Variants of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Vaccine Effectiveness
Abstract
1. Introduction
2. Structure and Genome Organization of SARS-CoV-2
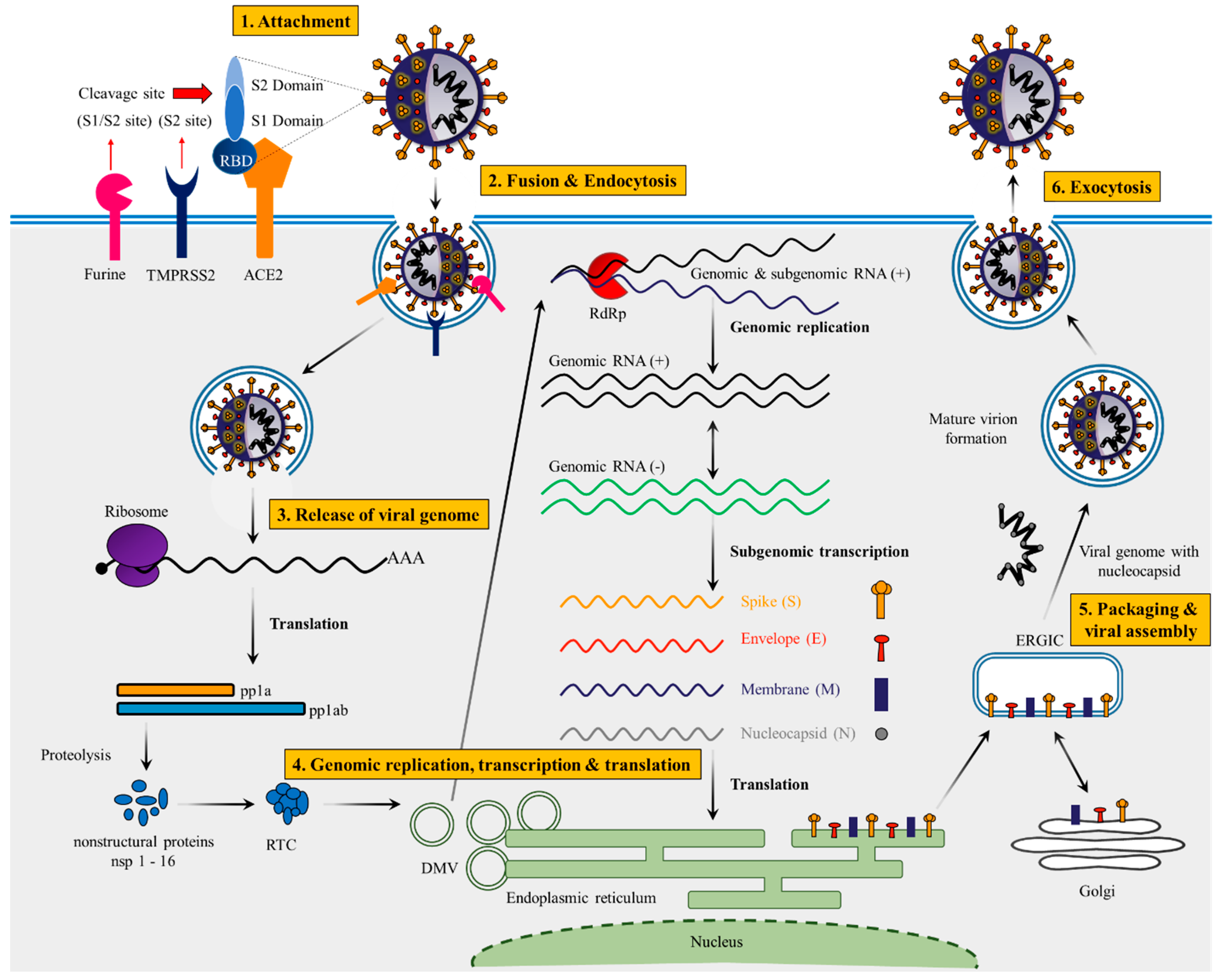
3. Lifecycle of SARS-CoV-2
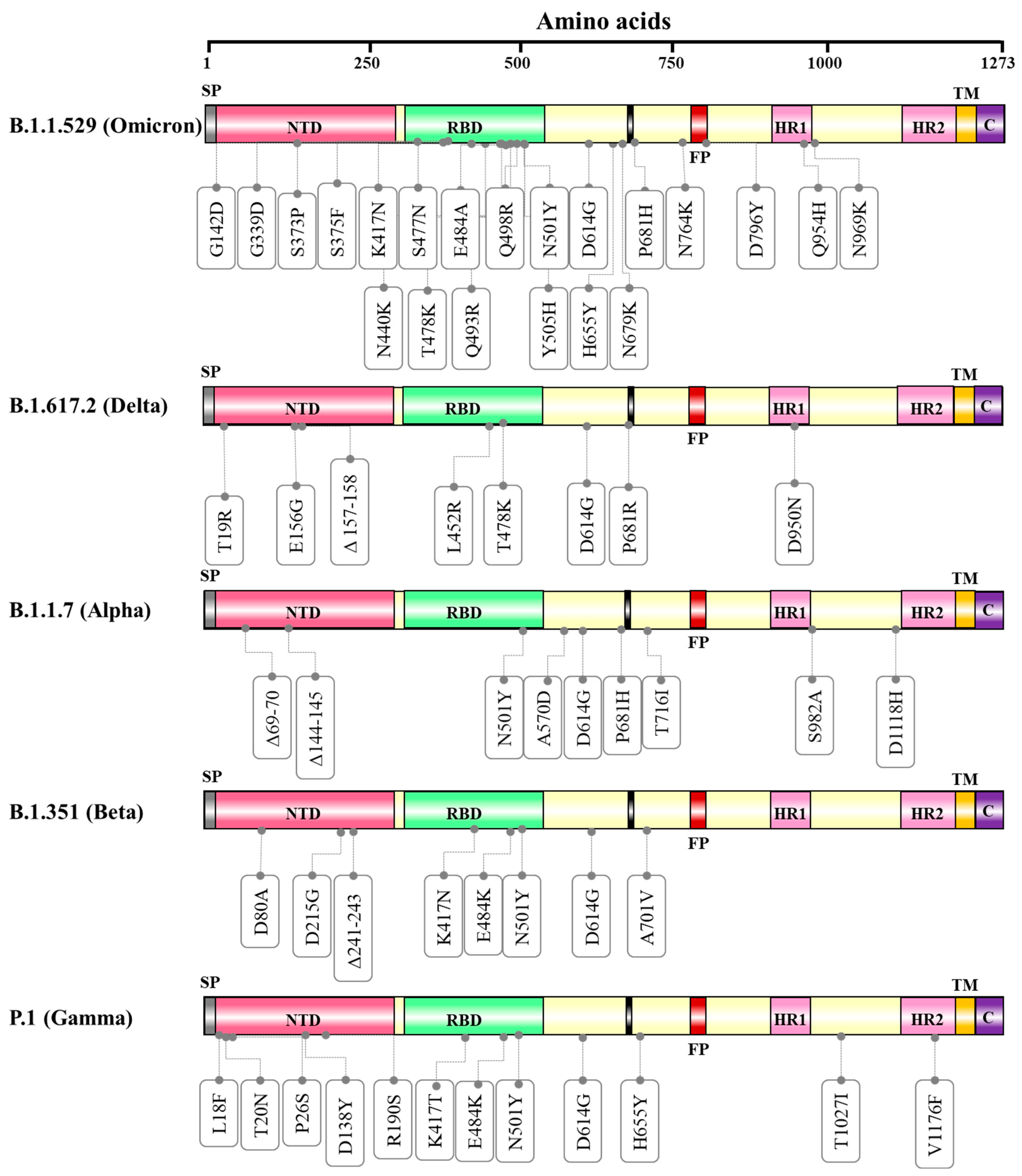
4. Variants of SARS-CoV-2
4.1. SARS-CoV-2 Variant of Concern
The B.1.1.529 (Omicron Variant)
4.2. SARS-CoV-2 Variants Being Monitored
B.1.617.2 Lineage (Delta Variant)
5. Vaccine Candidates Approved against SARS-CoV-2 Infection
5.1. Messenger RNA (mRNA) Vaccines
5.1.1. Pfizer–BioNTech Vaccine (Comirnaty (Formerly BNT162b2))
5.1.2. Moderna Vaccine (MV) (mRNA-1273)
5.2. Human Adenovirus Non-Replicating and Replicating Vector-Based Vaccines
5.2.1. Oxford–AstraZeneca Vaccine (AZD 1222)
5.2.2. Sputnik-V Vaccine (SVV)
5.2.3. AD5-nCoV Vaccine (PakVac, Ad5-nCoV)
5.2.4. Ad26.COV2.S/Janssen (JNJ-78436735; Ad26.COV2.S)
5.3. Inactivated Coronavirus Vaccines
5.3.1. Sinopharm Vaccine (BBIBP-CorV)
5.3.2. SARS-CoV-2 Vaccine (Vero Cells)
5.3.3. Sinopharm-Wuhan Vaccine (WIBP-CorV)
5.3.4. CoviVac
5.3.5. CoronaVac Vaccine
5.3.6. Inactivated SARS-CoV-2 Vaccine (Vero Cell)
5.3.7. Covaxin (BBV152)
5.3.8. QazVac (QazCovid-in)
5.3.9. COVIran
5.4. Recombinant Protein Subunit Vaccines
5.4.1. EpiVacCorona Vaccine (EVCV)
5.4.2. ZF2001 ((RBD Dimer) ZIFIVAX)
5.4.3. Abdala Vaccine (CIGB 66)
5.4.4. Soberana Vaccine
5.4.5. MVC COVID (MVC-COV1901)
5.4.6. Zycov-D
5.4.7. Spikogen
5.4.8. Fakhravac
5.4.9. Nuvaxovid
5.4.10. Turkovac
5.4.11. Carbovax
5.4.12. Covifenz
5.4.13. VLA2001
5.4.14. Noora
| S. No | Vaccine Name | Type of Vaccine | Primary Developers | Vaccine Efficacy on SARS-CoV-2 Wild Type or Variants | Primary End Point or Outcome | Number of Doses of Vaccination |
|---|---|---|---|---|---|---|
| 1 | BNT162b2 (Comirnaty) | mRNA-based vaccine | Pfizer, BioNTech; Fosun Pharma | SARS-CoV-2: 95% [76] Omicron: 70% [22] | Safety over a median of 2 months was similar to that of other viral vaccines | Two doses |
| 2 | mRNA-1273 (Spikevax) | mRNA-based vaccine | Moderna, BARDA, NIAID | SARS-CoV-2: 94.1% [130] Omicron: 85% [132] | The primary end point was prevention of COVID-19 illness with onset at least 14 days after the second injection in participants who had not previously been infected with SARS-CoV-2. | Two doses |
| 3 | AstraZeneca (AZD1222 also known as Vaxzevria and Covishield | Adenovirus vaccine | BARDA, OWS | 74% (Overall) and an efficacy of 83.5% in participants age 65 years and older [140] | Preventing the onset of symptomatic and severe coronavirus disease 2019 (COVID-19) 15 days or more after the second dose in adults, including older adults | Two doses |
| 4 | Sputnik V (Gam-COVID-Vac) | Recombinant adenovirus vaccine (rAd26 and rAd5) | Gamaleya Research Institute, Acellena Contract Drug Research and Development | 91.6% [143] | The primary outcome was the proportion of participants with PCR-confirmed COVID-19 from day 21 after receiving the first dose. | Two doses |
| 5 | Janssen (JNJ-78436735; Ad26.COV2.S) | Non-replicating viral vector | Janssen Vaccines (Johnson & Johnson) | 52.9% against moderate or severe-to-critical COVID-19 and 41.7% against any infection [150] | The primary end points were vaccine efficacy against moderate to severe–critical COVID-19 with onset at least 14 days after administration | Single dose |
