Advances in Next-Generation Coronavirus Vaccines in Response to Future Virus Evolution
Abstract
1. Introduction
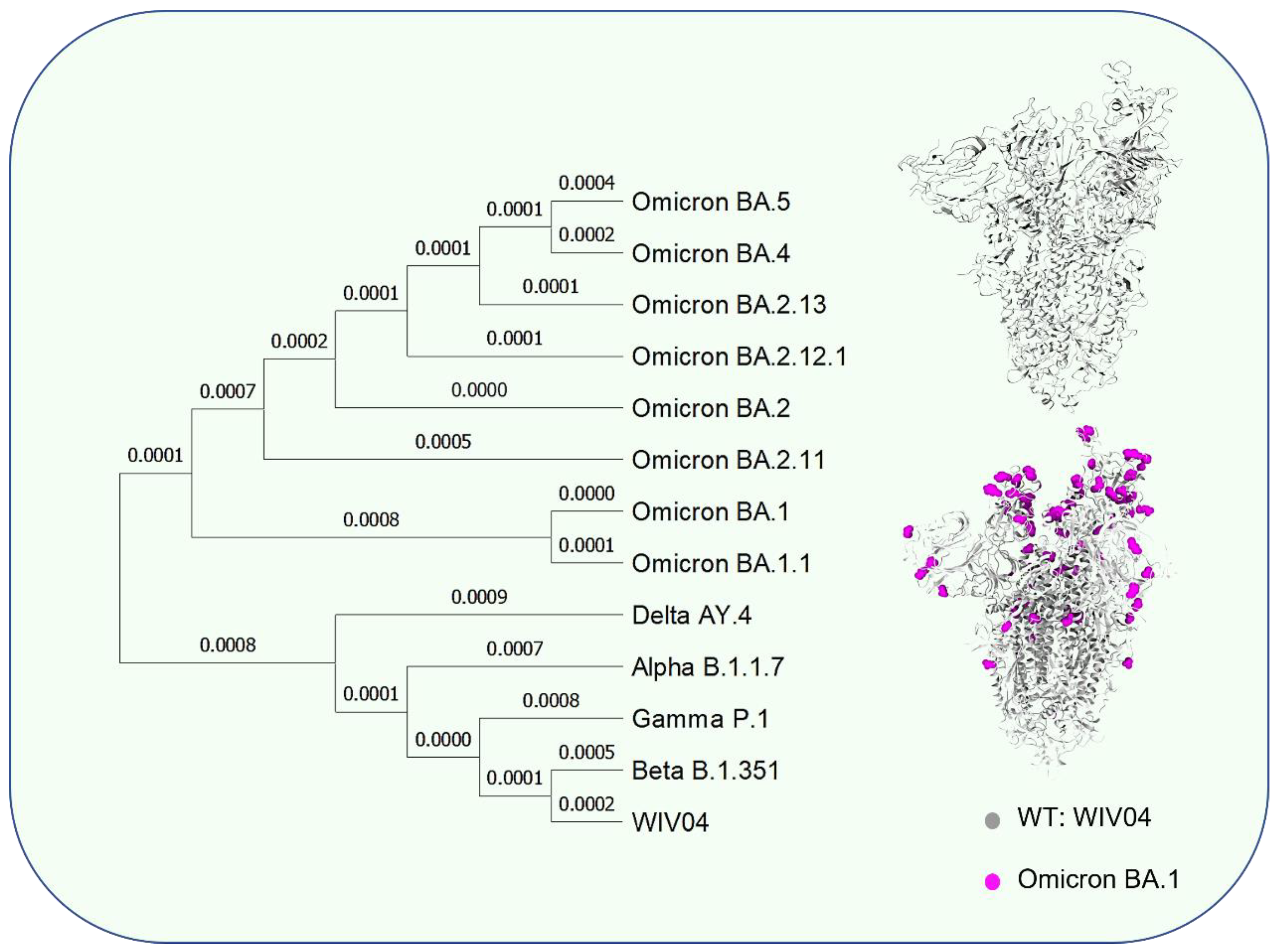
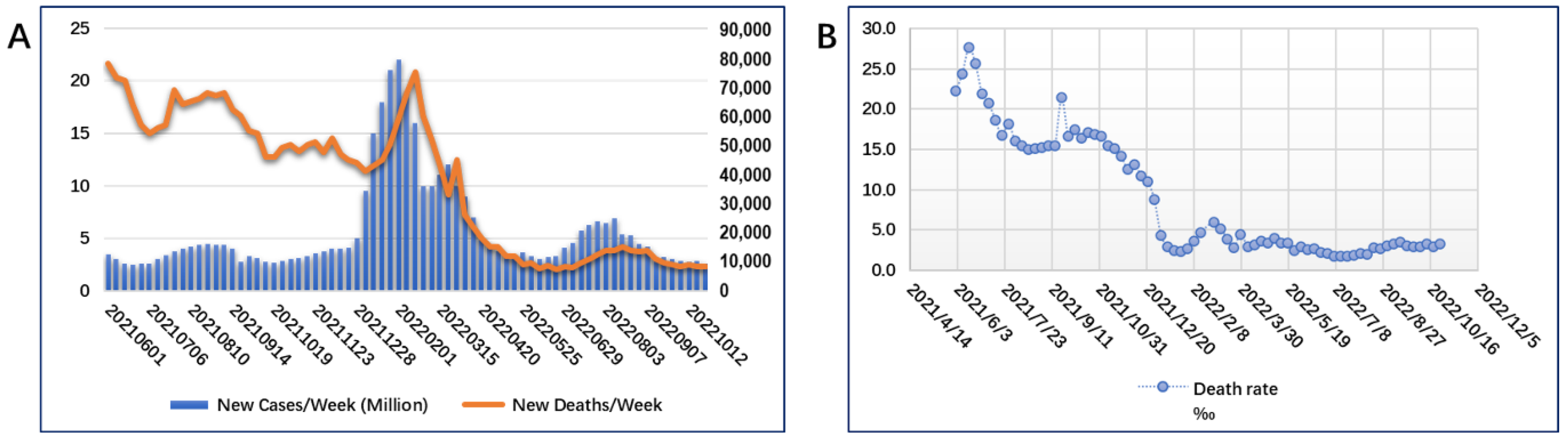
2. New Situation Caused by SARS-CoV-2 Evolution
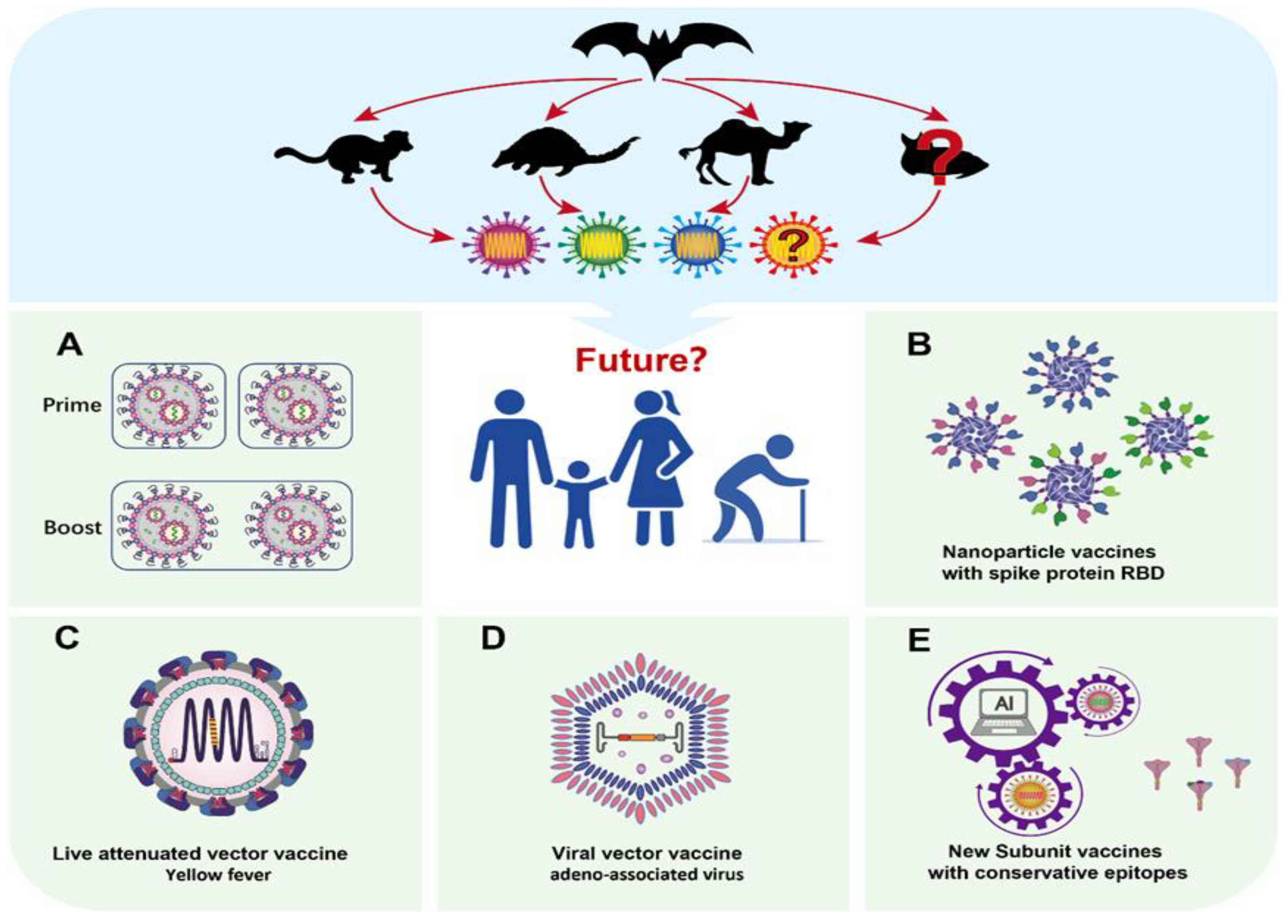
3. Next-Generation Vaccine Design against Mutated Viruses
4. Strategy One: Booster Immunization with Approved Vaccines
5. Strategy Two: Vaccines with More Complex Sources of Spike Protein RBD
6. Strategy Three: Live Attenuated Viral Vaccine Delivered SARS-CoV-2 Antigen
7. Strategy Four: Subunit Vaccines with T-Cell Epitopes
8. Strategy Five: More Cutting-Edge Vaccine Design Using AI Technology
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Howley, P.M.; Knipe, D.M.; Whelan, S. Fields Virology: Emerging Viruses, 7th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2020; ISBN 1975112547/978-1975112547. [Google Scholar]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020, 21, 224. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Sridhar, S.; Chiu, K.H.; Hung, D.L.; Li, X.; Hung, I.F.; Tam, A.R.; Chung, T.W.; Chan, J.F.; Zhang, A.J.; et al. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg. Microbes Infect. 2021, 10, 507–535. [Google Scholar] [CrossRef] [PubMed]
- WHO. Virtual Press Conference on COVID-19. 2020. Available online: https://www.who.int/docs/default-source/coronaviruse/transcripts/who-audio-emergencies-coronavirus-press-conference-full-and-final-11mar2020.pdf?sfvrsn=cb432bb3_2 (accessed on 26 October 2022).
- Ella, R.; Reddy, S.; Blackwelder, W.; Potdar, V.; Yadav, P.; Sarangi, V.; Aileni, V.K.; Kanungo, S.; Rai, S.; Reddy, P.; et al. Efficacy, safety, and lot-to-lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): Interim results of a randomised, double-blind, controlled, phase 3 trial. Lancet 2021, 398, 2173–2184. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Voysey, M.; Costa Clemens, S.A.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: A pooled analysis of four randomised trials. Lancet 2021, 397, 881–891. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 COVID-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- WHO. Coronavirus Disease (COVID-19): Vaccines. 2022. Available online: https://www.who.int/news-room/questions-and-answers/item/coronavirus-disease-(covid-19)-vaccines (accessed on 30 October 2022).
