. [Google Scholar] [CrossRef]
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef]
- Mertens, P.P.; Sangar, D.V. Analysis of the Terminal Sequences of the Genome Segments of Four Orbiviruses. Prog. Clin. Biol. Res. 1985, 178, 371–387. [Google Scholar] [CrossRef]
- Grimes, J.M.; Burroughs, J.N.; Gouet, P.; Diprose, J.M.; Malby, R.; Ziéntara, S.; Mertens, P.P.; Stuart, D.I. The Atomic Structure of the Bluetongue Virus Core. Nature 1998, 395, 470–478. [Google Scholar] [CrossRef]
- Huismans, H.; Erasmus, B.J. Identification of the Serotype-Specific and Group-Specific Antigens of Bluetongue Virus. Onderstepoort J. Vet. Res. 1981, 48, 51–58. [Google Scholar]
- Hassan, S.H.; Wirblich, C.; Forzan, M.; Roy, P. Expression and Functional Characterization of Bluetongue Virus VP5 Protein: Role in Cellular Permeabilization. J. Virol. 2001, 75, 8356–8367. [Google Scholar] [CrossRef]
- Boyce, M.; Wehrfritz, J.; Noad, R.; Roy, P. Purified Recombinant Bluetongue Virus VP1 Exhibits RNA Replicase Activity. J. Virol. 2004, 78, 3994–4002. [Google Scholar] [CrossRef] [PubMed]
- Ramadevi, N.; Burroughs, N.J.; Mertens, P.P.C.; Jones, I.M.; Roy, P. Capping and Methylation of MRNA by Purified Recombinant VP4 Protein of Bluetongue Virus. Proc. Natl. Acad. Sci. USA 1998, 95, 13537–13542. [Google Scholar] [CrossRef] [PubMed]
- Stäuber, N.; Martinez-Costas, J.; Sutton, G.; Monastyrskaya, K.; Roy, P. Bluetongue Virus VP6 Protein Binds ATP and Exhibits an RNA-Dependent ATPase Function and a Helicase Activity That Catalyze the Unwinding of Double-Stranded RNA Substrates. J. Virol. 1997, 71, 7220–7226. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Utrilla-Trigo, S.; Calvo-Pinilla, E.; Jiménez-Cabello, L.; Ortego, J.; Nogales, A. Inhibition of Orbivirus Replication by Aurintricarboxylic Acid. Int. J. Mol. Sci. 2020, 21, 7294. [Google Scholar] [CrossRef]
- Mohd Jaafar, F.; Monsion, B.; Belhouchet, M.; Mertens, P.P.C.; Attoui, H. Inhibition of Orbivirus Replication by Fluvastatin and Identification of the Key Elements of the Mevalonate Pathway Involved. Viruses 2021, 13, 1437. [Google Scholar] [CrossRef]
- John, L.; Vernersson, C.; Kwon, H.; Elling, U.; Penninger, J.M.; Mirazimi, A. Redirecting Imipramine against Bluetongue Virus Infection: Insights from a Genome-Wide Haploid Screening Study. Pathogens 2022, 11, 602. [Google Scholar] [CrossRef]
- Jiménez-Cabello, L.; Utrilla-Trigo, S.; Calvo-Pinilla, E.; Moreno, S.; Nogales, A.; Ortego, J.; Marín-López, A. Viral Vector Vaccines against Bluetongue Virus. Microorganisms 2020, 9, 42. [Google Scholar] [CrossRef]
- Van Rijn, P.A. Prospects of Next-Generation Vaccines for Bluetongue. Front. Vet. Sci. 2019, 6, 407. [Google Scholar] [CrossRef]
- Calvo-Pinilla, E.; Marín-López, A.; Utrilla-Trigo, S.; Jiménez-Cabello, L.; Ortego, J. Reverse Genetics Approaches: A Novel Strategy for African Horse Sickness Virus Vaccine Design. Curr. Opin. Virol. 2020, 44, 49–56. [Google Scholar] [CrossRef]
- Savini, G.; Afonso, A.; Mellor, P.; Aradaib, I.; Yadin, H.; Sanaa, M.; Wilson, W.; Monaco, F.; Domingo, M. Epizootic Heamorragic Disease. Res. Vet. Sci. 2011, 91, 1–17. [Google Scholar] [CrossRef]
- Qiao, C.; Liu, J.; Yang, J.; Li, Y.; Weng, J.; Shao, Y.; Zhang, X. Enhanced Non-Inflammasome Mediated Immune Responses by Mannosylated Zwitterionic-Based Cationic Liposomes for HIV DNA Vaccines. Biomaterials 2016, 85, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA COVID-19 Vaccine. N Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Wui, S.R.; Kim, K.S.; Ryu, J.I.; Ko, A.; Do, H.T.T.; Lee, Y.J.; Kim, H.J.; Lim, S.J.; Park, S.A.; Cho, Y.J.; et al. Efficient Induction of Cell-Mediated Immunity to Varicella-Zoster Virus Glycoprotein E Co-Lyophilized with a Cationic Liposome-Based Adjuvant in Mice. Vaccine 2019, 37, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- Rosenkrands, I.; Vingsbo-Lundberg, C.; Bundgaard, T.J.; Lindenstrøm, T.; Enouf, V.; van der Werf, S.; Andersen, P.; Agger, E.M. Enhanced Humoral and Cell-Mediated Immune Responses after Immunization with Trivalent Influenza Vaccine Adjuvanted with Cationic Liposomes. Vaccine 2011, 29, 6283–6291. [Google Scholar] [CrossRef] [PubMed]
- Senchi, K.; Matsunaga, S.; Hasegawa, H.; Kimura, H.; Ryo, A. Development of Oligomannose-Coated Liposome-Based Nasal Vaccine against Human Parainfluenza Virus Type 3. Front. Microbiol. 2013, 4, 346. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Taneichi, M.; Kasai, M.; Kakiuchi, T.; Uchida, T. Liposome-Coupled Antigens Are Internalized by Antigen-Presenting Cells via Pinocytosis and Cross-Presented to CD8 T Cells. PLoS ONE 2010, 5, e15225. [Google Scholar] [CrossRef]
- Wei, L.; Zhao, T.; Zhang, J.; Mao, Q.; Gong, G.; Sun, Y.; Chen, Y.; Wang, M.; Tan, D.; Gong, Z.; et al. Efficacy and Safety of a Nanoparticle Therapeutic Vaccine in Patients with Chronic Hepatitis B: A Randomized Clinical Trial. Hepatology 2022, 75, 182–195. [Google Scholar] [CrossRef]
- Storm, G.; Crommelin, D.J.A. Liposomes: Quo Vadis? Pharm. Sci. Technol. Today 1998, 1, 19–31. [Google Scholar] [CrossRef]
- Cappellano, G.; Abreu, H.; Casale, C.; Dianzani, U.; Chiocchetti, A. Nano-Microparticle Platforms in Developing Next-Generation Vaccines. Vaccines 2021, 9, 606. [Google Scholar] [CrossRef]
- Ho, H.-M.; Huang, C.-Y.; Cheng, Y.-J.; Shen, K.-Y.; Tzeng, T.-T.; Liu, S.-J.; Chen, H.-W.; Huang, C.-H.; Huang, M.-H. Assessment of Adjuvantation Strategy of Lipid Squalene Nanoparticles for Enhancing the Immunogenicity of a SARS-CoV-2 Spike Subunit Protein against COVID-19. Int. J. Pharm. 2021, 607, 121024. [Google Scholar] [CrossRef] [PubMed]
- Sia, Z.R.; Miller, M.S.; Lovell, J.F. Engineered Nanoparticle Applications for Recombinant Influenza Vaccines. Mol. Pharm. 2021, 18, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, Z.; Li, H.; Tian, J.; Chen, M.; Liu, T. Preparation and Immune Effect of HEV ORF2 P206@PLGA Nanoparticles. Nanomaterials 2022, 12, 595. [Google Scholar] [CrossRef] [PubMed]
- Danhier, F.; Ansorena, E.; Silva, J.M.; Coco, R.; Le Breton, A.; Préat, V. PLGA-Based Nanoparticles: An Overview of Biomedical Applications. J. Control. Release 2012, 161, 505–522. [Google Scholar] [CrossRef] [PubMed]
- Chaikhumwang, P.; Madapong, A.; Saeng-Chuto, K.; Nilubol, D.; Tantituvanont, A. Intranasal Delivery of Inactivated PRRSV Loaded Cationic Nanoparticles Coupled with Enterotoxin Subunit B Induces PRRSV-Specific Immune Responses in Pigs. Sci. Rep. 2022, 12, 3725. [Google Scholar] [CrossRef]
- Piperno, A.; Sciortino, M.T.; Giusto, E.; Montesi, M.; Panseri, S.; Scala, A. Recent Advances and Challenges in Gene Delivery Mediated by Polyester-Based Nanoparticles. Int. J. Nanomed. 2021, 16, 5981–6002. [Google Scholar] [CrossRef]
- Roopngam, P.; Liu, K.; Mei, L.; Zheng, Y.; Zhu, X.; Tsai, H.-I.; Huang, L. Hepatitis C Virus E2 Protein Encapsulation into Poly d, l-Lactic-Co-Glycolide Microspheres Could Induce Mice Cytotoxic T-Cell Response. Int. J. Nanomed. 2016, 11, 5361–5370. [Google Scholar] [CrossRef][Green Version]
- Huang, S.; Li, I.-H.; Hong, P.; Yeh, M. Evaluation of Protective Efficacy Using a Nonstructural Protein NS1 in DNA Vaccine-Loaded Microspheres against Dengue 2 Virus. Int. J. Nanomed. 2013, 8, 3161–3169. [Google Scholar] [CrossRef][Green Version]
- Bolhassani, A.; Javanzad, S.; Saleh, T.; Hashemi, M.; Aghasadeghi, M.R.; Sadat, S.M. Polymeric Nanoparticles. Hum. Vaccin. Immunother. 2014, 10, 321–332. [Google Scholar] [CrossRef]
- Nguyen, Q.T.; Kwak, C.; Lee, W.S.; Kim, J.; Jeong, J.; Sung, M.H.; Yang, J.; Poo, H. Poly-γ-Glutamic Acid Complexed With Alum Induces Cross-Protective Immunity of Pandemic H1N1 Vaccine. Front. Immunol. 2019, 10, 1604. [Google Scholar] [CrossRef]
- Akagi, T.; Watanabe, K.; Kim, H.; Akashi, M. Stabilization of Polyion Complex Nanoparticles Composed of Poly(Amino Acid) Using Hydrophobic Interactions. Langmuir 2010, 26, 2406–2413. [Google Scholar] [CrossRef] [PubMed]