| 6 | CoronaVac | Inactivated vaccine (formalin with alum adjuvant) | Sinovac | 65.9% for the prevention of COVID-19 and 87.5% for the prevention of hospitalization, 90.3% for the prevention of ICU admission, and 86.3% for the prevention of COVID-19–related death [161] | Estimated the change in the hazard ratio associated with partial immunization (≥14 days after receipt of the first dose and before receipt of the second dose) and full immunization (≥14 days after receipt of the second dose) | Both Single and Double dose |
| 7 | BBIBP-CorV (Vero Cells) also called as Covilo | Inactivated vaccine | Beijing Institute of Biological Products; China National Pharmaceutical Group (Sinopharm) | 78.1% [155] | Primary outcome was efficacy against laboratory-confirmed symptomatic COVID-19 14 days following a second vaccine dose among participants who had no virologic evidence of SARS-CoV-2 infection at randomization. The secondary outcome was efficacy against severe COVID-19. | Two Doses |
| 8 | Convidicea (PakVac, Ad5-nCoV) | Recombinant vaccine (adenovirus type 5 vector) | CanSino Biologics | 57.5% [148] | The primary efficacy objective evaluated Ad5-nCoV in preventing symptomatic, PCR-confirmed COVID-19 infection occurring at least 28 days after vaccination in all participants who were at least 28 days postvaccination | Single dose |
| 9 | Covaxin (BBV152) | Inactivated vaccine | Bharat Biotech, ICMR; Ocugen; ViroVax | 77.8% [165] | The primary outcome was the efficacy of the BBV152 vaccine in preventing a first occurrence of laboratory-confirmed (RT-PCR-positive) symptomatic COVID-19 (any severity), occurring at least 14 days after the second dose in the per-protocol population. | Two Doses |
| 10 | WIBP-CorV | Inactivated vaccine | Wuhan Institute of Biological Products; China National Pharmaceutical Group (Sinopharm) | 72.8% [155] | Primary outcome was efficacy against laboratory-confirmed symptomatic COVID-19 14 days following a second vaccine dose among participants who had no virologic evidence of SARS-CoV-2 infection at randomization. The secondary outcome was efficacy against severe COVID-19. | Two Doses |
| 11 | Zycov-D | DNA vaccine (plasmid) | Zydus Cadila | 66.6% [186] | The primary outcome was the number of participants with first occurrence of symptomatic RT-PCR-positive COVID-19 28 days after the third dose | Three doses |
| 12 | Nuvaxovid (Covovax in India; previously NVX-CoV2373) | Recombinant nanoparticle vaccine | Novavax CEPI, Serum Institute of India | 89.7% [190] | The primary efficacy end point was virologically confirmed mild, moderate, or severe SARS-CoV-2 infection with an onset at least 7 days after the second injection in participants who were serologically negative at baseline. | Two doses |
| 13 | Covifenz (CoVLP) | Plant-based adjuvant vaccine | Medicago; GSK; Dynavax | 78.8% [195] | The primary objective of the trial was to determine the efficacy of the CoVLP+AS03 vaccine in preventing symptomatic coronavirus disease 2019 (COVID-19) beginning at least 7 days after the second injection | Two Doses |
| 14 | Soberana 02/Soberana Plus | Conjugate vaccine | Finlay Institute of Vaccines; Pasteur Institute | 92.4% [182] | Study endpoints are vaccine efficacy (VE) evaluated through confirmed symptomatic COVID-19 and safety | Three doses |
6. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 29 August 2021).