- COVID-19 Vaccine Tacker. Available online: https://vac-lshtmshinyappsio/ncov_vaccine_landscape/ (accessed on 30 October 2022).
- Ourworldindata. Coronavirus (COVID-19) Vaccinations. 2022. Available online: https://ourworldindata.org/covid-vaccinations (accessed on 30 October 2022).
- Feikin, D.R.; Higdon, M.M.; Abu-Raddad, L.J.; Andrews, N.; Araos, R.; Goldberg, Y.; Groome, M.J.; Huppert, A.; O’Brien, K.L.; Smith, P.G.; et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: Results of a systematic review and meta-regression. Lancet 2022, 399, 924–944. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, X.; Song, J.; Wu, J.; Zhu, Y.; Li, M.; Cui, Y.; Chen, Y.; Yang, L.; Liu, J.; et al. Homologous or heterologous booster of inactivated vaccine reduces SARS-CoV-2 Omicron variant escape from neutralizing antibodies. Emerg. Microbes Infect. 2022, 11, 477–481. [Google Scholar] [CrossRef]
- Ai, J.; Zhang, H.; Zhang, Y.; Lin, K.; Zhang, Y.; Wu, J.; Wan, Y.; Huang, Y.; Song, J.; Fu, Z.; et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2022, 11, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Lyke, K.E.; Atmar, R.L.; Islas, C.D.; Posavad, C.M.; Szydlo, D.; Paul Chourdhury, R.; Deming, M.E.; Eaton, A.; Jackson, L.A.; Branche, A.R.; et al. Rapid decline in vaccine-boosted neutralizing antibodies against SARS-CoV-2 Omicron variant. Cell Rep. Med. 2022, 3, 100679. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature reviews Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef]
- Hirabara, S.M.; Serdan, T.D.A.; Gorjao, R.; Masi, L.N.; Pithon-Curi, T.C.; Covas, D.T.; Curi, R.; Durigon, E.L. SARS-CoV-2 Variants: Differences and Potential of Immune Evasion. Front. Cell. Infect. Microbiol. 2021, 11, 781429. [Google Scholar] [CrossRef]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef]
- Papanikolaou, V.; Chrysovergis, A.; Ragos, V.; Tsiambas, E.; Katsinis, S.; Manoli, A.; Papouliakos, S.; Roukas, D.; Mastronikolis, S.; Peschos, D.; et al. From delta to Omicron: S1-RBD/S2 mutation/deletion equilibrium in SARS-CoV-2 defined variants. Gene 2022, 814, 146134. [Google Scholar] [CrossRef]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011, 8, 270–279. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2-What Do They Mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef]
- Mourier, T.; Sadykov, M.; Carr, M.J.; Gonzalez, G.; Hall, W.W.; Pain, A. Host-directed editing of the SARS-CoV-2 genome. Biochem. Biophys. Res. Commun. 2021, 538, 35–39. [Google Scholar] [CrossRef]
- Wang, R.; Hozumi, Y.; Zheng, Y.H.; Yin, C.; Wei, G.W. Host Immune Response Driving SARS-CoV-2 Evolution. Viruses 2020, 12, 1095. [Google Scholar] [CrossRef]
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Farinholt, T.; Doddapaneni, H.; Qin, X.; Menon, V.; Meng, Q.; Metcalf, G.; Chao, H.; Gingras, M.C.; Avadhanula, V.; Farinholt, P.; et al. Transmission event of SARS-CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med. 2021, 19, 255. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.M.; Vostok, J.; Johnson, H.; Burns, M.; Gharpure, R.; Sami, S.; Sabo, R.T.; Hall, N.; Foreman, A.; Schubert, P.L.; et al. Outbreak of SARS-CoV-2 Infections, Including COVID-19 Vaccine Breakthrough Infections, Associated with Large Public Gatherings—Barnstable County, Massachusetts, July 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1059–1062. [Google Scholar] [CrossRef] [PubMed]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Rocklöv, J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J. Travel Med. 2021, 28, taab124. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.; Chan, J.F.; Hu, B.; Chai, Y.; Yuen, T.T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.C.; Liu, H.; et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022, 603, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Then, E.; Lucas, C.; Monteiro, V.S.; Miric, M.; Brache, V.; Cochon, L.; Vogels, C.B.F.; Malik, A.A.; De la Cruz, E.; Jorge, A.; et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022, 28, 481–485. [Google Scholar] [CrossRef]
- Edara, V.V.; Manning, K.E.; Ellis, M.; Lai, L.; Moore, K.M.; Foster, S.L.; Floyd, K.; Davis-Gardner, M.E.; Mantus, G.; Nyhoff, L.E.; et al. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep. Med. 2022, 3, 100529. [Google Scholar] [CrossRef]
- Huang, Z.; Xu, S.; Liu, J.; Wu, L.; Qiu, J.; Wang, N.; Ren, J.; Li, Z.; Guo, X.; Tao, F.; et al. Effectiveness of inactivated and Ad5-nCoV COVID-19 vaccines against SARS-CoV-2 Omicron BA. 2 variant infection, severe illness, and death. BMC Med. 2022, 20, 400. [Google Scholar] [CrossRef]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.M.; et al. COVID-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef]
- Zeng, B.; Gao, L.; Zhou, Q.; Yu, K.; Sun, F. Effectiveness of COVID-19 vaccines against SARS-CoV-2 variants of concern: A systematic review and meta-analysis. BMC Med. 2022, 20, 200. [Google Scholar] [CrossRef]