- Peleteiro, M.; Presas, E.; González-Aramundiz, J.V.; Sánchez-Correa, B.; Simón-Vázquez, R.; Csaba, N.; Alonso, M.J.; González-Fernández, Á. Polymeric Nanocapsules for Vaccine Delivery: Influence of the Polymeric Shell on the Interaction With the Immune System. Front. Immunol. 2018, 9, 791. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, N.; Wang, Z.; Gao, J.; Zhang, H.; Li, M.; Du, Y.; Gao, X.; Zheng, A. Development and Optimization of Chitosan Nanoparticle-Based Intranasal Vaccine Carrier. Molecules 2021, 27, 204. [Google Scholar] [CrossRef] [PubMed]
- Niculescu, A.-G.; Grumezescu, A.M. Applications of Chitosan-Alginate-Based Nanoparticles—An Up-to-Date Review. Nanomaterials 2022, 12, 186. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Campbell, K.; Painter, G.F.; Young, S.L.; Walker, G.F. Nanoparticle System Based on Amino-Dextran as a Drug Delivery Vehicle: Immune-Stimulatory CpG-Oligonucleotide Loading and Delivery. Pharmaceutics 2020, 12, 1150. [Google Scholar] [CrossRef]
- Liu, Q.; Zheng, X.; Zhang, C.; Shao, X.; Zhang, X.; Zhang, Q.; Jiang, X. Conjugating Influenza a (H1N1) Antigen to n-Trimethylaminoethylmethacrylate Chitosan Nanoparticles Improves the Immunogenicity of the Antigen after Nasal Administration. J. Med. Virol. 2015, 87, 1807–1815. [Google Scholar] [CrossRef]
- Singh, M.; Chakrapani, A.; O’Hagan, D. Nanoparticles and Microparticles as Vaccine-Delivery Systems. Expert. Rev. Vaccines 2007, 6, 797–808. [Google Scholar] [CrossRef]
- Chowdhury, S.; Toth, I.; Stephenson, R.J. Dendrimers in Vaccine Delivery: Recent Progress and Advances. Biomaterials 2022, 280, 121303. [Google Scholar] [CrossRef]
- Chahal, J.S.; Khan, O.F.; Cooper, C.L.; McPartlan, J.S.; Tsosie, J.K.; Tilley, L.D.; Sidik, S.M.; Lourido, S.; Langer, R.; Bavari, S.; et al. Dendrimer-RNA Nanoparticles Generate Protective Immunity against Lethal Ebola, H1N1 Influenza, and Toxoplasma Gondii Challenges with a Single Dose. Proc. Natl. Acad. Sci. USA 2016, 113, E4133–E4142. [Google Scholar] [CrossRef]
- Cañas-Arranz, R.; de León, P.; Defaus, S.; Torres, E.; Forner, M.; Bustos, M.J.; Andreu, D.; Blanco, E.; Sobrino, F. Immunogenicity of Foot-and-Mouth Disease Virus Dendrimer Peptides: Need for a T-Cell Epitope and Ability to Elicit Heterotypic Responses. Molecules 2021, 26, 4714. [Google Scholar] [CrossRef]
- Bahadoran, A.; Moeini, H.; Bejo, M.H.; Hussein, M.Z.; Omar, A.R. Development of Tat-Conjugated Dendrimer for Transdermal DNA Vaccine Delivery. J. Pharm. Pharm. Sci. 2016, 19, 325–338. [Google Scholar] [CrossRef]
- Pati, R.; Shevtsov, M.; Sonawane, A. Nanoparticle Vaccines Against Infectious Diseases. Front. Immunol. 2018, 9, 2224. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Hurst, B.L.; Shakya, A.K.; Uddin, M.J.; Ingrole, R.S.J.; Hernandez-Sanabria, M.; Arya, R.P.; Bimler, L.; Paust, S.; Tarbet, E.B.; et al. Consensus M2e Peptide Conjugated to Gold Nanoparticles Confers Protection against H1N1, H3N2 and H5N1 Influenza A Viruses. Antivir. Res. 2017, 141, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Mateu Ferrando, R.; Lay, L.; Polito, L. Gold Nanoparticle-Based Platforms for Vaccine Development. Drug Discov. Today Technol. 2020, 38, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Joyce, M.G.; Chen, W.-H.; Sankhala, R.S.; Hajduczki, A.; Thomas, P.V.; Choe, M.; Martinez, E.J.; Chang, W.C.; Peterson, C.E.; Morrison, E.B.; et al. SARS-CoV-2 Ferritin Nanoparticle Vaccines Elicit Broad SARS Coronavirus Immunogenicity. Cell Rep. 2021, 37, 110143. [Google Scholar] [CrossRef]
- Hou, F.; Teng, Z.; Ru, J.; Liu, H.; Li, J.; Zhang, Y.; Sun, S.; Guo, H. Flower-like Mesoporous Silica Nanoparticles as an Antigen Delivery Platform to Promote Systemic Immune Response. Nanomedicine 2022, 42, 102541. [Google Scholar] [CrossRef]
- Wang, T.; Zou, M.; Jiang, H.; Ji, Z.; Gao, P.; Cheng, G. Synthesis of a Novel Kind of Carbon Nanoparticle with Large Mesopores and Macropores and Its Application as an Oral Vaccine Adjuvant. Eur. J. Pharm. Sci. 2011, 44, 653–659. [Google Scholar] [CrossRef]
- Flenniken, M.L.; Willits, D.A.; Harmsen, A.L.; Liepold, L.O.; Harmsen, A.G.; Young, M.J.; Douglas, T. Melanoma and Lymphocyte Cell-Specific Targeting Incorporated into a Heat Shock Protein Cage Architecture. Chem. Biol. 2006, 13, 161–170. [Google Scholar] [CrossRef]
- Sasaki, E.; Hilvert, D. Self-Assembly of Proteinaceous Multishell Structures Mediated by a Supercharged Protein. J. Phys. Chem. B 2016, 120, 6089–6095. [Google Scholar] [CrossRef]
- Lee, L.A.; Wang, Q. Adaptations of Nanoscale Viruses and Other Protein Cages for Medical Applications. Nanomed. Nanotechnol. Biol. Med. 2006, 2, 137–149. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Wei, C.-J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.R.; Rao, S.S.; Kong, W.-P.; Wang, L.; Nabel, G.J. Self-Assembling Influenza Nanoparticle Vaccines Elicit Broadly Neutralizing H1N1 Antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Zeltins, A. Construction and Characterization of Virus-Like Particles: A Review. Mol. Biotechnol. 2013, 53, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Chiu, W. Structures of Virus and Virus-like Particles. Curr. Opin. Struct. Biol. 2000, 10, 229–235. [Google Scholar] [CrossRef]
- Grgacic, E.V.L.; Anderson, D.A. Virus-like Particles: Passport to Immune Recognition. Methods 2006, 40, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Pushko, P.; Pumpens, P.; Grens, E. Development of Virus-like Particle Technology from Small Highly Symmetric to Large Complex Virus-like Particle Structures. Intervirology 2013, 56, 141–165. [Google Scholar] [CrossRef]
- Garcea, R.L.; Gissmann, L. Virus-like Particles as Vaccines and Vessels for the Delivery of Small Molecules. Curr. Opin. Biotechnol. 2004, 15, 513–517. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Dai, S.; Wang, H.; Deng, F. Advances and Challenges in Enveloped Virus-like Particle (VLP)-Based Vaccines. J. Immunol. Sci. 2018, 2, 36–41. [Google Scholar]
- Fort, M.; Sibila, M.; Allepuz, A.; Mateu, E.; Roerink, F.; Segalés, J. Porcine Circovirus Type 2 (PCV2) Vaccination of Conventional Pigs Prevents Viremia against PCV2 Isolates of Different Genotypes and Geographic Origins. Vaccine 2008, 26, 1063–1071. [Google Scholar] [CrossRef]
- Rutkowska, D.A.; Mokoena, N.B.; Tsekoa, T.L.; Dibakwane, V.S.; O’Kennedy, M.M. Plant-Produced Chimeric Virus-like Particles—A New Generation Vaccine against African Horse Sickness. BMC Vet. Res. 2019, 15, 432. [Google Scholar] [CrossRef]
- Kang, Y.-M.; Cho, H.-K.; Kim, J.H.; Lee, S.J.; Park, S.-J.; Kim, D.-Y.; Kim, S.Y.; Park, J.-W.; Lee, M.-H.; Kim, M.-C.; et al. Single Dose of Multi-Clade Virus-like Particle Vaccine Protects Chickens against Clade 2.3.2.1 and Clade 2.3.4.4 Highly Pathogenic Avian Influenza Viruses. Sci. Rep. 2021, 11, 13786. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Dubois, E.; Sailleau, C.; Bréard, E.; Viarouge, C.; Desprat, A.; Thiéry, R.; Zientara, S.; Roy, P. Bluetongue Virus Serotype 8 Virus-like Particles Protect Sheep against Virulent Virus Infection as a Single or Multi-Serotype Cocktail Immunogen. Vaccine 2013, 31, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.-C.; Sun, S.-Q.; Jin, Y.; Yang, S.-L.; Wei, Y.-Q.; Sun, D.-H.; Yin, S.-H.; Ma, J.-W.; Liu, Z.-X.; Guo, J.-H.; et al. Foot-and-Mouth Disease Virus-like Particles Produced by a SUMO Fusion Protein System in Escherichia Coli Induce Potent Protective Immune Responses in Guinea Pigs, Swine and Cattle. Vet. Res. 2013, 44, 48. [Google Scholar] [CrossRef]
- Park, J.-K.; Lee, D.-H.; Yuk, S.-S.; Tseren-Ochir, E.-O.; Kwon, J.-H.; Noh, J.-Y.; Kim, B.-Y.; Choi, S.-W.; Kang, S.-M.; Lee, J.-B.; et al. Virus-like Particle Vaccine Confers Protection against a Lethal Newcastle Disease Virus Challenge in Chickens and Allows a Strategy of Differentiating Infected from Vaccinated Animals. Clin. Vaccine Immunol. 2014, 21, 360–365. [Google Scholar] [CrossRef]