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2-What Do They Mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Almubaid, Z.; Al-Mubaid, H. Analysis and comparison of genetic variants and mutations of the novel coronavirus SARS-CoV-2. Gene Rep. 2021, 23, 101064. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Gribble, J.; Stevens, L.J.; Agostini, M.L.; Anderson-Daniels, J.; Chappell, J.D.; Lu, X.; Pruijssers, A.J.; Routh, A.L.; Denison, M.R. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021, 17, e1009226. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e819. [Google Scholar] [CrossRef]
- World Health Organization. An Update on SARS-CoV-2 Virus Mutations & Variants. Available online: https://www.who.int/docs/default-source/coronaviruse/risk-comms-updates/update47-sars-cov-2-variants.pdf?sfvrsn=f2180835_4 (accessed on 25 March 2021).
- Centers for Disease Control and Prevention. About Variants of the Virus that Causes COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant.html (accessed on 25 March 2021).
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Chakraborty, S. Evolutionary and structural analysis elucidates mutations on SARS-CoV2 spike protein with altered human ACE2 binding affinity. Biochem. Biophys. Res. Commun. 2021, 534, 374–380. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H., 3rd; Leist, S.R.; Schafer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef]
- Burki, T. Understanding variants of SARS-CoV-2. Lancet 2021, 397, 462. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fvariants%2Fvariant-info.html (accessed on 25 August 2021).
- World Health Organization. Tracking SARS-CoV-2 variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 25 August 2021).
- Rambaut, A.; Holmes, E.C.; O’Toole, A.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Eng. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Shinde, V.; Bhikha, S.; Hoosain, Z.; Archary, M.; Bhorat, Q.; Fairlie, L.; Lalloo, U.; Masilela, M.S.; Moodley, D.; Hanley, S. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B. 1.351 variant. N. Eng. J. Med. 2021, 384, 1899–1909. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B. Safety and efficacy of single-dose Ad26. COV2. S vaccine against Covid-19. N. Eng. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef] [PubMed]
- Abu-Raddad, L.J.; Chemaitelly, H.; Butt, A.A.; National Study Group for COVID-19 Vaccination. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. N. Eng. J. Med. 2021, 385, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: An observational study using national surveillance data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef]
- Collie, S.; Champion, J.; Moultrie, H.; Bekker, L.-G.; Gray, G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N. Eng. J. Med. 2022, 386, 494–496. [Google Scholar] [CrossRef]
- Lauring, A.S.; Tenforde, M.W.; Chappell, J.D.; Gaglani, M.; Ginde, A.A.; McNeal, T.; Ghamande, S.; Douin, D.J.; Talbot, H.K.; Casey, J.D.; et al. Clinical Severity and mRNA Vaccine Effectiveness for Omicron, Delta, and Alpha SARS-CoV-2 Variants in the United States: A Prospective Observational Study. medRxiv 2022. [Google Scholar] [CrossRef]
- Li, X.; Zai, J.; Wang, X.; Li, Y. Potential of large "first generation" human-to-human transmission of 2019-nCoV. J. Med. Virol. 2020, 92, 448–454. [Google Scholar] [CrossRef]
- Gralinski, L.E.; Menachery, V.D. Return of the Coronavirus: 2019-nCoV. Viruses 2020, 12, 135. [Google Scholar] [CrossRef]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Renia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. Author Correction: A new coronavirus associated with human respiratory disease in China. Nature 2020, 580, E7. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Temmam, S.; Montagutelli, X.; Hérate, C.; Donati, F.; Regnault, B.; Attia, M.; Salazar, E.B.; Chrétien, D.; Conquet, L.; Jouvion, G. SARS-CoV-2-Related Bat Virus in Human Relevant Models Sheds Light on the Proximal Origin of COVID-19. Res. Sq. 2022. preprint. Available online: https://assets.researchsquare.com/files/rs-1803095/v1_covered.pdf?c=1657179682 (accessed on 1 July 2022).
- Jiang, X.; Wang, R. Wildlife trade is likely the source of SARS-CoV-2. Science 2022, 377, 925–926. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Li, F. Receptor recognition mechanisms of coronaviruses: A decade of structural studies. J. Virol. 2015, 89, 1954–1964. [Google Scholar] [CrossRef]
- Li, W.; Greenough, T.C.; Moore, M.J.; Vasilieva, N.; Somasundaran, M.; Sullivan, J.L.; Farzan, M.; Choe, H. Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 2004, 78, 11429–11433. [Google Scholar] [CrossRef]
- Li, W.; Zhang, C.; Sui, J.; Kuhn, J.H.; Moore, M.J.; Luo, S.; Wong, S.K.; Huang, I.C.; Xu, K.; Vasilieva, N.; et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005, 24, 1634–1643. [Google Scholar] [CrossRef]
- McCray, P.B., Jr.; Pewe, L.; Wohlford-Lenane, C.; Hickey, M.; Manzel, L.; Shi, L.; Netland, J.; Jia, H.P.; Halabi, C.; Sigmund, C.D.; et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007, 81, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 2019, 105, 93–116. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Sun, J.; Yan, Z.; Zhang, J.; Zhao, J.; Zhao, Z.; Gao, Q.; He, W.T.; Veit, M.; Su, S. Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.A. Properties of Coronavirus and SARS-CoV-2. Malays J. Pathol. 2020, 42, 3–11. [Google Scholar] [PubMed]
- Nakagawa, K.; Lokugamage, K.G.; Makino, S. Viral and Cellular mRNA Translation in Coronavirus-Infected Cells. Adv. Virus Res. 2016, 96, 165–192. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef] [PubMed]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Denison, M.R.; Perlman, S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J. Virol. 1986, 60, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Kamitani, W.; Narayanan, K.; Huang, C.; Lokugamage, K.; Ikegami, T.; Ito, N.; Kubo, H.; Makino, S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 2006, 103, 12885–12890. [Google Scholar] [CrossRef] [PubMed]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.A.; Leibundgut, M.; Thiel, V.; Muhlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020, 27, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Sims, A.C.; Ostermann, J.; Denison, M.R. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 2000, 74, 5647–5654. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar] [CrossRef]
- V’Kovski, P.; Gerber, M.; Kelly, J.; Pfaender, S.; Ebert, N.; Braga Lagache, S.; Simillion, C.; Portmann, J.; Stalder, H.; Gaschen, V.; et al. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife 2019, 8. [Google Scholar] [CrossRef]
- Oudshoorn, D.; Rijs, K.; Limpens, R.; Groen, K.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Barcena, M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Knoops, K.; Kikkert, M.; Worm, S.H.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.J.; Hawes, P.C.; Cottam, E.M.; Mantell, J.; Verkade, P.; Monaghan, P.; Wileman, T.; Britton, P. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio 2013, 4, e00801–e00813. [Google Scholar] [CrossRef]
- Snijder, E.J.; Limpens, R.; de Wilde, A.H.; de Jong, A.W.M.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.; Koster, A.J.; Barcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef]
- Ulasli, M.; Verheije, M.H.; de Haan, C.A.; Reggiori, F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010, 12, 844–861. [Google Scholar] [CrossRef]
- Ma, H.C.; Fang, C.P.; Hsieh, Y.C.; Chen, S.C.; Li, H.C.; Lo, S.Y. Expression and membrane integration of SARS-CoV M protein. J. Biomed. Sci. 2008, 15, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014, 5, 296. [Google Scholar] [CrossRef] [PubMed]
- Krijnse-Locker, J.; Ericsson, M.; Rottier, P.J.; Griffiths, G. Characterization of the budding compartment of mouse hepatitis virus: Evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 1994, 124, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Tooze, J.; Tooze, S.; Warren, G. Replication of coronavirus MHV-A59 in sac- cells: Determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 1984, 33, 281–293. [Google Scholar] [PubMed]
- de Haan, C.A.; Rottier, P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005, 64, 165–230. [Google Scholar] [CrossRef]
- Graham, R.L.; Becker, M.M.; Eckerle, L.D.; Bolles, M.; Denison, M.R.; Baric, R.S. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012, 18, 1820–1826. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Eng. J. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Eng. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). SARS-CoV-2 Variant Classifications and Definitions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html (accessed on 1 July 2022).