- Collie, S.; Champion, J.; Moultrie, H.; Bekker, L.G.; Gray, G. Effectiveness of BNT162b2 Vaccine against Omicron Variant in South Africa. N. Engl. J. Med. 2022, 386, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Tuekprakhon, A.; Nutalai, R.; Dijokaite-Guraliuc, A.; Zhou, D.; Ginn, H.M.; Selvaraj, M.; Liu, C.; Mentzer, A.J.; Supasa, P.; Duyvesteyn, H.M.E.; et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433.e13. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, C.; Mayer, C.K.; Claassen, M.; Maponga, T.; Burgers, W.A.; Keeton, R.; Riou, C.; Sutherland, A.D.; Suliman, T.; Shaw, M.L.; et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 2022, 399, 625–626. [Google Scholar] [CrossRef] [PubMed]
- Carreño, J.M.; Alshammary, H.; Tcheou, J.; Singh, G.; Raskin, A.J.; Kawabata, H.; Sominsky, L.A.; Clark, J.J.; Adelsberg, D.C.; Bielak, D.A.; et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 2022, 602, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef]
- Looi, M.K. The BMJ Interview: WHO chief scientist optimistic for a pan-coronavirus vaccine in two years. BMJ 2022, 377, o1003. [Google Scholar] [CrossRef] [PubMed]
- Lavine, J.S.; Bjornstad, O.N.; Antia, R. Immunological characteristics govern the transition of COVID-19 to endemicity. Science 2021, 371, 741–745. [Google Scholar] [CrossRef]
- Luo, R.; Delaunay-Moisan, A.; Timmis, K.; Danchin, A. SARS-CoV-2 biology and variants: Anticipation of viral evolution and what needs to be done. Environ. Microbiol. 2021, 23, 2339–2363. [Google Scholar] [CrossRef]
- Sandor, A.M.; Sturdivant, M.S.; Ting, J.P.Y. Influenza Virus and SARS-CoV-2 Vaccines. J. Immunol. 2021, 206, 2509–2520. [Google Scholar] [CrossRef]
- Martin, W.R.; Cheng, F. A rational design of a multi-epitope vaccine against SARS-CoV-2 which accounts for the glycan shield of the spike glycoprotein. J. Biomol. Struct. Dyn. 2022, 40, 7099–7113. [Google Scholar] [CrossRef]
- Kibria, K.M.K.; Faruque, M.O.; Islam, M.S.B.; Ullah, H.; Mahmud, S.; Miah, M.; Saleh, A.A. A conserved subunit vaccine designed against SARS-CoV-2 variants showed evidence in neutralizing the virus. Appl. Microbiol. Biotechnol. 2022, 106, 4091–4114. [Google Scholar] [CrossRef] [PubMed]
- Ong, E.; Wong, M.U.; Huffman, A.; He, Y. COVID-19 Coronavirus Vaccine Design Using Reverse Vaccinology and Machine Learning. Front. Immunol. 2020, 11, 1581. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, J.; Li, S.; Sun, J.; Teng, Y.; Wu, M.; Li, J.; Li, Y.; Hu, N.; Wang, H.; et al. Epitope-Based Vaccine Target Screening against Highly Pathogenic MERS-CoV: An In Silico Approach Applied to Emerging Infectious Diseases. PLoS ONE 2015, 10, e0144475. [Google Scholar] [CrossRef]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.G.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022, 602, 654–656. [Google Scholar] [CrossRef]
- Muik, A.; Lui, B.G.; Wallisch, A.K.; Bacher, M.; Mühl, J.; Reinholz, J.; Ozhelvaci, O.; Beckmann, N.; Güimil Garcia, R.C.; Poran, A.; et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science 2022, 375, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Mok, B.W.Y.; Chen, L.L.; Chan, J.M.C.; Tsang, O.T.Y.; Lam, B.H.S.; Chuang, V.W.M.; Chu, A.W.H.; Chan, W.M.; Ip, J.D.; et al. Neutralization of Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant by Sera from BNT162b2 or CoronaVac Vaccine Recipients. Clin. Infect. Dis. 2022, 75, e822–e826. [Google Scholar] [CrossRef]
- Edridge, A.W.D.; Kaczorowska, J.; Hoste, A.C.R.; Bakker, M.; Klein, M.; Loens, K.; Jebbink, M.F.; Matser, A.; Kinsella, C.M.; Rueda, P.; et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020, 26, 1691–1693. [Google Scholar] [CrossRef]
- Lustig, Y.; Gonen, T.; Meltzer, L.; Gilboa, M.; Indenbaum, V.; Cohen, C.; Amit, S.; Jaber, H.; Doolman, R.; Asraf, K.; et al. Superior immunogenicity and effectiveness of the third compared to the second BNT162b2 vaccine dose. Nat. Immunol. 2022, 23, 940–946. [Google Scholar] [CrossRef]
- Accorsi, E.K.; Britton, A.; Fleming-Dutra, K.E.; Smith, Z.R.; Shang, N.; Derado, G.; Miller, J.; Schrag, S.J.; Verani, J.R. Association Between 3 Doses of mRNA COVID-19 Vaccine and Symptomatic Infection Caused by the SARS-CoV-2 Omicron and Delta Variants. JAMA 2022, 327, 639–651. [Google Scholar] [CrossRef]
- Costa Clemens, S.A.; Weckx, L.; Clemens, R.; Almeida Mendes, A.V.; Ramos Souza, A.; Silveira, M.B.V.; da Guarda, S.N.F.; de Nobrega, M.M.; de Moraes Pinto, M.I.; Gonzalez, I.G.S.; et al. Heterologous versus homologous COVID-19 booster vaccination in previous recipients of two doses of CoronaVac COVID-19 vaccine in Brazil (RHH-001): A phase 4, non-inferiority, single blind, randomised study. Lancet 2022, 399, 521–529. [Google Scholar] [CrossRef]
- Regev-Yochay, G.; Gonen, T.; Gilboa, M.; Mandelboim, M.; Indenbaum, V.; Amit, S.; Meltzer, L.; Asraf, K.; Cohen, C.; Fluss, R.; et al. Efficacy of a Fourth Dose of COVID-19 mRNA Vaccine against Omicron. N. Engl. J. Med. 2022, 386, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Chalkias, S.; Harper, C.; Vrbicky, K.; Walsh, S.R.; Essink, B.; Brosz, A.; McGhee, N.; Tomassini, J.E.; Chen, X.; Chang, Y.; et al. A Bivalent Omicron-Containing Booster Vaccine against COVID-19. N. Engl. J. Med. 2022, 387, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Most Vulnerable Could Be Offered Booster COVID-19 Vaccines from September. Available online: https://www.gov.uk/government/news/most-vulnerable-could-be-offered-booster-covid-19-vaccines-from-september (accessed on 30 October 2022).