- Hua, T.; Zhang, D.; Tang, B.; Chang, C.; Liu, G.; Zhang, X. The Immunogenicity of the Virus-like Particles Derived from the VP2 Protein of Porcine Parvovirus. Vet. Microbiol. 2020, 248, 108795. [Google Scholar] [CrossRef] [PubMed]
- Näslund, J.; Lagerqvist, N.; Habjan, M.; Lundkvist, A.; Evander, M.; Ahlm, C.; Weber, F.; Bucht, G. Vaccination with Virus-like Particles Protects Mice from Lethal Infection of Rift Valley Fever Virus. Virology 2009, 385, 409–415. [Google Scholar] [CrossRef]
- Liu, F.; Ge, S.; Li, L.; Wu, X.; Liu, Z.; Wang, Z. Virus-like Particles: Potential Veterinary Vaccine Immunogens. Res. Vet. Sci. 2012, 93, 553–559. [Google Scholar] [CrossRef]
- Crisci, E.; Bárcena, J.; Montoya, M. Virus-like Particle-Based Vaccines for Animal Viral Infections. Inmunologia 2013, 32, 102–116. [Google Scholar] [CrossRef]
- Chung, Y.H.; Church, D.; Koellhoffer, E.C.; Osota, E.; Shukla, S.; Rybicki, E.P.; Pokorski, J.K.; Steinmetz, N.F. Integrating Plant Molecular Farming and Materials ReseArch. for Next-Generation Vaccines. Nat. Rev. Mater 2022, 7, 372–388. [Google Scholar] [CrossRef]
- Chan, J.C.; Chan, A.T. Biologics and Biosimilars: What, Why and How? ESMO Open 2017, 2, e000180. [Google Scholar] [CrossRef]
- Goeddel, D.V.; Kleid, D.G.; Bolivar, F.; Heyneker, H.L.; Yansura, D.G.; Crea, R.; Hirose, T.; Kraszewski, A.; Itakura, K.; Riggs, A.D. Expression in Escherichia Coli of Chemically Synthesized Genes for Human Insulin. Proc. Natl. Acad. Sci. USA 1979, 76, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Overton, T.W. Recombinant Protein Production in Bacterial Hosts. Drug Discov. Today 2014, 19, 590–601. [Google Scholar] [CrossRef]
- Makrides, S.C. Strategies for Achieving High-Level Expression of Genes in Escherichia Coli. Microbiol. Rev. 1996, 60, 512–538. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, H.-J. Yeast as an Expression System for Producing Virus-like Particles: What Factors Do We Need to Consider? Lett. Appl. Microbiol. 2017, 64, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Jefferis, R. Posttranslational Modifications and the Immunogenicity of Biotherapeutics. J. Immunol. Res. 2016, 2016, 5358272. [Google Scholar] [CrossRef] [PubMed]
- Rybicki, E.P. Plant Molecular Farming of Virus-like Nanoparticles as Vaccines and Reagents. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1587. [Google Scholar] [CrossRef]
- Zahmanova, G.; Takova, K.; Valkova, R.; Toneva, V.; Minkov, I.; Andonov, A.; Lukov, G.L. Plant-Derived Recombinant Vaccines against Zoonotic Viruses. Life 2022, 12, 156. [Google Scholar] [CrossRef]
- Scotti, N.; Rybicki, E.P. Virus-like Particles Produced in Plants as Potential Vaccines. Expert Rev. Vaccines 2013, 12, 211–224. [Google Scholar] [CrossRef]
- Walwyn, D.R.; Huddy, S.M.; Rybicki, E.P. Techno-Economic Analysis of Horseradish Peroxidase Production Using a Transient Expression System in Nicotiana benthamiana. Appl. Biochem. Biotechnol. 2015, 175, 841–854. [Google Scholar] [CrossRef]
- Ward, B.J.; Makarkov, A.; Séguin, A.; Pillet, S.; Trépanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, Immunogenicity, and Safety of a Plant-Derived, Quadrivalent, Virus-like Particle Influenza Vaccine in Adults (18–64 Years) and Older Adults (≥65 Years): Two Multicentre, Randomised Phase 3 Trials. Lancet 2020, 396, 1491–1503. [Google Scholar] [CrossRef]
- Kyriakidis, N.C.; López-Cortés, A.; González, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 Vaccines Strategies: A Comprehensive Review of Phase 3 Candidates. npj Vaccines 2021, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, N.; Robinson, M.K.; Bernard, C.; Kawaguchi, Y.; Koujin, Y.; Koen, A.; Madhi, S.; Polasek, T.M.; McNeal, M.; Dargis, M.; et al. Safety and Immunogenicity of a Plant-Derived Rotavirus-like Particle Vaccine in Adults, Toddlers and Infants. Vaccine 2021, 39, 5513–5523. [Google Scholar] [CrossRef] [PubMed]
- Jordan, L.E.; Mayor, H.D. The Fine Structure of Reovirus, a New Member of the Icosahedral Series. Virology 1962, 17, 597–599. [Google Scholar] [CrossRef]
- Silverstein, S.C.; Schur, P.H. Immunofluorescent Localization of Double-Stranded RNA in Reovirus-Infected Cells. Virology 1970, 41, 564–566. [Google Scholar] [CrossRef]
- Touris-Otero, F.; Martínez-Costas, J.; Vakharia, V.N.; Benavente, J. Avian Reovirus Nonstructural Protein MicroNS Forms Viroplasm-like Inclusions and Recruits Protein SigmaNS to These Structures. Virology 2004, 319, 94–106. [Google Scholar] [CrossRef]
- Tourís-Otero, F.; Cortez-San Martín, M.; Martínez-Costas, J.; Benavente, J. Avian Reovirus Morphogenesis Occurs within Viral Factories and Begins with the Selective Recruitment of SigmaNS and LambdaA to MicroNS Inclusions. J. Mol. Biol. 2004, 341, 361–374. [Google Scholar] [CrossRef]
- Brandariz-Nuñez, A.; Menaya-Vargas, R.; Benavente, J.; Martinez-Costas, J. Avian Reovirus MicroNS Protein Forms Homo-Oligomeric Inclusions in a Microtubule-Independent Fashion, Which Involves Specific Regions of Its C-Terminal Domain. J. Virol. 2010, 84, 4289–4301. [Google Scholar] [CrossRef]
- Brandariz-Nuñez, A.; Menaya-Vargas, R.; Benavente, J.; Martinez-Costas, J. A Versatile Molecular Tagging Method for Targeting Proteins to Avian Reovirus MuNS Inclusions. Use in Protein Immobilization and Purification. PLoS ONE 2010, 5, e13961. [Google Scholar] [CrossRef]
- Brandariz-Nuñez, A.; Menaya-Vargas, R.; Benavente, J.; Martinez-Costas, J. IC-Tagging and Protein Relocation to ARV MuNS Inclusions: A Method to Study Protein-Protein Interactions in the Cytoplasm or Nucleus of Living Cells. PLoS ONE 2010, 5, e13785. [Google Scholar] [CrossRef]
- Barreiro-Piñeiro, N.; Lostalé-Seijo, I.; Varela-Calviño, R.; Benavente, J.; Martínez-Costas, J.M. IC-Tagging Methodology Applied to the Expression of Viral Glycoproteins and the Difficult-to-Express Membrane-Bound IGRP Autoantigen. Sci. Rep. 2018, 8, 16286. [Google Scholar] [CrossRef]
- Pose-Boirazian, T.; Eibes, G.; Barreiro-Piñeiro, N.; Díaz-Jullien, C.; Lema, J.M.; Martínez-Costas, J. Chemical and Thermal Stabilization of CotA Laccase via a Novel One-Step Expression and Immobilization in MuNS-Mi Nanospheres. Sci. Rep. 2021, 11, 2802. [Google Scholar] [CrossRef] [PubMed]
- Bastús, N.G.; Sánchez-Tilló, E.; Pujals, S.; Farrera, C.; Kogan, M.J.; Giralt, E.; Celada, A.; Lloberas, J.; Puntes, V. Peptides Conjugated to Gold Nanoparticles Induce Macrophage Activation. Mol. Immunol. 2009, 46, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction Between Virus-Like Particles (VLPs) and Pattern Recognition Receptors (PRRs) From Dendritic Cells (DCs): Toward Better Engineering of VLPs. Front Immunol. 2020, 11, 1100. [Google Scholar] [CrossRef]
- Fromen, C.A.; Rahhal, T.B.; Robbins, G.R.; Kai, M.P.; Shen, T.W.; Luft, J.C.; DeSimone, J.M. Nanoparticle Surface Charge Impacts Distribution, Uptake and Lymph Node Trafficking by Pulmonary Antigen-Presenting Cells. Nanomedicine 2016, 12, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Benne, N.; van Duijn, J.; Kuiper, J.; Jiskoot, W.; Slütter, B. Orchestrating Immune Responses: How Size, Shape and Rigidity Affect the Immunogenicity of Particulate Vaccines. J. Control. Release 2016, 234, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Zinkhan, S.; Ogrina, A.; Balke, I.; Reseviča, G.; Zeltins, A.; de Brot, S.; Lipp, C.; Chang, X.; Zha, L.; Vogel, M.; et al. The Impact of Size on Particle Drainage Dynamics and Antibody Response. J. Control. Release 2021, 331, 296–308. [Google Scholar] [CrossRef]
- Reddy, S.T.; Rehor, A.; Schmoekel, H.G.; Hubbell, J.A.; Swartz, M.A. In Vivo Targeting of Dendritic Cells in Lymph Nodes with Poly(Propylene Sulfide) Nanoparticles. J. Control. Release 2006, 112, 26–34. [Google Scholar] [CrossRef]
- Smith, D.M.; Simon, J.K.; Baker Jr, J.R. Applications of Nanotechnology for Immunology. Nat. Rev. Immunol. 2013, 13, 592–605. [Google Scholar] [CrossRef]