- World Health Organization. COVID-19 vaccine tracker and landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 18 January 2022).
- World Health Organization. Enhancing response to Omicron SARS-CoV-2 variant. Available online: https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states (accessed on 21 January 2022).
- Qin, S.; Cui, M.; Sun, S.; Zhou, J.; Du, Z.; Cui, Y.; Fan, H. Genome characterization and potential risk assessment of the novel SARS-CoV-2 variant Omicron (B. 1.1. 529). Zoonoses 2021, 1, 1–5. [Google Scholar]
- Torjesen, I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ 2021, 375, 2943. [Google Scholar]
- Espenhain, L.; Funk, T.; Overvad, M.; Edslev, S.M.; Fonager, J.; Ingham, A.C.; Rasmussen, M.; Madsen, S.L.; Espersen, C.H.; Sieber, R.N.; et al. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark, December 2021. Eurosurveillance 2021, 26. [Google Scholar] [CrossRef]
- Callaway, E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021, 600, 21. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Science Brief: Omicron (B.1.1.529) variant. Available online: https://www.ncbi.nlm.nih.gov/books/NBK575856/ (accessed on 4 December 2021).
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Greasley, S.E.; Noell, S.; Plotnikova, O.; Ferre, R.A.; Liu, W.; Bolanos, B.; Fennell, K.F.; Nicki, J.; Craig, T.; Zhu, Y. Structural basis for Nirmatrelvir in vitro efficacy against the Omicron variant of SARS-CoV-2. BioRxiv 2022. [Google Scholar]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.M.; et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Eng. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Pulliam, J.R.; van Schalkwyk, C.; Govender, N.; von Gottberg, A.; Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science 2022, 376, eabn4947. [Google Scholar] [CrossRef]
- Hossain, M.G.; Tang, Y.D.; Akter, S.; Zheng, C. Roles of the polybasic furin cleavage site of spike protein in SARS-CoV-2 replication, pathogenesis, and host immune responses and vaccination. J. Med. Virol. 2022, 94, 1815–1820. [Google Scholar] [CrossRef]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhou, L.; Mo, M.; Li, Y.; Han, J.; Li, J.; Yang, Y.; Zhang, X.; Gong, C.; Lu, K. The effect of the multiple mutations in Omicron RBD on its binding to human ACE2 receptor and immune evasion: An investigation of molecular dynamics simulations. 2021. 2021. Available online: https://chemrxiv.org/engage/chemrxiv/article-details/61acb409bc299c44248a2b12 (accessed on 16 May 2022).
- Outbreak.info. Omicron Variant Report. Available online: https://outbreak.info/situation-reports/omicron?loc=ZAF&loc=GBR&loc=USA&selected=ZAF (accessed on 16 May 2022).
- Wang, L.; Cheng, G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J. Med. Virol. 2022, 94, 1728–1733. [Google Scholar] [CrossRef] [PubMed]
- Maxmen, A. Why call it BA. 2.12. 1? A guide to the tangled Omicron family. Nature 2022. [Google Scholar] [CrossRef] [PubMed]
- Yamasoba, D.; Kimura, I.; Nasser, H.; Morioka, Y.; Nao, N.; Ito, J.; Uriu, K.; Tsuda, M.; Zahradnik, J.; Shirakawa, K. Virological characteristics of the SARS-CoV-2 Omicron BA. 2 spike. Cell 2022. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Karuppanan, K.; Subramaniam, G. Omicron (BA. 1) and Sub-Variants (BA. 1.1, BA. 2 and BA. 3) of SARS-CoV-2 Spike Infectivity and Pathogenicity: A Comparative Sequence and Structural-based Computational Assessment. J. Med. Virol. 2022, 94, 4780–4791. [Google Scholar]
- Desingu, P.A.; Nagarajan, K.; Dhama, K. Emergence of Omicron third lineage BA. 3 and its importance. J. Med. Virol. 2022, 94, 1808–1810. [Google Scholar] [CrossRef]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X. BA. 2.12. 1, BA. 4 and BA. 5 escape antibodies elicited by Omicron infection. Nature 2022, 1–10. [Google Scholar]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.G. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022, 602, 654–656. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, L.; Mo, M.; Liu, T.; Wu, C.; Gong, C.; Lu, K.; Gong, L.; Zhu, W.; Xu, Z. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Target. Ther. 2022, 7, 1–3. [Google Scholar] [CrossRef]
- Bruel, T.; Hadjadj, J.; Maes, P.; Planas, D.; Seve, A.; Staropoli, I.; Guivel-Benhassine, F.; Porrot, F.; Bolland, W.-H.; Nguyen, Y. Serum neutralization of SARS-CoV-2 Omicron sublineages BA. 1 and BA. 2 in patients receiving monoclonal antibodies. Nat. Med. 2022, 1–6. [Google Scholar] [CrossRef]
- Saied, A.A.; Metwally, A.A.; Dhawan, M.; Choudhary, O.P.; Aiash, H. Strengthening vaccines and medicines manufacturing capabilities in Africa: Challenges and perspectives. EMBO Molec. Med. 2022, 14, e16287. [Google Scholar] [CrossRef]
- Callaway, E. What Omicron’s BA. 4 and BA. 5 variants mean for the pandemic. Nature 2022. [Google Scholar] [CrossRef]
- Quandt, J.; Muik, A.; Salisch, N.; Lui, B.G.; Lutz, S.; Krüger, K.; Wallisch, A.-K.; Adams-Quack, P.; Bacher, M.; Finlayson, A. Omicron BA. 1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2021, 7, eabq2427. [Google Scholar]
- Park, Y.-J.; Pinto, D.; Walls, A.C.; Liu, Z.; De Marco, A.; Benigni, F.; Zatta, F.; Silacci-Fregni, C.; Bassi, J.; Sprouse, K.R. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. bioRxiv 2022. [Google Scholar]
- Dhawan, M.; Saied, A.A.; Emran, T.B.; Choudhary, O.P. Emergence of omicron variant’s sublineages BA. 4 and BA. 5: Risks assessment and possible countermeasures. N. Microbes N.Infect. 2022, 48, 100997. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, R.K.; Kandi, V.; Sarangi, A.K.; Verma, S.; Tuli, H.S.; Chakraborty, S.; Chakraborty, C.; Dhama, K. The recently emerged BA. 4 and BA. 5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic–Correspondence. Int. J. Surg. 2022, 103, 106698. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Delta coronavirus variant: Scientists brace for impact. Nature 2021, 595, 17–18. [Google Scholar] [CrossRef]
- Peacock, T.P.; Brown, J.C.; Zhou, J.; Thakur, N.; Newman, J.; Kugathasan, R.; Sukhova, K.; Kaforou, M.; Bailey, D.; Barclay, W.S. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. BioRxiv 2012. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9, 1542. [Google Scholar] [CrossRef]
- Singanayagam, A.; Hakki, S.; Dunning, J.; Madon, K.J.; Crone, M.A.; Koycheva, A.; Derqui-Fernandez, N.; Barnett, J.L.; Whitfield, M.G.; Varro, R. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B. 1.617. 2) variant in vaccinated and unvaccinated individuals in the UK: A prospective, longitudinal, cohort study. Lancet Infect. Dis. 2022, 22, 183–195. [Google Scholar] [CrossRef]
- Danner, C.; Rosa-Aquino, P. What we know about the dangerous COVID B. 1.617. 2 (Delta) variant NY Intelligencer. 2021. Available online: https://nymag.com/intelligencer/article/covid-b-1-617-2-delta-variant-what-we-know.html. (accessed on 22 May 2022).