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef] [PubMed]
- McLean, G.; Kamil, J.; Lee, B.; Moore, P.; Schulz, T.F.; Muik, A.; Sahin, U.; Türeci, Ö.; Pather, S. The Impact of Evolving SARS-CoV-2 Mutations and Variants on COVID-19 Vaccines. mBio 2022, 13, e0297921. [Google Scholar] [CrossRef] [PubMed]
- El-Shabasy, R.M.; Nayel, M.A.; Taher, M.M.; Abdelmonem, R.; Shoueir, K.R.; Kenawy, E.R. Three waves changes, new variant strains, and vaccination effect against COVID-19 pandemic. Int. J. Biol. Macromol. 2022, 204, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, L.; Park, Y.J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042.e21. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; McMahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef]
- Mercado, N.B.; Zahn, R.; Wegmann, F.; Loos, C.; Chandrashekar, A.; Yu, J.; Liu, J.; Peter, L.; McMahan, K.; Tostanoski, L.H.; et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 2020, 586, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Joyce, M.G.; Gillespie, R.A.; Gallagher, J.R.; Andrews, S.F.; Yassine, H.M.; Wheatley, A.K.; Fisher, B.E.; Ambrozak, D.R.; Creanga, A.; et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019, 20, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Darricarrère, N.; Qiu, Y.; Kanekiyo, M.; Creanga, A.; Gillespie, R.A.; Moin, S.M.; Saleh, J.; Sancho, J.; Chou, T.H.; Zhou, Y.; et al. Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates. Sci. Transl. Med. 2021, 13, eabe5449. [Google Scholar] [CrossRef]
- Pati, R.; Shevtsov, M.; Sonawane, A. Nanoparticle Vaccines Against Infectious Diseases. Front. Immunol. 2018, 9, 2224. [Google Scholar] [CrossRef]
- Yamashita, I.; Iwahori, K.; Kumagai, S. Ferritin in the field of nanodevices. Biochim. Biophys. Acta 2010, 1800, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Worwood, M. Ferritin. Blood Rev. 1990, 4, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Chakraborti, S.; Chakrabarti, P. Self-Assembly of Ferritin: Structure, Biological Function and Potential Applications in Nanotechnology. Adv. Exp. Med. Biol. 2019, 1174, 313–329. [Google Scholar] [PubMed]
- Lua, L.H.; Connors, N.K.; Sainsbury, F.; Chuan, Y.P.; Wibowo, N.; Middelberg, A.P. Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 2014, 111, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Joyce, M.G.; Chen, W.H.; Sankhala, R.S.; Hajduczki, A.; Thomas, P.V.; Choe, M.; Martinez, E.J.; Chang, W.C.; Peterson, C.E.; Morrison, E.B.; et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 2021, 37, 110143. [Google Scholar] [CrossRef] [PubMed]
- Wuertz, K.M.; Barkei, E.K.; Chen, W.H.; Martinez, E.J.; Lakhal-Naouar, I.; Jagodzinski, L.L.; Paquin-Proulx, D.; Gromowski, G.D.; Swafford, I.; Ganesh, A.; et al. A SARS-CoV-2 spike ferritin nanoparticle vaccine protects hamsters against Alpha and Beta virus variant challenge. NPJ Vaccines 2021, 6, 129. [Google Scholar] [CrossRef] [PubMed]
- Joyce, M.G.; King, H.A.D.; Elakhal-Naouar, I.; Ahmed, A.; Peachman, K.K.; Macedo Cincotta, C.; Subra, C.; Chen, R.E.; Thomas, P.V.; Chen, W.H.; et al. A SARS-CoV-2 ferritin nanoparticle vaccine elicits protective immune responses in nonhuman primates. Sci. Transl. Med. 2022, 14, eabi5735. [Google Scholar] [CrossRef] [PubMed]
- Bruun, T.U.J.; Andersson, A.C.; Draper, S.J.; Howarth, M. Engineering a Rugged Nanoscaffold to Enhance Plug-and-Display Vaccination. ACS Nano 2018, 12, 8855–8866. [Google Scholar] [CrossRef]
- Cohen, A.A.; Gnanapragasam, P.N.P.; Lee, Y.E.; Hoffman, P.R.; Ou, S.; Kakutani, L.M.; Keeffe, J.R.; Wu, H.J.; Howarth, M.; West, A.P.; et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 2021, 371, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; van Doremalen, N.; Greaney, A.J.; Andersen, H.; Sharma, A.; Starr, T.N.; Keeffe, J.R.; Fan, C.; Schulz, J.E.; Gnanapragasam, P.N.P.; et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science 2022, 377, eabq0839. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Bivalent Vaccine Boosters. Available online: https://wwwfdagov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-bivalent-vaccine-boosters (accessed on 31 October 2022).