- Cruz, L.J.; Tacken, P.J.; Fokkink, R.; Joosten, B.; Stuart, M.C.; Albericio, F.; Torensma, R.; Figdor, C.G. Targeted PLGA Nano- but Not Microparticles Specifically Deliver Antigen to Human Dendritic Cells via DC-SIGN in Vitro. J. Control. Release 2010, 144, 118–126. [Google Scholar] [CrossRef]
- Blank, F.; Stumbles, P.A.; Seydoux, E.; Holt, P.G.; Fink, A.; Rothen-Rutishauser, B.; Strickland, D.H.; von Garnier, C. Size-Dependent Uptake of Particles by Pulmonary Antigen-Presenting Cell Populations and Trafficking to Regional Lymph Nodes. Am. J. Respir. Cell Mol. Biol. 2013, 49, 67–77. [Google Scholar] [CrossRef]
- Dacoba, T.G.; Olivera, A.; Torres, D.; Crecente-Campo, J.; Alonso, M.J. Modulating the Immune System through Nanotechnology. Semin. Immunol. 2017, 34, 78–102. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, P.; White, D.; Sulchek, T. Effects of Microparticle Size and Fc Density on Macrophage Phagocytosis. PLoS ONE 2013, 8, e60989. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hu, Y.; Yin, L.; Tang, C.; Yin, C. Effects of Particle Size and Surface Charge on Cellular Uptake and Biodistribution of Polymeric Nanoparticles. Biomaterials 2010, 31, 3657–3666. [Google Scholar] [CrossRef] [PubMed]
- Beduneau, A.; Ma, Z.; Grotepas, C.B.; Kabanov, A.; Rabinow, B.E.; Gong, N.; Mosley, R.L.; Dou, H.; Boska, M.D.; Gendelman, H.E. Facilitated Monocyte-Macrophage Uptake and Tissue Distribution of Superparmagnetic Iron-Oxide Nanoparticles. PLoS ONE 2009, 4, e4343. [Google Scholar] [CrossRef]
- Storni, T.; Bachmann, M.F. Loading of MHC Class I and II Presentation Pathways by Exogenous Antigens: A Quantitative in Vivo Comparison. J. Immunol. 2004, 172, 6129–6135. [Google Scholar] [CrossRef]
- Barth, H.; Ulsenheimer, A.; Pape, G.R.; Diepolder, H.M.; Hoffmann, M.; Neumann-Haefelin, C.; Thimme, R.; Henneke, P.; Klein, R.; Paranhos-Baccalà, G.; et al. Uptake and Presentation of Hepatitis C Virus-like Particles by Human Dendritic Cells. Blood 2005, 105, 3605–3614. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, M.; Zhao, H.; Wang, P.; Ma, W.; Zhang, Y.; Wu, W.; Peng, C. Induction of Robust and Specific Humoral and Cellular Immune Responses by Bovine Viral Diarrhea Virus Virus-Like Particles (BVDV-VLPs) Engineered with Baculovirus Expression Vector System. Vaccines 2021, 9, 350. [Google Scholar] [CrossRef]
- Morón, V.G.; Rueda, P.; Sedlik, C.; Leclerc, C. In Vivo, Dendritic Cells Can Cross-Present Virus-like Particles Using an Endosome-to-Cytosol Pathway. J. Immunol. 2003, 171, 2242–2250. [Google Scholar] [CrossRef]
- Joffre, O.P.; Segura, E.; Savina, A.; Amigorena, S. Cross-Presentation by Dendritic Cells. Nat. Rev. Immunol. 2012, 12, 557–569. [Google Scholar] [CrossRef]
- Jeggo, M.H.; Wardley, R.C.; Brownlie, J. A Study of the Role of Cell-Mediated Immunity in Bluetongue Virus Infection in Sheep, Using Cellular Adoptive Transfer Techniques. Immunology 1984, 52, 403–410. [Google Scholar]
- Umeshappa, C.S.; Singh, K.P.; Pandey, A.B.; Singh, R.P.; Nanjundappa, R.H. Cell-Mediated Immune Response and Cross-Protective Efficacy of Binary Ethylenimine-Inactivated Bluetongue Virus Serotype-1 Vaccine in Sheep. Vaccine 2010, 28, 2522–2531. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordón, P.J.; Pérez de Diego, A.C.; Gómez-Villamandos, J.C.; Sánchez-Vizcaíno, J.M.; Pleguezuelos, F.J.; Garfia, B.; del Carmen, P.; Pedrera, M. Comparative Analysis of Cellular Immune Responses and Cytokine Levels in Sheep Experimentally Infected with Bluetongue Virus Serotype 1 and 8. Vet. Microbiol. 2015, 177, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Martín, V.; Pascual, E.; Avia, M.; Peña, L.; Valcárcel, F.; Sevilla, N. Protective Efficacy in Sheep of Adenovirus-Vectored Vaccines against Bluetongue Virus Is Associated with Specific T Cell Responses. PLoS ONE 2015, 10, e0143273. [Google Scholar] [CrossRef] [PubMed]
- Utrilla-Trigo, S.; Jiménez-Cabello, L.; Alonso-Ravelo, R.; Calvo-Pinilla, E.; Marín-López, A.; Moreno, S.; Lorenzo, G.; Benavides, J.; Gilbert, S.; Nogales, A.; et al. Heterologous Combination of ChAdOx1 and MVA Vectors Expressing Protein NS1 as Vaccination Strategy to Induce Durable and Cross-Protective CD8+ T Cell Immunity to Bluetongue Virus. Vaccines 2020, 8, 346. [Google Scholar] [CrossRef] [PubMed]
- Utrilla-Trigo, S.; Jiménez-Cabello, L.; Calvo-Pinilla, E.; Marín-López, A.; Lorenzo, G.; Sánchez-Cordón, P.; Moreno, S.; Benavides, J.; Gilbert, S.; Nogales, A.; et al. The Combined Expression of the Non-Structural Protein NS1 and the N-Terminal Half of NS2 (NS21-180) by ChAdOx1 and MVA Confers Protection against Clinical Disease in Sheep upon Bluetongue Virus Challenge. J. Virol. 2021, 96, e01614–e01621. [Google Scholar] [CrossRef]
- Anderson, J.; Hägglund, S.; Bréard, E.; Comtet, L.; Lövgren Bengtsson, K.; Pringle, J.; Zientara, S.; Valarcher, J.F. Evaluation of the Immunogenicity of an Experimental Subunit Vaccine That Allows Differentiation between Infected and Vaccinated Animals against Bluetongue Virus Serotype 8 in Cattle. Clin. Vaccine Immunol. 2013, 20, 1115–1122. [Google Scholar] [CrossRef]
- Marín-López, A.; Calvo-Pinilla, E.; Barriales, D.; Lorenzo, G.; Brun, A.; Anguita, J.; Ortego, J. CD8 T Cell Responses to an Immunodominant Epitope within the Nonstructural Protein NS1 Provide Wide Immunoprotection against Bluetongue Virus in IFNAR −/− Mice. J. Virol. 2018, 92, e00938-18. [Google Scholar] [CrossRef]
- Marín-López, A.; Barreiro-Piñeiro, N.; Utrilla-Trigo, S.; Barriales, D.; Benavente, J.; Nogales, A.; Martínez-Costas, J.; Ortego, J.; Calvo-Pinilla, E. Cross-Protective Immune Responses against African Horse Sickness Virus after Vaccination with Protein NS1 Delivered by Avian Reovirus MuNS Microspheres and Modified Vaccinia Virus Ankara. Vaccine 2020, 38, 882–889. [Google Scholar] [CrossRef]
- Fearon, S.H.; Dennis, S.J.; Hitzeroth, I.I.; Rybicki, E.P.; Meyers, A.E. Humoral and Cell-Mediated Immune Responses to Plant-Produced African Horse Sickness Virus VP7 Quasi-Crystals. Virus Res. 2021, 294, 198284. [Google Scholar] [CrossRef]
- Rojas, J.M.; Peña, L.; Martín, V.; Sevilla, N. Ovine and Murine T Cell Epitopes from the Non-Structural Protein 1 (NS1) of Bluetongue Virus Serotype 8 (BTV-8) Are Shared among Viral Serotypes. Vet. Res. 2014, 45, 30. [Google Scholar] [CrossRef]
- Calvo-Pinilla, E.; Marín-López, A.; Moreno, S.; Lorenzo, G.; Utrilla-Trigo, S.; Jiménez-Cabello, L.; Benavides, J.; Nogales, A.; Blasco, R.; Brun, A.; et al. A Protective Bivalent Vaccine against Rift Valley Fever and Bluetongue. npj Vaccines 2020, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Storni, T.; Lechner, F.; Erdmann, I.; Bächi, T.; Jegerlehner, A.; Dumrese, T.; Kündig, T.M.; Ruedl, C.; Bachmann, M.F. Critical Role for Activation of Antigen-Presenting Cells in Priming of Cytotoxic T Cell Responses after Vaccination with Virus-like Particles. J. Immunol. 2002, 168, 2880–2886. [Google Scholar] [CrossRef] [PubMed]
- Selby, L.I.; Cortez-Jugo, C.M.; Such, G.K.; Johnston, A.P.R. Nanoescapology: Progress toward Understanding the Endosomal Escape of Polymeric Nanoparticles. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1452. [Google Scholar] [CrossRef] [PubMed]
- Mant, A.; Chinnery, F.; Elliott, T.; Williams, A.P. The Pathway of Cross-Presentation Is Influenced by the Particle Size of Phagocytosed Antigen. Immunology 2012, 136, 163–175. [Google Scholar] [CrossRef]
- Silva, A.L.; Rosalia, R.A.; Varypataki, E.; Sibuea, S.; Ossendorp, F.; Jiskoot, W. Poly-(Lactic-Co-Glycolic-Acid)-Based Particulate Vaccines: Particle Uptake by Dendritic Cells Is a Key Parameter for Immune Activation. Vaccine 2015, 33, 847–854. [Google Scholar] [CrossRef]
- Hirai, T.; Yoshioka, Y.; Takahashi, H.; Ichihashi, K.; Yoshida, T.; Tochigi, S.; Nagano, K.; Abe, Y.; Kamada, H.; Tsunoda, S.; et al. Amorphous Silica Nanoparticles Enhance Cross-Presentation in Murine Dendritic Cells. Biochem. Biophys. Res. Commun 2012, 427, 553–556. [Google Scholar] [CrossRef]