- Elbanna, A.; Goldenfeld, N. Frequency of surveillance testing necessary to reduce transmission of the Delta variant of SARS-CoV-2. medRxiv 2021. [Google Scholar]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv 2021, arXiv:10.1016/j.celrep.2022.110829. [Google Scholar]
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.-H.; Porrot, F.; Staropoli, I.; Lemoine, F. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Sharma, A.R. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: Their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev. Med. Virol. 2022, 32, e2270. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Uriu, K.; Kimura, I.; Shirakawa, K.; Takaori-Kondo, A.; Nakada, T.-a.; Kaneda, A.; Nakagawa, S.; Sato, K. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N. Eng. J. Med. 2021, 385, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J.L.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G. Effectiveness of Covid-19 vaccines against the B. 1.617. 2 (Delta) variant. N. Eng. J. Med. 2021, 385, 585–594. [Google Scholar]
- Mullard, A. COVID-19 vaccine development pipeline gears up. Lancet 2020, 395, 1751–1752. [Google Scholar] [CrossRef]
- Wang, F.; Kream, R.M.; Stefano, G.B. An Evidence Based Perspective on mRNA-SARS-CoV-2 Vaccine Development. Med. Sci. Monit. 2020, 26, e924700. [Google Scholar] [CrossRef]
- Alfagih, I.M.; Aldosari, B.; AlQuadeib, B.; Almurshedi, A.; Alfagih, M.M. Nanoparticles as Adjuvants and Nanodelivery Systems for mRNA-Based Vaccines. Pharmaceutics 2020, 13, 45. [Google Scholar] [CrossRef]
- Amit, S.; Regev-Yochay, G.; Afek, A.; Kreiss, Y.; Leshem, E. Early rate reductions of SARS-CoV-2 infection and COVID-19 in BNT162b2 vaccine recipients. Lancet 2021, 397, 875–877. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Eng. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Price, A.M.; Olson, S.M.; Newhams, M.M.; Halasa, N.B.; Boom, J.A.; Sahni, L.C.; Pannaraj, P.S.; Irby, K.; Bline, K.E.; Maddux, A.B.; et al. BNT162b2 Protection against the Omicron Variant in Children and Adolescents. N. Eng. J. Med. 2022, 386, 1899–1909. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Eng. J. Med. 2020. [Google Scholar] [CrossRef]
- Grannis, S.J.; Rowley, E.A.; Ong, T.C.; Stenehjem, E.; Klein, N.P.; DeSilva, M.B.; Naleway, A.L.; Natarajan, K.; Thompson, M.G.; Network, V. Interim estimates of COVID-19 vaccine effectiveness against COVID-19–associated emergency department or urgent care clinic encounters and hospitalizations among adults during SARS-CoV-2 B. 1.617. 2 (Delta) variant predominance—nine states, June–August 2021. Morb. Mortal. Weekly Rep. 2021, 70, 1291. [Google Scholar] [CrossRef] [PubMed]
- Pajon, R.; Doria-Rose, N.A.; Shen, X.; Schmidt, S.D.; O’Dell, S.; McDanal, C.; Feng, W.; Tong, J.; Eaton, A.; Maglinao, M. SARS-CoV-2 Omicron variant neutralization after mRNA-1273 booster vaccination. N. Eng. J. of Med. 2022, 386, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Werner, A.P.; Koch, M.; Choi, A.; Narayanan, E.; Stewart-Jones, G.B.E.; Colpitts, T.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Eng. J. Med. 2021, 384, 1468–1470. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K. Application of Viral Vectors for Vaccine Development with a Special Emphasis on COVID-19. Viruses 2020, 12, 1324. [Google Scholar] [CrossRef]
- Zhu, F.C.; Li, Y.H.; Guan, X.H.; Hou, L.H.; Wang, W.J.; Li, J.X.; Wu, S.P.; Wang, B.S.; Wang, Z.; Wang, L.; et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Kyriakidis, N.C.; Lopez-Cortes, A.; Gonzalez, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. NPJ Vaccines 2021, 6, 28. [Google Scholar] [CrossRef]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Wise, J. Covid-19: The E484K mutation and the risks it poses. BMJ 2021, 372, n359. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Falsey, A.R.; Sobieszczyk, M.E.; Hirsch, I.; Sproule, S.; Robb, M.L.; Corey, L.; Neuzil, K.M.; Hahn, W.; Hunt, J.; Mulligan, M.J. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 vaccine. N. Eng. J. Med. 2021, 385, 2348–2360. [Google Scholar] [CrossRef]
- Emary, K.R.; Golubchik, T.; Aley, P.K.; Ariani, C.V.; Angus, B.; Bibi, S.; Blane, B.; Bonsall, D.; Cicconi, P.; Charlton, S. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B. 1.1. 7): An exploratory analysis of a randomised controlled trial. The Lancet 2021, 397, 1351–1362. [Google Scholar] [CrossRef]
- Jones, I.; Roy, P. Sputnik V COVID-19 vaccine candidate appears safe and effective. Lancet 2021, 397, 642–643. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatulin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- Rossi, A.H.; Ojeda, D.S.; Varese, A.; Sanchez, L.; Ledesma, M.M.G.L.; Mazzitelli, I.; Juliá, A.A.; Rouco, S.O.; Pallarés, H.M.; Navarro, G.S.C. Sputnik V vaccine elicits seroconversion and neutralizing capacity to SARS-CoV-2 after a single dose. Cell Rep. Med. 2021, 2, 100359. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, V.A.; Dolzhikova, I.V.; Shchetinin, A.M.; Odintsova, A.S.; Siniavin, A.E.; Nikiforova, M.A.; Pochtovyi, A.A.; Shidlovskaya, E.V.; Kuznetsova, N.A.; Burgasova, O.A. Neutralizing activity of sera from Sputnik V-vaccinated people against variants of concern (VOC: B. 1.1. 7, B. 1.351, P. 1, B. 1.617. 2, B. 1.617. 3) and Moscow endemic SARS-CoV-2 variants. Vaccines 2021, 9, 779. [Google Scholar] [CrossRef]
- Zhu, F.C.; Guan, X.H.; Li, Y.H.; Huang, J.Y.; Jiang, T.; Hou, L.H.; Li, J.X.; Yang, B.F.; Wang, L.; Wang, W.J.; et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Halperin, S.A.; Ye, L.; MacKinnon-Cameron, D.; Smith, B.; Cahn, P.E.; Ruiz-Palacios, G.M.; Ikram, A.; Lanas, F.; Guerrero, M.L.; Navarro, S.R.l.M.o. Final efficacy analysis, interim safety analysis, and immunogenicity of a single dose of recombinant novel coronavirus vaccine (adenovirus type 5 vector) in adults 18 years and older: An international, multicentre, randomised, double-blinded, placebo-controlled phase 3 trial. Lancet 2022, 399, 237–248. [Google Scholar]
- Livingston, E.H.; Malani, P.N.; Creech, C.B. The Johnson & Johnson Vaccine for COVID-19. JAMA 2021, 325, 1575. [Google Scholar] [CrossRef]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Van Dromme, I.; Spiessens, B. Final analysis of efficacy and safety of single-dose Ad26. COV2. S. N. Eng. J. Med. 2022, 386, 847–860. [Google Scholar] [CrossRef]