- Martinez, D.R.; Schäfer, A.; Leist, S.R.; De la Cruz, G.; West, A.; Atochina-Vasserman, E.N.; Lindesmith, L.C.; Pardi, N.; Parks, R.; Barr, M.; et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science 2021, 373, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Krugman, S.; Katz, S.L. Attenuated smallpox vaccine. N. Engl. J. Med. 1970, 282, 344. [Google Scholar] [PubMed]
- Yokote, H.; Shinmura, Y.; Kanehara, T.; Maruno, S.; Kuranaga, M.; Matsui, H.; Hashizume, S. Safety of attenuated smallpox vaccine LC16m8 in immunodeficient mice. Clin. Vaccine Immunol. 2014, 21, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Gershon, A.A. Live-attenuated varicella vaccine. Infect. Dis. Clin. N. Am. 2001, 15, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Gershon, A.A.; Gershon, M.D.; Shapiro, E.D. Live Attenuated Varicella Vaccine: Prevention of Varicella and of Zoster. J. Infect. Dis. 2021, 224 (Suppl. 2), S387–S397. [Google Scholar] [CrossRef]
- Davis, E.H.; Beck, A.S.; Li, L.; White, M.M.; Greenberg, M.B.; Thompson, J.K.; Widen, S.G.; Barrett, A.D.T.; Bourne, N. Japanese encephalitis virus live attenuated vaccine strains display altered immunogenicity, virulence and genetic diversity. NPJ Vaccines 2021, 6, 112. [Google Scholar] [CrossRef]
- Ngwe Tun, M.M.; Kyaw, A.K.; Nwe, K.M.; Inoue, S.; Thant, K.Z.; Morita, K. Effectiveness of the SA 14-14-2 Live-Attenuated Japanese Encephalitis Vaccine in Myanmar. Vaccines 2021, 9, 568. [Google Scholar] [CrossRef]
- Barrett, A.D.T. Yellow fever live attenuated vaccine: A very successful live attenuated vaccine but still we have problems controlling the disease. Vaccine 2017, 35, 5951–5955. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.D.; Barrett, A.D. Live Attenuated Yellow Fever 17D Vaccine: A Legacy Vaccine Still Controlling Outbreaks in Modern Day. Curr. Infect. Dis. Rep. 2017, 19, 14. [Google Scholar] [CrossRef] [PubMed]
- Jones, T. IMVAMUNE, an attenuated modified vaccinia Ankara virus vaccine for smallpox infection. Curr. Opin. Mol. Ther. 2008, 10, 407–417. [Google Scholar] [PubMed]
- Sricharoenchai, S.; Lapphra, K.; Chuenkitmongkol, S.; Phongsamart, W.; Bouckenooghe, A.; Wittawatmongkol, O.; Rungmaitree, S.; Chokephaibulkit, K. Immunogenicity of a Live Attenuated Chimeric Japanese Encephalitis Vaccine as a Booster Dose After Primary Vaccination with Live Attenuated SA14-14-2 Vaccine: A Phase IV Study in Thai Children. Pediatr. Infect. Dis. J. 2017, 36, e45–e47. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhang, R.; Guan, Q.; Mu, Q.; He, P.; Ye, X.; Wang, W.; Quan, J.; Li, J.; Liang, L.; et al. Immunogenicity and safety of a live attenuated varicella vaccine in children 1–12 years of age: A randomized, blinded, controlled, non-inferiority phase 3 clinical trial. Contemp. Clin. Trials 2021, 107, 106489. [Google Scholar] [CrossRef]
- Verheust, C.; Goossens, M.; Pauwels, K.; Breyer, D. Biosafety aspects of modified vaccinia virus Ankara (MVA)-based vectors used for gene therapy or vaccination. Vaccine 2012, 30, 2623–2632. [Google Scholar] [CrossRef]
- Lin, I.Y.; Van, T.T.; Smooker, P.M. Live-Attenuated Bacterial Vectors: Tools for Vaccine and Therapeutic Agent Delivery. Vaccines 2015, 3, 940–972. [Google Scholar] [CrossRef]
- Toussaint, B.; Chauchet, X.; Wang, Y.; Polack, B.; Le Gouëllec, A. Live-attenuated bacteria as a cancer vaccine vector. Expert Rev. Vaccines 2013, 12, 1139–1154. [Google Scholar] [CrossRef]
- Remy-Ziller, C.; Thioudellet, C.; Hortelano, J.; Gantzer, M.; Nourtier, V.; Claudepierre, M.C.; Sansas, B.; Préville, X.; Bendjama, K.; Quemeneur, E.; et al. Sequential administration of MVA-based vaccines and PD-1/PD-L1-blocking antibodies confers measurable benefits on tumor growth and survival: Preclinical studies with MVA-βGal and MVA-MUC1 (TG4010) in a murine tumor model. Hum. Vaccines Immunother. 2018, 14, 140–145. [Google Scholar] [CrossRef]
- Li, L.L.; Wang, H.R.; Zhou, Z.Y.; Luo, J.; Xiao, X.Q.; Wang, X.L.; Li, J.T.; Zhou, Y.B.; Zeng, Y. One-prime multi-boost strategy immunization with recombinant DNA, adenovirus, and MVA vector vaccines expressing HPV16 L1 induces potent, sustained, and specific immune response in mice. Antivir. Res. 2016, 128, 20–27. [Google Scholar] [CrossRef]
- Brett, D.L.C.L.M.; Heinz-Jürgen, T.; Rice, C.M. Flaviviridae. In Fields Virology, 6th ed.; Knipe, D.M., Ed.; Lippincott Willims & Wilkins: Philadelphia, PA, USA, 2013; p. 712. [Google Scholar]
- Wieten, R.W.; Jonker, E.F.; van Leeuwen, E.M.; Remmerswaal, E.B.; Ten Berge, I.J.; de Visser, A.W.; van Genderen, P.J.; Goorhuis, A.; Visser, L.G.; Grobusch, M.P.; et al. A Single 17D Yellow Fever Vaccination Provides Lifelong Immunity; Characterization of Yellow-Fever-Specific Neutralizing Antibody and T-Cell Responses after Vaccination. PLoS ONE 2016, 11, e0149871. [Google Scholar] [CrossRef]
- Grobusch, M.P.; van Aalst, M.; Goorhuis, A. Yellow fever vaccination—Once in a lifetime? Travel Med. Infect. Dis. 2017, 15, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.; Li, W.; Qing, F.; Stoyanov, C.T.; Moran, T.; Rice, C.M.; Ho, D.D. Evaluation of yellow fever virus 17D strain as a new vector for HIV-1 vaccine development. Vaccine 2010, 28, 5676–5685. [Google Scholar] [CrossRef] [PubMed]
- Blyth, D.M.; Liang, Z.; Williams, M.; Murray, C.K. Immune interference revisited: Impact of live-attenuated influenza vaccine prior to yellow fever vaccination. Vaccine 2022, 40, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Boudewijns, R.; Ma, J.; Neyts, J.; Dallmeier, K. A novel therapeutic HBV vaccine candidate induces strong polyfunctional cytotoxic T cell responses in mice. JHEP Rep. 2021, 3, 100295. [Google Scholar] [CrossRef] [PubMed]