- Rodríguez-Martín, D.; Louloudes-Lázaro, A.; Avia, M.; Martín, V.; Rojas, J.M.; Sevilla, N. The Interplay between Bluetongue Virus Infections and Adaptive Immunity. Viruses 2021, 13, 1511. [Google Scholar] [CrossRef]
- French, T.J.; Marshall, J.J.; Roy, P. Assembly of Double-Shelled, Viruslike Particles of Bluetongue Virus by the Simultaneous Expression of Four Structural Proteins. J. Virol. 1990, 64, 5695–5700. [Google Scholar] [CrossRef]
- Martinez-Torrecuadrada, J.L.; Iwata, H.; Venteo, A.; Casal, I.; Roy, P. Expression and Characterization of the Two Outer Capsid Proteins of African Horsesickness Virus: The Role of VP2 in Virus Neutralization. Virology 1994, 202, 348–359. [Google Scholar] [CrossRef]
- Sailleau, C.; Breard, E.; Viarouge, C.; Belbis, G.; Lilin, T.; Vitour, D.; Zientara, S. Experimental Infection of Calves with Seven Serotypes of Epizootic Hemorrhagic Disease Virus: Production and Characterization of Reference Sera. Vet. Ital. 2019, 55, 339–346. [Google Scholar] [CrossRef]
- Jeggo, M.H.; Wardley, R.C.; Taylor, W.P. Role of Neutralising Antibody in Passive Immunity to Bluetongue Infection. Res. Vet. Sci. 1984, 36, 81–86. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Coffman, R.L. TH1 and TH2 Cells: Different Patterns of Lymphokine Secretion Lead to Different Functional Properties. Annu. Rev. Immunol. 1989, 7, 145–173. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.K.; Murphy, K.M.; Sher, A. Functional Diversity of Helper T Lymphocytes. Nature 1996, 383, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Zhang, R.; Guo, L.; Li, M.; Chen, C. Th Cell-Independent Immune Responses to Chimeric Hemagglutinin/Simian Human Immunodeficiency Virus-like Particles Vaccine. J. Immunol. 2004, 173, 1951–1958. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, K.; Wahid, R.; Richardson, C.; Bargatze, R.F.; El-Kamary, S.S.; Sztein, M.B.; Pasetti, M.F. Intranasal Vaccination with an Adjuvanted Norwalk Virus-like Particle Vaccine Elicits Antigen-Specific B Memory Responses in Human Adult Volunteers. Clin. Immunol. 2012, 144, 98–108. [Google Scholar] [CrossRef]
- Schmidt, M.R.; McGinnes, L.W.; Kenward, S.A.; Willems, K.N.; Woodland, R.T.; Morrison, T.G. Long-Term and Memory Immune Responses in Mice against Newcastle Disease Virus-like Particles Containing Respiratory Syncytial Virus Glycoprotein Ectodomains. J. Virol. 2012, 86, 11654–11662. [Google Scholar] [CrossRef]
- Giannini, S.L.; Hanon, E.; Moris, P.; Van Mechelen, M.; Morel, S.; Dessy, F.; Fourneau, M.A.; Colau, B.; Suzich, J.; Losonksy, G.; et al. Enhanced Humoral and Memory B Cellular Immunity Using HPV16/18 L1 VLP Vaccine Formulated with the MPL/Aluminium Salt Combination (AS04) Compared to Aluminium Salt Only. Vaccine 2006, 24, 5937–5949. [Google Scholar] [CrossRef]
- Zabel, F.; Mohanan, D.; Bessa, J.; Link, A.; Fettelschoss, A.; Saudan, P.; Kündig, T.M.; Bachmann, M.F. Viral Particles Drive Rapid Differentiation of Memory B Cells into Secondary Plasma Cells Producing Increased Levels of Antibodies. J. Immunol. 2014, 192, 5499–5508. [Google Scholar] [CrossRef]
- Jiao, Y.-Y.; Fu, Y.-H.; Yan, Y.-F.; Hua, Y.; Ma, Y.; Zhang, X.-J.; Song, J.-D.; Peng, X.-L.; Huang, J.; Hong, T.; et al. A Single Intranasal Administration of Virus-like Particle Vaccine Induces an Efficient Protection for Mice against Human Respiratory Syncytial Virus. Antivir. Res. 2017, 144, 57–69. [Google Scholar] [CrossRef]
- Landry, N.; Pillet, S.; Favre, D.; Poulin, J.-F.; Trépanier, S.; Yassine-Diab, B.; Ward, B.J. Influenza Virus-like Particle Vaccines Made in Nicotiana benthamiana Elicit Durable, Poly-Functional and Cross-Reactive T Cell Responses to Influenza HA Antigens. Clin. Immunol. 2014, 154, 164–177. [Google Scholar] [CrossRef]
- Karuturi, B.V.K.; Tallapaka, S.B.; Yeapuri, P.; Curran, S.M.; Sanderson, S.D.; Vetro, J.A. Encapsulation of an EP67-Conjugated CTL Peptide Vaccine in Nanoscale Biodegradable Particles Increases the Efficacy of Respiratory Immunization and Affects the Magnitude and Memory Subsets of Vaccine-Generated Mucosal and Systemic CD8+ T Cells in a Diameter-Dependent Manner. Mol. Pharm. 2017, 14, 1469–1481. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, P.; Bhatia, E.; Sharma, S.; Ahamad, N.; Banerjee, R. Advancements in Prophylactic and Therapeutic Nanovaccines. Acta Biomater. 2020, 108, 1–21. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.; Lefkowitz, E.; Adams, M.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 978-0-12-384685-3. [Google Scholar]
- Du Toit, R.M. The Transmission of Blue-Tongue and Horse-Sickness by Culicoides. J. Vet. Sci. Anim. Ind. 1944, 19, 7–16. [Google Scholar]
- Maclachlan, N.J.; Drew, C.P.; Darpel, K.E.; Worwa, G. The Pathology and Pathogenesis of Bluetongue. J. Comp. Pathol. 2009, 141, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Drolet, B.S.; Reister, L.M.; Rigg, T.D.; Nol, P.; Podell, B.K.; Mecham, J.O.; VerCauteren, K.C.; van Rijn, P.A.; Wilson, W.C.; Bowen, R.A. Experimental Infection of White-Tailed Deer (Odocoileus virginianus) with Northern European Bluetongue Virus Serotype 8. Vet. Microbiol. 2013, 166, 347–355. [Google Scholar] [CrossRef]
- Erasmus, B.J. Bluetongue in Sheep and Goats. Aust. Vet. J. 1975, 51, 165–170. [Google Scholar] [CrossRef]
- Rushton, J.; Lyons, N. Economic Impact of Bluetongue: A Review of the Effects on Production. Vet. Ital. 2015, 51, 401–406. [Google Scholar] [CrossRef]
- Gethmann, J.; Probst, C.; Conraths, F.J. Economic Impact of a Bluetongue Serotype 8 Epidemic in Germany. Front. Vet. Sci. 2020, 7, 65. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Samuel, A.R.; Rao, S.; Attoui, H.; Mertens, P.P.C. Analysis and Phylogenetic Comparisons of Full-Length VP2 Genes of the 24 Bluetongue Virus Serotypes. J. Gen. Virol. 2007, 88, 621–630. [Google Scholar] [CrossRef]
- Saminathan, M.; Singh, K.P.; Khorajiya, J.H.; Dinesh, M.; Vineetha, S.; Maity, M.; Rahman, A.F.; Misri, J.; Malik, Y.S.; Gupta, V.K.; et al. An Updated Review on Bluetongue Virus: Epidemiology, Pathobiology, and Advances in Diagnosis and Control with Special Reference to India. Vet. Q. 2020, 40, 258–321. [Google Scholar] [CrossRef]
- French, T.J.; Roy, P. Synthesis of Bluetongue Virus (BTV) Corelike Particles by a Recombinant Baculovirus Expressing the Two Major Structural Core Proteins of BTV. J. Virol. 1990, 64, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Loudon, P.T.; Hirasawa, T.; Oldfield, S.; Murphy, M.; Roy, P. Expression of the Outer Capsid Protein VP5 of Two Bluetongue Viruses, and Synthesis of Chimeric Double-Shelled Virus-like Particles Using Combinations of Recombinant Baculoviruses. Virology 1991, 182, 793–801. [Google Scholar] [CrossRef]
- Thuenemann, E.C.; Meyers, A.E.; Verwey, J.; Rybicki, E.P.; Lomonossoff, G.P. A Method for Rapid Production of Heteromultimeric Protein Complexes in Plants: Assembly of Protective Bluetongue Virus-like Particles. Plant Biotechnol. J. 2013, 11, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Mokoena, N.B.; Moetlhoa, B.; Rutkowska, D.A.; Mamputha, S.; Dibakwane, V.S.; Tsekoa, T.L.; O’Kennedy, M.M. Plant-Produced Bluetongue Chimaeric VLP Vaccine Candidates Elicit Serotype-Specific Immunity in Sheep. Vaccine 2019, 37, 6068–6075. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Dovas, C.I.; Chatzinasiou, E.; Athmaram, T.N.; Papanastassopoulou, M.; Papadopoulos, O.; Roy, P. Protective Efficacy of Bluetongue Virus-like and Subvirus-like Particles in Sheep: Presence of the Serotype-Specific VP2, Independent of Its Geographic Lineage, Is Essential for Protection. Vaccine 2012, 30, 2131–2139. [Google Scholar] [CrossRef]
- Wade-Evans, A.M.; Romero, C.H.; Mellor, P.; Takamatsu, H.; Anderson, J.; Thevasagayam, J.; Fleming, M.J.; Mertens, P.P.; Black, D.N. Expression of the Major Core Structural Protein (VP7) of Bluetongue Virus, by a Recombinant Capripox Virus, Provides Partial Protection of Sheep against a Virulent Heterotypic Bluetongue Virus Challenge. Virology 1996, 220, 227–231. [Google Scholar] [CrossRef]