- Barouch, D.H.; Stephenson, K.E.; Sadoff, J.; Yu, J.; Chang, A.; Gebre, M.; McMahan, K.; Liu, J.; Chandrashekar, A.; Patel, S. Durable humoral and cellular immune responses 8 months after Ad26. COV2. S vaccination. N. Eng. J. Med. 2021, 385, 951–953. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mendonca, L.; Yang, Y.; Gao, Y.; Shen, C.; Liu, J.; Ni, T.; Ju, B.; Liu, C.; Tang, X.; et al. The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET. Structure 2020, 28, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Duan, K.; Zhang, Y.; Zhao, D.; Zhang, H.; Xie, Z.; Li, X.; Peng, C.; Zhang, Y.; Zhang, W.; et al. Effect of an Inactivated Vaccine Against SARS-CoV-2 on Safety and Immunogenicity Outcomes: Interim Analysis of 2 Randomized Clinical Trials. JAMA 2020, 324, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.F.; Tan, W.; Wu, G.; Xu, M.; Lou, Z. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021, 21, 39–51. [Google Scholar] [CrossRef]
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J. Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: A randomized clinical trial. Jama 2021, 326, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Jiang, Q.; Wang, Y.; Yang, J.; Du, T.; Yi, H.; Li, C.; Li, Y.; Fan, S.; Liao, Y. SARS-CoV-2 inactivated vaccine (Vero cells) shows good safety in repeated administration toxicity test of Sprague Dawley rats. Food Chem. Toxicol. 2021, 152, 112239. [Google Scholar] [CrossRef]
- Barrett, P.N.; Terpening, S.J.; Snow, D.; Cobb, R.R.; Kistner, O. Vero cell technology for rapid development of inactivated whole virus vaccines for emerging viral diseases. Expert Rev. Vaccines 2017, 16, 883–894. [Google Scholar] [CrossRef]
- Pu, J.; Yu, Q.; Yin, Z.; Zhang, Y.; Li, X.; Yin, Q.; Chen, H.; Long, R.; Zhao, Z.; Mou, T. The safety and immunogenicity of an inactivated SARS-CoV-2 vaccine in Chinese adults aged 18–59 years: A phase I randomized, double-blinded, controlled trial. Vaccine 2021, 39, 2746–2754. [Google Scholar] [CrossRef]
- Kozlovskaya, L.I.; Gordeychuk, I.; Piniaeva, A.; Kovpak, A.; Shishova, A.; Lunin, A.; Shustova, E.; Apolokhov, V.; Fominykh, K.; Ivin, Y. CoviVac vaccination induces production of neutralizing antibodies against Delta and Omicron variants of SARS-CoV-2. medRxiv 2022. [Google Scholar]
- Wang, G.-L.; Wang, Z.-Y.; Duan, L.-J.; Meng, Q.-C.; Jiang, M.-D.; Cao, J.; Yao, L.; Zhu, K.-L.; Cao, W.-C.; Ma, M.-J. Susceptibility of circulating SARS-CoV-2 variants to neutralization. N. Eng. J. Med. 2021, 384, 2354–2356. [Google Scholar] [CrossRef]
- Jara, A.; Undurraga, E.A.; González, C.; Paredes, F.; Fontecilla, T.; Jara, G.; Pizarro, A.; Acevedo, J.; Leo, K.; Leon, F. Effectiveness of an inactivated SARS-CoV-2 vaccine in Chile. N. Eng. J. Med. 2021, 385, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Reuters. Kangtai Biological’s COVID-19 vaccine gets emergency use approval in China. Available online: https://www.reuters.com/article/us-health-coronavirus-vaccine-kangtai-idUSKBN2CV1F6 (accessed on 12 June 2022).
- Ella, R.; Reddy, S.; Jogdand, H.; Sarangi, V.; Ganneru, B.; Prasad, S.; Das, D.; Raju, D.; Praturi, U.; Sapkal, G.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: Interim results from a double-blind, randomised, multicentre, phase 2 trial, and 3-month follow-up of a double-blind, randomised phase 1 trial. Lancet Infect. Dis. 2021, 21, 950–961. [Google Scholar] [CrossRef]
- Thiagarajan, K. What do we know about India’s Covaxin vaccine? BMJ 2021, 373, n997. [Google Scholar] [CrossRef] [PubMed]
- Ella, R.; Reddy, S.; Blackwelder, W.; Potdar, V.; Yadav, P.; Sarangi, V.; Aileni, V.K.; Kanungo, S.; Rai, S.; Reddy, P. Efficacy, safety, and lot-to-lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): Interim results of a randomised, double-blind, controlled, phase 3 trial. Lancet 2021, 398, 2173–2184. [Google Scholar] [CrossRef]
- Yadav, P.D.; Sapkal, G.N.; Abraham, P.; Ella, R.; Deshpande, G.; Patil, D.Y.; Nyayanit, D.A.; Gupta, N.; Sahay, R.R.; Shete, A.M. Neutralization of variant under investigation B. 1.617. 1 with sera of BBV152 vaccinees. Clinical Infect. Dis. 2022, 74, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Desai, D.; Khan, A.R.; Soneja, M.; Mittal, A.; Naik, S.; Kodan, P.; Mandal, A.; Maher, G.T.; Kumar, R.; Agarwal, A. Effectiveness of an inactivated virus-based SARS-CoV-2 vaccine, BBV152, in India: A test-negative, case-control study. Lancet Infect. Dis. 2022, 22, 349–356. [Google Scholar] [CrossRef]
- Zhugunissov, K.; Zakarya, K.; Khairullin, B.; Orynbayev, M.; Abduraimov, Y.; Kassenov, M.; Sultankulova, K.; Kerimbayev, A.; Nurabayev, S.; Myrzakhmetova, B. Development of the inactivated QazCovid-in vaccine: Protective efficacy of the vaccine in Syrian hamsters. Front. Microbiol. 2021, 2765. [Google Scholar] [CrossRef]
- Khoshnood, S.; Arshadi, M.; Akrami, S.; Koupaei, M.; Ghahramanpour, H.; Shariati, A.; Sadeghifard, N.; Heidary, M. An overview on inactivated and live-attenuated SARS-CoV-2 vaccines. J. Clin. Lab. Anal. 2022, 36, e24418. [Google Scholar] [CrossRef] [PubMed]
- Salehi, M.; Hosseini, H.; Jamshidi, H.R.; Jalili, H.; Tabarsi, P.; Mohraz, M.; Karimi, H.; Lotfinia, M.; Aalizadeh, R.; Mohammadi, M. Assessment of BIV1-CovIran inactivated vaccine–elicited neutralizing antibody against the emerging SARS-CoV-2 variants of concern. Clinical Microbiol. and Infect. 2022, 28, e881–e882.e887. [Google Scholar] [CrossRef] [PubMed]
- Abdoli, A.; Aalizadeh, R.; Aminianfar, H.; Kianmehr, Z.; Teimoori, A.; Azimi, E.; Emamipour, N.; Eghtedardoost, M.; Siavashi, V.; Jamshidi, H. Safety and potency of BIV1-CovIran inactivated vaccine candidate for SARS-CoV-2: A preclinical study. Rev. Med. Virol. 2021, e2305. [Google Scholar] [CrossRef] [PubMed]
- Mohraz, M.; Salehi, M.; Tabarsi, P.; Abbasi-Kangevari, M.; Ghamari, S.-H.; Ghasemi, E.; Pouya, M.A.; Rezaei, N.; Ahmadi, N.; Heidari, K. Safety and immunogenicity of an inactivated virus particle vaccine for SARS-CoV-2, BIV1-CovIran: Findings from double-blind, randomised, placebo-controlled, phase I and II clinical trials among healthy adults. BMJ Open 2022, 12, e056872. [Google Scholar] [CrossRef] [PubMed]
- Pollet, J.; Chen, W.H.; Strych, U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv. Drug Deliv. Rev. 2021, 170, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.S.; Walls, A.C.; Golden, N.; Atyeo, C.; Fischinger, S.; Li, C.; Aye, P.; Navarro, M.J.; Lai, L.; Edara, V.V.; et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 2021, 594, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Reuters. Russia’s second vaccine ’100% effective’, watchdog tells media. Available online: https://www.reuters.com/article/us-health-coronavirus-russia-vaccine-vec-idUSKBN29O151 (accessed on 26 March 2021).
- ClinicalTrials.gov. Study of the Safety, Reactogenicity and Immunogenicity of "EpiVacCorona" Vaccine for the Prevention of COVID-19 (EpiVacCorona). Available online: https://clinicaltrials.gov/ct2/show/NCT04527575 (accessed on 1 March 2021).