- Kwek, S.S.; Watanabe, S.; Chan, K.R.; Ong, E.Z.; Tan, H.C.; Ng, W.C.; Nguyen, M.T.X.; Gan, E.S.; Zhang, S.L.; Chan, K.W.K.; et al. A systematic approach to the development of a safe live attenuated Zika vaccine. Nat. Commun. 2018, 9, 1031. [Google Scholar] [CrossRef]
- Oreshkova, N.; Myeni, S.K.; Mishra, N.; Albulescu, I.C.; Dalebout, T.J.; Snijder, E.J.; Bredenbeek, P.J.; Dallmeier, K.; Kikkert, M. A Yellow Fever 17D Virus Replicon-Based Vaccine Platform for Emerging Coronaviruses. Vaccines 2021, 9, 1492. [Google Scholar] [CrossRef]
- Sanchez-Felipe, L.; Vercruysse, T.; Sharma, S.; Ma, J.; Lemmens, V.; Van Looveren, D.; Arkalagud Javarappa, M.P.; Boudewijns, R.; Malengier-Devlies, B.; Liesenborghs, L.; et al. A single-dose live-attenuated YF17D-vectored SARS-CoV-2 vaccine candidate. Nature 2021, 590, 320–325. [Google Scholar] [CrossRef]
- Chiuppesi, F.; Salazar, M.D.; Contreras, H.; Nguyen, V.H.; Martinez, J.; Park, Y.; Nguyen, J.; Kha, M.; Iniguez, A.; Zhou, Q.; et al. Development of a multi-antigenic SARS-CoV-2 vaccine candidate using a synthetic poxvirus platform. Nat. Commun. 2020, 11, 6121. [Google Scholar] [CrossRef]
- García-Arriaza, J.; Garaigorta, U.; Pérez, P.; Lázaro-Frías, A.; Zamora, C.; Gastaminza, P.; Del Fresno, C.; Casasnovas, J.M.; Sorzano CÓ, S.; Sancho, D.; et al. COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike induce robust T- and B-cell immune responses and full efficacy in mice. J. Virol. 2021, 95, e02260-20. [Google Scholar] [CrossRef]
- Routhu, N.K.; Cheedarla, N.; Gangadhara, S.; Bollimpelli, V.S.; Boddapati, A.K.; Shiferaw, A.; Rahman, S.A.; Sahoo, A.; Edara, V.V.; Lai, L.; et al. A modified vaccinia Ankara vector-based vaccine protects macaques from SARS-CoV-2 infection, immune pathology, and dysfunction in the lungs. Immunity 2021, 54, 542–556.e9. [Google Scholar] [CrossRef]
- Liu, R.; Americo, J.L.; Cotter, C.A.; Earl, P.L.; Erez, N.; Peng, C.; Moss, B. MVA Vector Vaccines Inhibit SARS CoV-2 Replication in Upper and Lower Respiratory Tracts of Transgenic Mice and Prevent Lethal Disease. bioRxiv 2021. [Google Scholar] [CrossRef]
- Tscherne, A.; Schwarz, J.H.; Rohde, C.; Kupke, A.; Kalodimou, G.; Limpinsel, L.; Okba, N.M.A.; Bošnjak, B.; Sandrock, I.; Odak, I.; et al. Immunogenicity and efficacy of the COVID-19 candidate vector vaccine MVA-SARS-2-S in preclinical vaccination. Proc. Natl. Acad. Sci. USA 2021, 118, e2026207118. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.W.; Moran, E.; Adams, R.J.; Sievers, R.E.; Hauer, D.; Godin, S.; Griffin, D.E. A durable protective immune response to wild-type measles virus infection of macaques is due to viral replication and spread in lymphoid tissues. Sci. Transl. Med. 2020, 12, eaax7799. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.E. Measles Vaccine. Viral Immunol. 2018, 31, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Dravid, P.; Zhang, Y.; Trivedi, S.; Li, A.; Harder, O.; Kc, M.; Chaiwatpongsakorn, S.; Zani, A.; Kenney, A.; et al. A safe and highly efficacious measles virus-based vaccine expressing SARS-CoV-2 stabilized prefusion spike. Proc. Natl. Acad. Sci. USA 2021, 118, e2026153118. [Google Scholar] [CrossRef]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef]
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef]
- Li, X.; Cao, H.; Wang, Q.; Di, B.; Wang, M.; Lu, J.; Pan, L.; Yang, L.; Mei, M.; Pan, X.; et al. Novel AAV-based genetic vaccines encoding truncated dengue virus envelope proteins elicit humoral immune responses in mice. Microbes Infect. 2012, 14, 1000–1007. [Google Scholar] [CrossRef]
- Zhu, F.; Wang, Y.; Xu, Z.; Qu, H.; Zhang, H.; Niu, L.; Xue, H.; Jing, D.; He, H. Novel adeno-associated virus-based genetic vaccines encoding hepatitis C virus E2 glycoprotein elicit humoral immune responses in mice. Mol. Med. Rep. 2019, 19, 1016–1023. [Google Scholar] [CrossRef]
- Lin, J.; Calcedo, R.; Vandenberghe, L.H.; Bell, P.; Somanathan, S.; Wilson, J.M. A new genetic vaccine platform based on an adeno-associated virus isolated from a rhesus macaque. J. Virol. 2009, 83, 12738–12750. [Google Scholar] [CrossRef]
- Zabaleta, N.; Dai, W.; Bhatt, U.; Hérate, C.; Maisonnasse, P.; Chichester, J.A.; Sanmiguel, J.; Estelien, R.; Michalson, K.T.; Diop, C.; et al. An AAV-based, room-temperature-stable, single-dose COVID-19 vaccine provides durable immunogenicity and protection in non-human primates. Cell Host Microbe 2021, 29, 1437–1453.e8. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Luo, S.; Zhang, Y.; Ou, Y.; Wang, H.; Guo, Z.; He, C.; Bai, S.; He, P.; Jiang, M.; et al. Single-shot AAV-vectored vaccine against SARS-CoV-2 with fast and long-lasting immunity. Acta Pharm. Sin. B, 2022; in press. [Google Scholar]
- Ahmed, R.; Gray, D. Immunological memory and protective immunity: Understanding their relation. Science 1996, 272, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, J.; Perlman, S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010, 84, 9318–9325. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, N.; Kuo, H.H.; Boucau, J.; Farmer, J.R.; Allard-Chamard, H.; Mahajan, V.S.; Piechocka-Trocha, A.; Lefteri, K.; Osborn, M.; Bals, J.; et al. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell 2020, 183, 143–157.e13. [Google Scholar] [CrossRef]
- Seder, R.A.; Darrah, P.A.; Roederer, M. T-cell quality in memory and protection: Implications for vaccine design. Nat. Rev. Immunol. 2008, 8, 247–258. [Google Scholar] [CrossRef]
- Swain, S.L.; McKinstry, K.K.; Strutt, T.M. Expanding roles for CD4⁺ T cells in immunity to viruses. Nat. Rev. Immunol. 2012, 12, 136–148. [Google Scholar] [CrossRef]
- Wang, C.Y.; Lin, F.; Ding, S.; Peng, W.-J. Designer Peptides and Proteins for the Detection, Prevention and Treatment of Coronavirus Disease, 2019 (COVID 19). WO2021/168305A1.2020. WIPO. 2021. Available online: https://patentscope2.wipo.int/search/ja/detail.jsf?docId=WO2021168305 (accessed on 2 November 2022).