- Calvo-Pinilla, E.; Rodríguez-Calvo, T.; Sevilla, N.; Ortego, J. Heterologous Prime Boost Vaccination with DNA and Recombinant Modified Vaccinia Virus Ankara Protects IFNAR(−/−) Mice against Lethal Bluetongue Infection. Vaccine 2009, 28, 437–445. [Google Scholar] [CrossRef]
- Roy, P.; Bishop, D.H.; LeBlois, H.; Erasmus, B.J. Long-Lasting Protection of Sheep against Bluetongue Challenge after Vaccination with Virus-like Particles: Evidence for Homologous and Partial Heterologous Protection. Vaccine 1994, 12, 805–811. [Google Scholar] [CrossRef]
- Stewart, M.; Bhatia, Y.; Athmaran, T.N.; Noad, R.; Gastaldi, C.; Dubois, E.; Russo, P.; Thiéry, R.; Sailleau, C.; Bréard, E.; et al. Validation of a Novel Approach for the Rapid Production of Immunogenic Virus-like Particles for Bluetongue Virus. Vaccine 2010, 28, 3047–3054. [Google Scholar] [CrossRef]
- Pérez de Diego, A.C.; Athmaram, T.N.; Stewart, M.; Rodríguez-Sánchez, B.; Sánchez-Vizcaíno, J.M.; Noad, R.; Roy, P. Characterization of Protection Afforded by a Bivalent Virus-Like Particle Vaccine against Bluetongue Virus Serotypes 1 and 4 in Sheep. PLoS ONE 2011, 6, e26666. [Google Scholar] [CrossRef]
- Roy, P.; French, T.; Erasmus, B.J. Protective Efficacy of Virus-like Particles for Bluetongue Disease. Vaccine 1992, 10, 28–32. [Google Scholar] [CrossRef]
- Guy, B.; Barban, V.; Mantel, N.; Aguirre, M.; Gulia, S.; Pontvianne, J.; Jourdier, T.-M.; Ramirez, L.; Gregoire, V.; Charnay, C.; et al. Evaluation of Interferences between Dengue Vaccine Serotypes in a Monkey Model. Am. J. Trop. Med. Hyg. 2009, 80, 302–311. [Google Scholar] [CrossRef] [PubMed]
- McClain, D.J.; Pittman, P.R.; Ramsburg, H.H.; Nelson, G.O.; Rossi, C.A.; Mangiafico, J.A.; Schmaljohn, A.L.; Malinoski, F.J. Immunologic Interference from Sequential Administration of Live Attenuated Alphavirus Vaccines. J. Infect. Dis. 1998, 177, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Parker, E.P.K.; Kampmann, B.; Kang, G.; Grassly, N.C. Influence of Enteric Infections on Response to Oral Poliovirus Vaccine: A Systematic Review and Meta-Analysis. J. Infect. Dis. 2014, 210, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Roy, P. Sialic Acid Binding Sites in VP2 of Bluetongue Virus and Their Use during Virus Entry. J. Virol. 2022, 96, e0167721. [Google Scholar] [CrossRef]
- Dishlers, A.; Skrastina, D.; Renhofa, R.; Petrovskis, I.; Ose, V.; Lieknina, I.; Jansons, J.; Pumpens, P.; Sominskaya, I. The Hepatitis B Virus Core Variants That Expose Foreign C-Terminal Insertions on the Outer Surface of Virus-Like Particles. Mol. Biotechnol. 2015, 57, 1038–1049. [Google Scholar] [CrossRef]
- Gedvilaite, A.; Frömmel, C.; Sasnauskas, K.; Micheel, B.; Ozel, M.; Behrsing, O.; Staniulis, J.; Jandrig, B.; Scherneck, S.; Ulrich, R. Formation of Immunogenic Virus-like Particles by Inserting Epitopes into Surface-Exposed Regions of Hamster Polyomavirus Major Capsid Protein. Virology 2000, 273, 21–35. [Google Scholar] [CrossRef]
- Aston-Deaville, S.; Carlsson, E.; Saleem, M.; Thistlethwaite, A.; Chan, H.; Maharjan, S.; Facchetti, A.; Feavers, I.M.; Alistair Siebert, C.; Collins, R.F.; et al. An Assessment of the Use of Hepatitis B Virus Core Protein Virus-like Particles to Display Heterologous Antigens from Neisseria Meningitidis. Vaccine 2020, 38, 3201–3209. [Google Scholar] [CrossRef]
- Li, G.; Liu, L.; Xu, B.; Hu, J.; Kuang, H.; Wang, X.; Wang, L.; Cui, X.; Sun, H.; Rong, J. Displaying Epitope B and Epitope 7 of Porcine Reproductive and Respiratory Syndrome Virus on Virus like Particles of Porcine Circovirus Type 2 Provides Partial Protection to Pigs. J. Vet. Med. Sci. 2021, 83, 1263–1272. [Google Scholar] [CrossRef]
- Pascual, E.; Mata, C.P.; Gómez-Blanco, J.; Moreno, N.; Bárcena, J.; Blanco, E.; Rodríguez-Frandsen, A.; Nieto, A.; Carrascosa, J.L.; Castón, J.R. Structural Basis for the Development of Avian Virus Capsids That Display Influenza Virus Proteins and Induce Protective Immunity. J. Virol. 2015, 89, 2563–2574. [Google Scholar] [CrossRef]
- Roose, K.; De Baets, S.; Schepens, B.; Saelens, X. Hepatitis B Core-Based Virus-like Particles to Present Heterologous Epitopes. Expert Rev. Vaccines 2013, 12, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Nagesha, H.S.; Wang, L.F.; Hyatt, A.D. Virus-like Particles of Calicivirus as Epitope Carriers. Arch. Virol. 1999, 144, 2429–2439. [Google Scholar] [CrossRef] [PubMed]
- Belyaev, A.S.; Roy, P. Presentation of Hepatitis B Virus PreS2 Epitope on Bluetongue Virus Core-like Particles. Virology 1992, 190, 840–844. [Google Scholar] [CrossRef]
- Le Blois, H.; Roy, P. A Single Point Mutation in the VP7 Major Core Protein of Bluetongue Virus Prevents the Formation of Core-like Particles. J. Virol. 1993, 67, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Adler, S.; Reay, P.; Roy, P.; Klenk, H.D. Induction of T Cell Response by Bluetongue Virus Core-like Particles Expressing a T Cell Epitope of the M1 Protein of Influenza A Virus. Med. Microbiol. Immunol. 1998, 187, 91–96. [Google Scholar] [CrossRef]
- Tanaka, S.; Mikhailov, M.; Roy, P. Synthesis of Bluetongue Virus Chimeric VP3 Molecules and Their Interactions with VP7 Protein to Assemble into Virus Core-like Particles. Virology 1995, 214, 593–601. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ponndorf, D.; Meshcheriakova, Y.; Thuenemann, E.C.; Dobon Alonso, A.; Overman, R.; Holton, N.; Dowall, S.; Kennedy, E.; Stocks, M.; Lomonossoff, G.P.; et al. Plant-made Dengue Virus-like Particles Produced by Co-expression of Structural and Non-structural Proteins Induce a Humoral Immune Response in Mice. Plant Biotechnol. J. 2021, 19, 745–756. [Google Scholar] [CrossRef]
- Brillault, L.; Jutras, P.V.; Dashti, N.; Thuenemann, E.C.; Morgan, G.; Lomonossoff, G.P.; Landsberg, M.J.; Sainsbury, F. Engineering Recombinant Virus-like Nanoparticles from Plants for Cellular Delivery. ACS Nano 2017, 11, 3476–3484. [Google Scholar] [CrossRef]
- Jeggo, M.H.; Wardley, R.C.; Brownlie, J. Importance of Ovine Cytotoxic T Cells in Protection against Bluetongue Virus Infection. Prog. Clin. Biol. Res. 1985, 178, 477–487. [Google Scholar]
- Marín-López, A.; Otero-Romero, I.; de la Poza, F.; Menaya-Vargas, R.; Calvo-Pinilla, E.; Benavente, J.; Martínez-Costas, J.M.; Ortego, J. VP2, VP7, and NS1 Proteins of Bluetongue Virus Targeted in Avian Reovirus MuNS-Mi Microspheres Elicit a Protective Immune Response in IFNAR(-/-) Mice. Antivir. Res. 2014, 110, 42–51. [Google Scholar] [CrossRef]
- Marín-López, A.; Calvo-Pinilla, E.; Barriales, D.; Lorenzo, G.; Benavente, J.; Brun, A.; Martínez-Costas, J.M.; Ortego, J. Microspheres-Prime/RMVA-Boost Vaccination Enhances Humoral and Cellular Immune Response in IFNAR(−/−) Mice Conferring Protection against Serotypes 1 and 4 of Bluetongue Virus. Antivir. Res. 2017, 142, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Hewat, E.A.; Booth, T.F.; Wade, R.H.; Roy, P. 3-D Reconstruction of Bluetongue Virus Tubules Using Cryoelectron Microscopy. J. Struct. Biol. 1992, 108, 35–48. [Google Scholar] [CrossRef]
- Boyce, M.; Celma, C.C.P.; Roy, P. Bluetongue Virus Non-Structural Protein 1 Is a Positive Regulator of Viral Protein Synthesis. Virol. J. 2012, 9, 178. [Google Scholar] [CrossRef]
- Urakawa, T.; Roy, P. Bluetongue Virus Tubules Made in Insect Cells by Recombinant Baculoviruses: Expression of the NS1 Gene of Bluetongue Virus Serotype 10. J. Virol. 1988, 62, 3919–3927. [Google Scholar] [CrossRef] [PubMed]
- Huismans, H.; Els, H.J. Characterization of the Tubules Associated with the Replication of Three Different Orbiviruses. Virology 1979, 92, 397–406. [Google Scholar] [CrossRef]
- Kerviel, A.; Ge, P.; Lai, M.; Jih, J.; Boyce, M.; Zhang, X.; Zhou, Z.H.; Roy, P. Atomic Structure of the Translation Regulatory Protein NS1 of Bluetongue Virus. Nat. Microbiol. 2019, 4, 837–845. [Google Scholar] [CrossRef]