- Yang, S.; Li, Y.; Dai, L.; Wang, J.; He, P.; Li, C.; Fang, X.; Wang, C.; Zhao, X.; Huang, E. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: Two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021, 21, 1107–1119. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, A.; Li, D.; Zhang, R.; Sun, H.; Wang, Q.; Gao, G.F.; Han, P.; Dai, L. Neutralization of recombinant RBD-subunit vaccine ZF2001-elicited antisera to SARS-CoV-2 variants including Delta. bioRxiv 2021. [Google Scholar]
- Registro Público Cubano de Ensayos Clínicos. ABDALA Clinical Study. Available online: https://rpcec.sld.cu/print/trials/RPCEC00000346-En (accessed on 1 July 2022).
- Medicalxpress. Cuban COVID vaccine Abdala 92 percent ’effective’, maker says. Available online: https://medicalxpress.com/news/2021-06-cuban-covid-vaccine-abdala-percent.html (accessed on 1 July 2022).
- Medicalxpress. Cuba approves emergency use of home-grown COVID vaccine, Latin America’s first. Available online: https://medicalxpress.com/news/2021-07-cuba-emergency-home-grown-covid-vaccine.html (accessed on 1 July 2022).
- Toledo-Romani, M.E.; Garcia-Carmenate, M.; Silva, C.V.; Baldoquin-Rodriguez, W.; Perez, M.M.; Gonzalez, M.C.R.; Moreno, B.P.; Hernandez, I.M.; Romero, R.G.-M.; Tabio, O.S. Efficacy and Safety of SOBERANA 02, a COVID-19 conjugate vaccine in heterologous three doses combination. MedRxiv 2021. [Google Scholar]
- Hsieh, S.-M.; Liu, W.-D.; Huang, Y.-S.; Lin, Y.-J.; Hsieh, E.-F.; Lian, W.-C.; Chen, C.; Janssen, R.; Shih, S.-R.; Huang, C.-G. Safety and immunogenicity of a Recombinant Stabilized Prefusion SARS-CoV-2 Spike Protein Vaccine (MVCCOV1901) Adjuvanted with CpG 1018 and Aluminum Hydroxide in healthy adults: A Phase 1, dose-escalation study. eClinicalMedicine 2021, 38, 100989. [Google Scholar] [CrossRef]
- Hsieh, S.-M.; Liu, M.-C.; Chen, Y.-H.; Lee, W.-S.; Hwang, S.-J.; Cheng, S.-H.; Ko, W.-C.; Hwang, K.-P.; Wang, N.-C.; Lee, Y.-L. Safety and immunogenicity of CpG 1018 and aluminium hydroxide-adjuvanted SARS-CoV-2 S-2P protein vaccine MVC-COV1901: Interim results of a large-scale, double-blind, randomised, placebo-controlled phase 2 trial in Taiwan. Lancet Resp. Med. 2021, 9, 1396–1406. [Google Scholar] [CrossRef]
- Liu, L.T.C.; Chiu, C.-H.; Chiu, N.-C.; Tan, B.-F.; Lin, C.-Y.; Cheng, H.-Y.; Lin, M.-Y.; Lien, C.E.; Chen, C.; Huang, L.-M. Safety and immunogenicity of SARS-CoV-2 vaccine MVC-COV1901 in adolescents in Taiwan: A double-blind, randomized, placebo-controlled phase 2 trial. medRxiv 2022. [Google Scholar]
- Khobragade, A.; Bhate, S.; Ramaiah, V.; Deshpande, S.; Giri, K.; Phophle, H.; Supe, P.; Godara, I.; Revanna, R.; Nagarkar, R. Efficacy, safety, and immunogenicity of the DNA SARS-CoV-2 vaccine (ZyCoV-D): The interim efficacy results of a phase 3, randomised, double-blind, placebo-controlled study in India. Lancet 2022, 399, 1313–1321. [Google Scholar] [CrossRef]
- Li, L.; Honda-Okubo, Y.; Baldwin, J.; Bowen, R.; Bielefeldt-Ohmann, H.; Petrovsky, N. Covax-19/Spikogen® vaccine based on recombinant spike protein extracellular domain with Advax-CpG55. 2 adjuvant provides single dose protection against SARS-CoV-2 infection in hamsters. Vaccine 2022, 40, 3182–3192. [Google Scholar] [CrossRef] [PubMed]
- Parums, D.V. First Approval of the Protein-Based Adjuvanted Nuvaxovid (NVX-CoV2373) Novavax Vaccine for SARS-CoV-2 Could Increase Vaccine Uptake and Provide Immune Protection from Viral Variants. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2022, 28, e936523-1–e936523-3. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, H.; Tian, L.; Pang, Z.; Yang, Q.; Huang, T.; Fan, J.; Song, L.; Tong, Y.; Fan, H. COVID-19 vaccine development: Milestones, lessons and prospects. Signal Transduct. Target. Ther. 2022, 7, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N. Eng. J. Med. 2021, 385, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Dunkle, L.M.; Kotloff, K.L.; Gay, C.L.; Áñez, G.; Adelglass, J.M.; Barrat Hernández, A.Q.; Harper, W.L.; Duncanson, D.M.; McArthur, M.A.; Florescu, D.F. Efficacy and safety of NVX-CoV2373 in adults in the United States and Mexico. N. Eng. J. Med. 2022, 386, 531–543. [Google Scholar] [CrossRef]
- Stuart, A.S.; Shaw, R.H.; Liu, X.; Greenland, M.; Aley, P.K.; Andrews, N.J.; Cameron, J.; Charlton, S.; Clutterbuck, E.A.; Collins, A.M. Immunogenicity, safety, and reactogenicity of heterologous COVID-19 primary vaccination incorporating mRNA, viral-vector, and protein-adjuvant vaccines in the UK (Com-COV2): A single-blind, randomised, phase 2, non-inferiority trial. Lancet 2022, 399, 36–49. [Google Scholar] [CrossRef]
- Toback, S.; Galiza, E.; Cosgrove, C.; Galloway, J.; Goodman, A.L.; Swift, P.A.; Rajaram, S.; Graves-Jones, A.; Edelman, J.; Burns, F. Safety, immunogenicity, and efficacy of a COVID-19 vaccine (NVX-CoV2373) co-administered with seasonal influenza vaccines: An exploratory substudy of a randomised, observer-blinded, placebo-controlled, phase 3 trial. Lancet Respir. Med. 2022, 10, 167–179. [Google Scholar] [CrossRef]
- Thuluva, S.; Paradkar, V.; Turaga, K.; Gunneri, S.; Yerroju, V.; Mogulla, R.; Kyasani, M.; Manoharan, S.; Medigeshi, G.R.; Singh, J. Selection of optimum formulation of RBD-based protein sub-unit covid19 vaccine (Corbevax) based on safety and immunogenicity in an open-label, randomized Phase-1 and 2 clinical studies. MedRxiv 2022. [Google Scholar]
- Hager, K.J.; Pérez Marc, G.; Gobeil, P.; Diaz, R.S.; Heizer, G.; Llapur, C.; Makarkov, A.I.; Vasconcellos, E.; Pillet, S.; Riera, F. Efficacy and safety of a recombinant plant-based adjuvanted Covid-19 vaccine. N. Eng. J. Med. 2022, 386, 2084–2096. [Google Scholar] [CrossRef]
- Mahase, E. Covid-19: UK approves Valneva vaccine for adults under 50. 2022. Available online: https://www.bmj.com/content/377/bmj.o985.long (accessed on 13 June 2022).