- Wang, C.Y.; Hwang, K.P.; Kuo, H.K.; Peng, W.J.; Shen, Y.H.; Kuo, B.S.; Huang, J.H.; Liu, H.; Ho, Y.H.; Lin, F.; et al. A multitope SARS-CoV-2 vaccine provides long-lasting B cell and T cell immunity against Delta and Omicron variants. J. Clin. Investig. 2022, 132, e157707. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zeng, J.; Li, R.; Wen, Z.; Cai, Y.; Wallin, J.; Shu, Y.; Du, X.; Sun, C. Rational Design of a Pan-Coronavirus Vaccine Based on Conserved CTL Epitopes. Viruses 2021, 13, 333. [Google Scholar] [CrossRef]
- Prakash, S.; Srivastava, R.; Coulon, P.G.; Dhanushkodi, N.R.; Chentoufi, A.A.; Tifrea, D.F.; Edwards, R.A.; Figueroa, C.J.; Schubl, S.D.; Hsieh, L.; et al. Genome-Wide B Cell, CD4(+), and CD8(+) T Cell Epitopes That Are Highly Conserved between Human and Animal Coronaviruses, Identified from SARS-CoV-2 as Targets for Preemptive Pan-Coronavirus Vaccines. J. Immunol. 2021, 206, 2566–2582. [Google Scholar] [CrossRef]
- Atmar, R.L.; Lyke, K.E.; Deming, M.E.; Jackson, L.A.; Branche, A.R.; El Sahly, H.M.; Rostad, C.A.; Martin, J.M.; Johnston, C.; Rupp, R.E.; et al. Homologous and Heterologous COVID-19 Booster Vaccinations. N. Engl. J. Med. 2022, 386, 1046–1057. [Google Scholar] [CrossRef]
| Type of Vaccine | Vaccine | Sponsor | Properties | Status |
|---|---|---|---|---|
| Booster immunization with approved vaccines | Comirnaty | Pfizer-BioNTechr | Boost used a bivalent vaccine using the same technology of approved vaccine | approved by FDA |
| Spikevax | Moderna | approved by FDA | ||
| Vaccines with more complex sources of spike protein RBD | SpFN | U.S. Army Medical Research and Development Command | Ferritin nanoparticle with prefusion- stabilized spike antigens from the Wuhan strain of SARS- CoV-2 | Phase 1 |
| Mosaic-4a/4b/8 | California Institute of Technology | Ferritin nanoparticle with prefusion- spike antigens from different Coronavirus | Preclinical | |
| Chimera 1 /4 | University of North Carolina at Chapel Hill | Chimeric spike mRNA vaccines with epitops come from differet Coronavirus | Preclinical | |
| Live attenuated viral vaccine delivered SARS-CoV-2 antigen | YF-S0 | KU Leuven Department of Microbiology, Belgium. | live-attenuated yellow fever 17D (YF17D) vaccine as a vector to express a noncleavable prefusion form of the SARS-CoV-2 spike antigen. | Preclinical |
| MVA-SARS-2-S | Universitätsklinikum Hamburg-Eppendorf | recombinant MVA expressing full-length SARS-CoV-2 spike (S) protein | Phase 1 | |
| rMeV-preS | The Ohio State University | measles virus (rMeV) vaccine strain as the backbone expressing SARS-CoV-2 spike (S) protein and its receptor binding domain (RBD) | Preclinical | |
| Subunit vaccines with T-cell epitopes | UB-612 | United Biomedical Inc., Asia | recombinant subunit vaccine expressing the spike 1 (S1)-RBD fused with a single-chain Fc protein (S1-RBD-sFc), 5 Th cell, and CTL epitope peptides from the nucleocapsid (N), membrane (M), and S2 proteins of sarbecoviruses | Phase 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Wei, Y.; Yang, H.; Yan, J.; Li, X.; Li, Z.; Zhao, Y.; Liang, H.; Wang, H. Advances in Next-Generation Coronavirus Vaccines in Response to Future Virus Evolution. Vaccines 2022, 10, 2035. https://doi.org/10.3390/vaccines10122035
Li L, Wei Y, Yang H, Yan J, Li X, Li Z, Zhao Y, Liang H, Wang H. Advances in Next-Generation Coronavirus Vaccines in Response to Future Virus Evolution. Vaccines. 2022; 10(12):2035. https://doi.org/10.3390/vaccines10122035
Chicago/Turabian StyleLi, Lili, Yangyang Wei, Han Yang, Junyu Yan, Xin Li, Ziqian Li, Yuxiu Zhao, Hongyang Liang, and Hui Wang. 2022. "Advances in Next-Generation Coronavirus Vaccines in Response to Future Virus Evolution" Vaccines 10, no. 12: 2035. https://doi.org/10.3390/vaccines10122035
APA StyleLi, L., Wei, Y., Yang, H., Yan, J., Li, X., Li, Z., Zhao, Y., Liang, H., & Wang, H. (2022). Advances in Next-Generation Coronavirus Vaccines in Response to Future Virus Evolution. Vaccines, 10(12), 2035. https://doi.org/10.3390/vaccines10122035