- Koo, M.; Bendahmane, M.; Lettieri, G.A.; Paoletti, A.D.; Lane, T.E.; Fitchen, J.H.; Buchmeier, M.J.; Beachy, R.N. Protective Immunity against Murine Hepatitis Virus (MHV) Induced by Intranasal or Subcutaneous Administration of Hybrids of Tobacco Mosaic Virus That Carries an MHV Epitope. Proc. Natl. Acad. Sci. USA 1999, 96, 7774–7779. [Google Scholar] [CrossRef]
- Špakova, A.; Dalgėdienė, I.; Insodaitė, R.; Sasnauskienė, A.; Žvirblienė, A.; Petraitytė-Burneikienė, R. VB_EcoS_NBD2 Bacteriophage-Originated Polytubes as a Carrier for the Presentation of Foreign Sequences. Virus Res. 2020, 290, 198194. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological Applications of Bacteriophages: State of the Art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage Display as a Promising Approach for Vaccine Development. J. Biomed. Sci. 2016, 23, 66. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous Fusion Phage: Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, V.F.; Lal, A.A.; McCutchan, T.F. Immunogenicity and Epitope Mapping of Foreign Sequences via Genetically Engineered Filamentous Phage. J. Biol. Chem. 1988, 263, 4318–4322. [Google Scholar] [CrossRef]
- Larke, N.; Murphy, A.; Wirblich, C.; Teoh, D.; Estcourt, M.J.; McMichael, A.J.; Roy, P.; Hanke, T. Induction of Human Immunodeficiency Virus Type 1-Specific T Cells by a Bluetongue Virus Tubule-Vectored Vaccine Prime-Recombinant Modified Virus Ankara Boost Regimen. J. Virol. 2005, 79, 14822–14833. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marshall, J.J.A.; Fayard, B.; Roy, P. Biophysical Studies on the Morphology of Baculovirus-Expressed Bluetongue Virus Tubules. J. Gen. Virol. 1990, 71, 1839–1844. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.; Roy, P. Manipulation of the Bluetongue Virus Tubules for Immunogen Delivery. Future Microbiol. 2008, 3, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Monastyrskaya, K.; Gould, E.A.; Roy, P. Characterization and Modification of the Carboxy-Terminal Sequences of Bluetongue Virus Type 10 NS1 Protein in Relation to Tubule Formation and Location of an Antigenic Epitope in the Vicinity of the Carboxy Terminus of the Protein. J. Virol. 1995, 69, 2831–2841. [Google Scholar] [CrossRef]
- Mikhailov, M.; Monastyrskaya, K.; Bakker, T.; Roy, P. A New Form of Particulate Single and Multiple Immunogen Delivery System Based on Recombinant Bluetongue Virus-Derived Tubules. Virology 1996, 217, 323–331. [Google Scholar] [CrossRef][Green Version]
- Andrade, S.; Pinho, F.; Ribeiro, A.M.; Carreira, M.; Casanueva, F.F.; Roy, P.; Monteiro, M.P. Immunization Against Active Ghrelin Using Virus-Like Particles for Obesity Treatment. Curr. Pharm. Des 2013, 19, 6551–6558. [Google Scholar] [CrossRef]
- Ghosh, M.K.; Dériaud, E.; Saron, M.F.; Lo-Man, R.; Henry, T.; Jiao, X.; Roy, P.; Leclerc, C. Induction of Protective Antiviral Cytotoxic T Cells by a Tubular Structure Capable of Carrying Large Foreign Sequences. Vaccine 2002, 20, 1369–1377. [Google Scholar] [CrossRef]
- Ghosh, M.K.; Borca, M.V.; Roy, P. Virus-Derived Tubular Structure Displaying Foreign Sequences on the Surface Elicit CD4+ Th Cell and Protective Humoral Responses. Virology 2002, 302, 383–392. [Google Scholar] [CrossRef][Green Version]
- Ghosh, M.K.; Li, C.-L.; Fayolle, C.; Dadaglio, G.; Murphy, A.; Lemonnier, F.A.; Roy, P.; Leclerc, C. Induction of HLA-A2-Restricted CTL Responses by a Tubular Structure Carrying Human Melanoma Epitopes. Vaccine 2002, 20, 2463–2473. [Google Scholar] [CrossRef]
- Soi, R.K.; Rurangirwa, F.R.; McGuire, T.C.; Rwambo, P.M.; DeMartini, J.C.; Crawford, T.B. Protection of Sheep against Rift Valley Fever Virus and Sheep Poxvirus with a Recombinant Capripoxvirus Vaccine. Clin. Vaccine Immunol. 2010, 17, 1842–1849. [Google Scholar] [CrossRef] [PubMed]
- Ngichabe, C.K.; Wamwayi, H.M.; Barrett, T.; Ndungu, E.K.; Black, D.N.; Bostock, C.J. Trial of a Capripoxvirus-Rinderpest Recombinant Vaccine in African Cattle. Epidemiol. Infect. 1997, 118, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Romero, C.H.; Barrett, T.; Kitching, R.P.; Carn, V.M.; Black, D.N. Protection of Cattle against Rinderpest and Lumpy Skin Disease with a Recombinant Capripoxvirus Expressing the Fusion Protein Gene of Rinderpest Virus. Vet. Rec. 1994, 135, 152–154. [Google Scholar] [CrossRef]
- Ngichabe, C.K.; Wamwayi, H.M.; Ndungu, E.K.; Mirangi, P.K.; Bostock, C.J.; Black, D.N.; Barrett, T. Long Term Immunity in African Cattle Vaccinated with a Recombinant Capripox-Rinderpest Virus Vaccine. Epidemiol. Infect. 2002, 128, 343–349. [Google Scholar] [CrossRef]
- Romero, C.H.; Barrett, T.; Kitching, R.P.; Bostock, C.; Black, D.N. Protection of Goats against Peste Des Petits Ruminants with Recombinant Capripoxviruses Expressing the Fusion and Haemagglutinin Protein Genes of Rinderpest Virus. Vaccine 1995, 13, 36–40. [Google Scholar] [CrossRef]
- Mellor, P.S.; Hamblin, C. African Horse Sickness. Vet. Res. 2004, 35, 445–466. [Google Scholar] [CrossRef]
- Fassi-Fihri, O.; el Harrak, M.; Fassi-Fehri, M.M. Clinical, Virological and Immune Responses of Normal and Immunosuppressed Donkeys (Equus Asinus Africanus) after Inoculation with African Horse Sickness Virus. Arch. Virol. Suppl. 1998, 14, 49–56. [Google Scholar] [CrossRef]
- Becker, E.; Venter, G.J.; Greyling, T.; Molini, U.; van Hamburg, H. Evidence of African Horse Sickness Virus Infection of Equus Zebra Hartmannae in the South-Western Khomas Region, Namibia. Transbound. Emerg. Dis. 2018, 65, 278–280. [Google Scholar] [CrossRef]
- Weyer, C.T.; Grewar, J.D.; Burger, P.; Rossouw, E.; Lourens, C.; Joone, C.; le Grange, M.; Coetzee, P.; Venter, E.; Martin, D.P.; et al. African Horse Sickness Caused by Genome Reassortment and Reversion to Virulence of Live, Attenuated Vaccine Viruses, South Africa, 2004–2014. Emerg. Infect. Dis. 2016, 22, 2087–2096. [Google Scholar] [CrossRef]
- Molini, U.; Marucchella, G.; Maseke, A.; Ronchi, G.F.; Di Ventura, M.; Salini, R.; Scacchia, M.; Pini, A. Immunization of Horses with a Polyvalent Live-Attenuated African Horse Sickness Vaccine: Serological Response and Disease Occurrence under Field Conditions. Trials Vaccinol. 2015, 4, 24–28. [Google Scholar] [CrossRef]
- Dennis, S.J.; Meyers, A.E.; Hitzeroth, I.I.; Rybicki, E.P. African Horse Sickness: A Review of Current Understanding and Vaccine Development. Viruses 2019, 11, 844. [Google Scholar] [CrossRef]
- Maree, S.; Durbach, S.; Huismans, H. Intracellular Production of African Horsesickness Virus Core-like Particles by Expression of the Two Major Core Proteins, VP3 and VP7, in Insect Cells. J. Gen. Virol. 1998, 79 Pt 2, 333–337. [Google Scholar] [CrossRef][Green Version]
- Maree, S.; Maree, F.F.; Putterill, J.F.; de Beer, T.A.P.; Huismans, H.; Theron, J. Synthesis of Empty African Horse Sickness Virus Particles. Virus Res. 2016, 213, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, J.N.; O’Hara, R.S.; Smale, C.J.; Hamblin, C.; Walton, A.; Armstrong, R.; Mertens, P.P. Purification and Properties of Virus Particles, Infectious Subviral Particles, Cores and VP7 Crystals of African Horsesickness Virus Serotype 9. J. Gen. Virol. 1994, 75 Pt 8, 1849–1857. [Google Scholar] [CrossRef] [PubMed]
- Dennis, S.J.; Meyers, A.E.; Guthrie, A.J.; Hitzeroth, I.I.; Rybicki, E.P. Immunogenicity of Plant-produced African Horse Sickness Virus-like Particles: Implications for a Novel Vaccine. Plant Biotechnol. J. 2018, 16, 442–450. [Google Scholar] [CrossRef]
- Dennis, S.J.; O’Kennedy, M.M.; Rutkowska, D.; Tsekoa, T.; Lourens, C.W.; Hitzeroth, I.I.; Meyers, A.E.; Rybicki, E.P. Safety and Immunogenicity of Plant-Produced African Horse Sickness Virus-like Particles in Horses. Vet. Res. 2018, 49, 105. [Google Scholar] [CrossRef]
- Maree, F.F.; Huismans, H. Characterization of Tubular Structures Composed of Nonstructural Protein NS1 of African Horsesickness Virus Expressed in Insect Cells. J. Gen. Virol. 1997, 78 Pt 5, 1077–1082. [Google Scholar] [CrossRef][Green Version]
- De la Poza, F.; Marín-López, A.; Castillo-Olivares, J.; Calvo-Pinilla, E.; Ortego, J. Identification of CD8 T Cell Epitopes in VP2 and NS1 Proteins of African Horse Sickness Virus in IFNAR(-/-) Mice. Virus Res. 2015, 210, 149–153. [Google Scholar] [CrossRef]
- Yadin, H.; Brenner, J.; Bumbrov, V.; Oved, Z.; Stram, Y.; Klement, E.; Perl, S.; Anthony, S.; Maan, S.; Batten, C.; et al. Epizootic Haemorrhagic Disease Virus Type 7 Infection in Cattle in Israel. Vet. Rec. 2008, 162, 53–56. [Google Scholar] [CrossRef]
- Mahmoud, A.; Danzetta, M.L.; di Sabatino, D.; Spedicato, M.; Alkhatal, Z.; Dayhum, A.; Tolari, F.; Forzan, M.; Mazzei, M.; Savini, G. First Seroprevalence Investigation of Epizootic Haemorrhagic Disease Virus in Libya. Open Vet. J. 2021, 11, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.B.; Goekjian, V.H.; Potgieter, A.C.; Wilson, W.C.; Johnson, D.J.; Mertens, P.P.C.; Stallknecht, D.E. Detection of a Novel Reassortant Epizootic Hemorrhagic Disease Virus (EHDV) in the USA Containing RNA Segments Derived from Both Exotic (EHDV-6) and Endemic (EHDV-2) Serotypes. J. Gen. Virol. 2010, 91, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Brown-Joseph, T.; Rajko-Nenow, P.; Hicks, H.; Sahadeo, N.; Harrup, L.E.; Carrington, C.V.; Batten, C.; Oura, C.A.L. Identification and Characterization of Epizootic Hemorrhagic Disease Virus Serotype 6 in Cattle Co-Infected with Bluetongue Virus in Trinidad, West Indies. Vet. Microbiol. 2019, 229, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ruder, M.G.; Johnson, D.; Ostlund, E.; Allison, A.B.; Kienzle, C.; Phillips, J.E.; Poulson, R.L.; Stallknecht, D.E. The First 10 Years (2006-15) of Epizootic Hemorrhagic Disease Virus Serotype 6 in the USA. J. Wildl. Dis. 2017, 53, 901–905. [Google Scholar] [CrossRef][Green Version]
- Homan, E.J.; Taylor, W.P.; de Ruiz, H.L.; Yuill, T.M. Bluetongue Virus and Epizootic Haemorrhagic Disease of Deer Virus Serotypes in Northern Colombian Cattle. J. Hyg. 1985, 95, 165–172. [Google Scholar] [CrossRef]
- Viarouge, C.; Lancelot, R.; Rives, G.; Bréard, E.; Miller, M.; Baudrimont, X.; Doceul, V.; Vitour, D.; Zientara, S.; Sailleau, C. Identification of Bluetongue Virus and Epizootic Hemorrhagic Disease Virus Serotypes in French Guiana in 2011 and 2012. Vet. Microbiol. 2014, 174, 78–85. [Google Scholar] [CrossRef]
- Verdezoto, J.; Breard, E.; Viarouge, C.; Quenault, H.; Lucas, P.; Sailleau, C.; Zientara, S.; Augot, D.; Zapata, S. Novel Serotype of Bluetongue Virus in South America and First Report of Epizootic Haemorrhagic Disease Virus in Ecuador. Transbound. Emerg. Dis. 2018, 65, 244–247. [Google Scholar] [CrossRef]
- Vinueza, R.L.; Cruz, M.; Bréard, E.; Viarouge, C.; Zanella, G. Bluetongue Virus and Epizootic Hemorrhagic Disease Virus Survey in Cattle of the Galapagos Islands. J. Vet. Diagn. Investig. 2019, 31, 271–275. [Google Scholar] [CrossRef]
- Weir, R.P.; Harmsen, M.B.; Hunt, N.T.; Blacksell, S.D.; Lunt, R.A.; Pritchard, L.I.; Newberry, K.M.; Hyatt, A.D.; Gould, A.R.; Melville, L.F. EHDV-1, a New Australian Serotype of Epizootic Haemorrhagic Disease Virus Isolated from Sentinel Cattle in the Northern Territory. Vet. Microbiol. 1997, 58, 135–143. [Google Scholar] [CrossRef]
- Kedmi, M.; Van Straten, M.; Ezra, E.; Galon, N.; Klement, E. Assessment of the Productivity Effects Associated with Epizootic Hemorrhagic Disease in Dairy Herds. J. Dairy Sci. 2010, 93, 2486–2495. [Google Scholar] [CrossRef]
- Spedicato, M.; Carmine, I.; Teodori, L.; Leone, A.; Portanti, O.; Marini, V.; Pisciella, M.; Lorusso, A.; Savini, G. Innocuity of a Commercial Live Attenuated Vaccine for Epizootic Hemorrhagic Disease Virus Serotype 2 in Late-Term Pregnant Cows. Vaccine 2016, 34, 1430–1435. [Google Scholar] [CrossRef] [PubMed]
- Sunwoo, S.Y.; Noronha, L.E.; Morozov, I.; Trujillo, J.D.; Kim, I.J.; Schirtzinger, E.E.; Faburay, B.; Drolet, B.S.; Urbaniak, K.; McVey, D.S.; et al. Evaluation of A Baculovirus-Expressed VP2 Subunit Vaccine for the Protection of White-Tailed Deer (Odocoileus virginianus) from Epizootic Hemorrhagic Disease. Vaccines 2020, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, J.; Xu, Q.; Sun, E.; Li, J.; Lv, S.; Feng, Y.; Zhang, Q.; Wang, H.; Wang, H.; et al. Development of a Reverse Genetics System for Epizootic Hemorrhagic Disease Virus and Evaluation of Novel Strains Containing Duplicative Gene Rearrangements. J. Gen. Virol. 2015, 96, 2714–2720. [Google Scholar] [CrossRef][Green Version]
- Matsuo, E.; Saeki, K.; Roy, P.; Kawano, J. Development of Reverse Genetics for Ibaraki Virus to Produce Viable VP6-Tagged IBAV. FEBS Open Bio. 2015, 5, 445–453. [Google Scholar] [CrossRef]
- Alshaikhahmed, K.; Roy, P. Generation of Virus-like Particles for Emerging Epizootic Haemorrhagic Disease Virus: Towards the Development of Safe Vaccine Candidates. Vaccine 2016, 34, 1103–1108. [Google Scholar] [CrossRef]
- Mecham, J.O.; Stallknecht, D.; Wilson, W.C. The S7 Gene and VP7 Protein Are Highly Conserved among Temporally and Geographically Distinct American Isolates of Epizootic Hemorrhagic Disease Virus. Virus Res. 2003, 94, 129–133. [Google Scholar] [CrossRef]
- Forzan, M.; Maan, S.; Mazzei, M.; Belaganahalli, M.N.; Bonuccelli, L.; Calamari, M.; Carrozza, M.L.; Cappello, V.; Di Luca, M.; Bandecchi, P.; et al. Generation of Virus like Particles for Epizootic Hemorrhagic Disease Virus. Res. Vet. Sci. 2016, 107, 116–122. [Google Scholar] [CrossRef]
- De la Poza, F.; Calvo-Pinilla, E.; López-Gil, E.; Marín-López, A.; Mateos, F.; Castillo-Olivares, J.; Lorenzo, G.; Ortego, J. Ns1 Is a Key Protein in the Vaccine Composition to Protect Ifnar(−/−) Mice against Infection with Multiple Serotypes of African Horse Sickness Virus. PLoS ONE 2013, 8, e0070197. [Google Scholar] [CrossRef]
- Marín-Lopez, A.; Calvo-Pinilla, E.; Moreno, S.; Utrilla-Trigo, S.; Nogales, A.; Brun, A.; Fikrig, E.; Ortego, J. Modeling Arboviral Infection in Mice Lacking the Interferon Alpha/Beta Receptor. Viruses 2019, 11, 35. [Google Scholar] [CrossRef]
- Marín-López, A.; Bermúdez, R.; Calvo-Pinilla, E.; Moreno, S.; Brun, A.; Ortego, J. Pathological Characterization of IFNAR(−/−) Mice Infected With Bluetongue Virus Serotype 4. Int. J. Biol. Sci. 2016, 12, 1448–1460. [Google Scholar] [CrossRef]
- Calvo-Pinilla, E.; Rodríguez-Calvo, T.; Anguita, J.; Sevilla, N.; Ortego, J. Establishment of a Bluetongue Virus Infection Model in Mice That Are Deficient in the Alpha/Beta Interferon Receptor. PLoS ONE 2009, 4, e5171. [Google Scholar] [CrossRef] [PubMed]
- Eschbaumer, M.; Keller, M.; Beer, M.; Hoffmann, B. Epizootic Hemorrhagic Disease Virus Infection of Type I Interferon Receptor Deficient Mice. Vet. Microbiol. 2012, 155, 417–419. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Cabello, L.; Utrilla-Trigo, S.; Barreiro-Piñeiro, N.; Pose-Boirazian, T.; Martínez-Costas, J.; Marín-López, A.; Ortego, J. Nanoparticle- and Microparticle-Based Vaccines against Orbiviruses of Veterinary Importance. Vaccines 2022, 10, 1124. https://doi.org/10.3390/vaccines10071124
Jiménez-Cabello L, Utrilla-Trigo S, Barreiro-Piñeiro N, Pose-Boirazian T, Martínez-Costas J, Marín-López A, Ortego J. Nanoparticle- and Microparticle-Based Vaccines against Orbiviruses of Veterinary Importance. Vaccines. 2022; 10(7):1124. https://doi.org/10.3390/vaccines10071124
Chicago/Turabian StyleJiménez-Cabello, Luis, Sergio Utrilla-Trigo, Natalia Barreiro-Piñeiro, Tomás Pose-Boirazian, José Martínez-Costas, Alejandro Marín-López, and Javier Ortego. 2022. "Nanoparticle- and Microparticle-Based Vaccines against Orbiviruses of Veterinary Importance" Vaccines 10, no. 7: 1124. https://doi.org/10.3390/vaccines10071124
APA StyleJiménez-Cabello, L., Utrilla-Trigo, S., Barreiro-Piñeiro, N., Pose-Boirazian, T., Martínez-Costas, J., Marín-López, A., & Ortego, J. (2022). Nanoparticle- and Microparticle-Based Vaccines against Orbiviruses of Veterinary Importance. Vaccines, 10(7), 1124. https://doi.org/10.3390/vaccines10071124