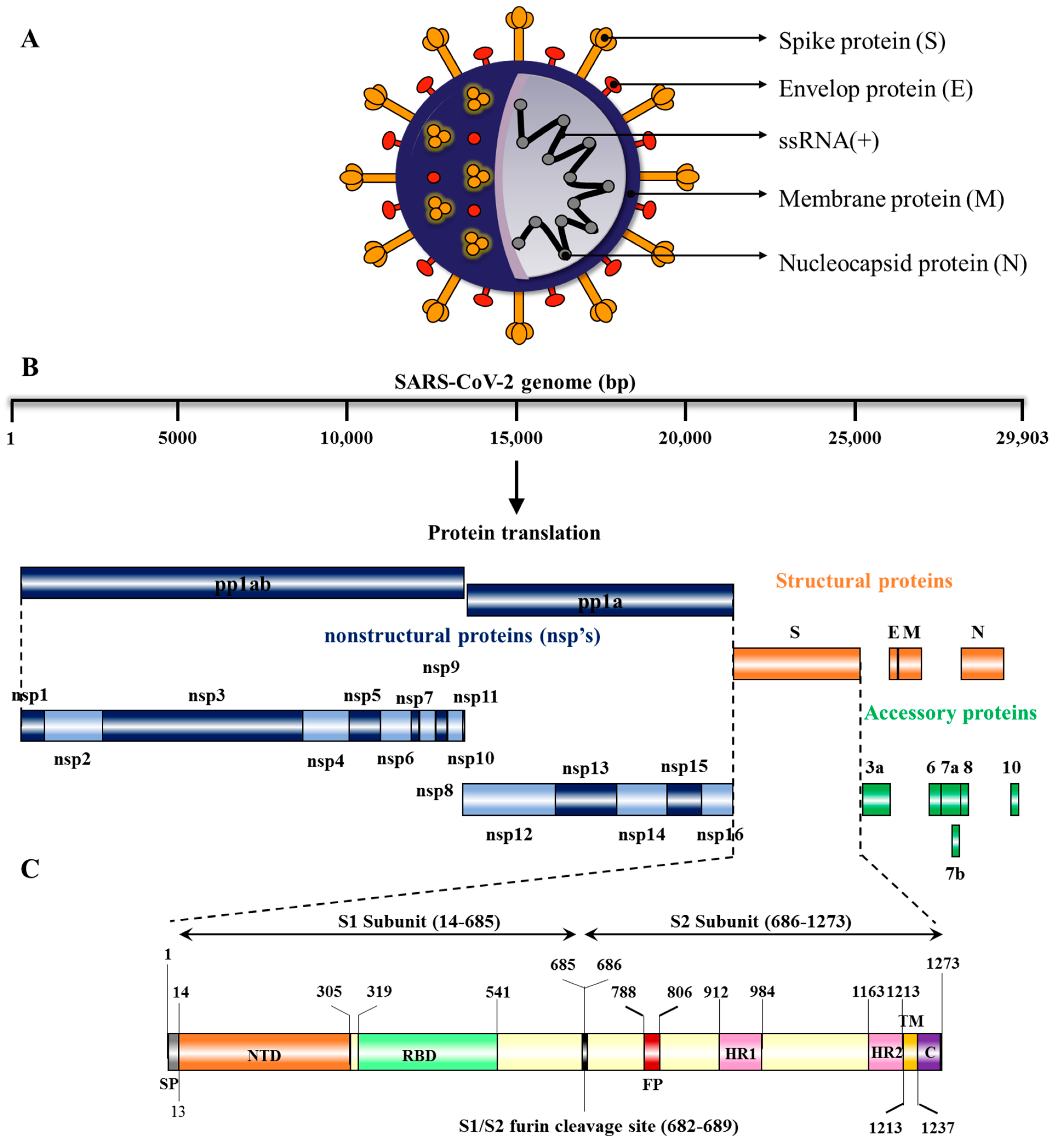
| Number | Gene | Nucleotide Location | Protein | Amino Acids |
|---|---|---|---|---|
| 1 | ORF1a | 266–13,483 | Polyprotein 1a | 4405 |
| 1 | ORF1ab | 266–21,555 | Polyprotein 1ab | 7096 |
| 2 | ORF2 | 21,563–25,384 | Surface glycoprotein or Spike (S protein) | 1273 |
| 3 | ORF3a | 25,393–26,220 | ORF3a protein | 275 |
| 4 | ORF4 | 26,245–26,472 | Envelope protein (E protein) | 75 |
| 5 | ORF5 | 26,523–27,191 | Membrane glycoprotein (M protein) | 222 |
| 6 | ORF6 | 27,202–27,387 | ORF 6 protein | 61 |
| 7 | ORF7a | 27,394–27,759 | ORF 7a protein | 121 |
| 8 | ORF7b | 27,756–27,887 | ORF 7b protein | 43 |
| 9 | ORF8 | 27,894–28,259 | ORF 8 protein | 121 |
| 10 | ORF9 | 28,274–29,533 | Nucleocapsid phosphoprotein (N protein) | 419 |
| 11 | ORF10 | 29,558–29,674 | ORF10 protein | 38 |
| Name | Function | Nucleotide Location | Amino Acids |
|---|---|---|---|
| NSP1 | Host mRNA degradation and translation inhibition | 266–805 | 180 |
| NSP2 | Unknown | 806–2719 | 638 |
| NSP3 | Polyprotein processing, de-ADP ribosylation, Deubiquitinating, Interferon antagonist, DMV (double-membrane vesicle) formation | 2720–8554 | 1945 |
| NSP4 | DMV formation | 8555–10,054 | 500 |
| NSP5 | Polyprotein processing, Inhibition of interferon signaling | 10,055–10,972 | 306 |
| NSP6 | DMV formation | 10,973–11,842 | 290 |
| NSP7 | Cofactor for RNA-dependent RNA polymerase | 11,843–12,091 | 83 |
| NSP8 | primase or 3′-terminal adenylyl transferase, cofactor for RNA-dependent RNA polymerase | 12,092–12,685 | 198 |
| NSP9 | Binding of single-stranded RNA | 12,686–13,024 | 113 |
| NSP10 | Cofactor for nsp14 and 16 | 13,025–13,441 | 139 |
| NSP11 | Unknown | 13,442–13,480 | 13 |
| NSP12 | RNA-dependent RNA polymerase, Nucleotidyl transferase | 13,442–16,236 | 932 |
| NSP13 | Helicase, RNA 5′ triphosphatase | 16,237–18,039 | 601 |
| NSP14 | 3′ to 5′ exoribonuclease, Proofreading, RNA cap formation, Guanosine N7-methyltransferase | 18,040–19,620 | 527 |
| NSP15 | Endoribonuclease, evasion of immune response | 19,621–20,658 | 346 |
| NSP16 | RNA cap formation, Ribose 2′ O-methyltransferase | 20,659–21,552 | 298 |
| S. No. | WHO Designation | Pango Lineage | Nextstrain Clade | GISAID Clade | Mutations Reported |
|---|---|---|---|---|---|
| 1 | Omicron | B.1.1.529 | 21K, 21L, 21M | GR/484A | ORF1a—T3255I, P3395H, P314L, I1566V S—G142D, G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K E—T9I M—Q19E, A63T ORF8—S84L N—P13L, del131/133, R203K, G204R |
| S. No. | WHO Designation | Pango Lineage | Nextstrain Clade | GISAID Clade | Mutations Reported |
|---|---|---|---|---|---|
| 1 | Alpha | B.1.1.7 | 20I (V1) | GRY | ORF1a—T1001I, A1708D, I2230T, del3675/3677 ORF1b—P314L S—del69/70, del144/145, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H ORF8—Q27 *, R52I, Y73C, S84L N—D3L, R203K, G204R, S235F |
| 2 | Beta | B.1.351 | 20H (V2) | GH/501Y.V2 | ORF1a—T265I, K1655N, K3353, del3675/3677 ORF1b—P314L S—D80A, D215G, del241/243, K417N, E484K, N501Y, D614G, A701V ORF3a—Q57H, S171L, E—P71L ORF8—S84L N—T205I |
| 3 | Gamma | P.1 | 20J (V3) | GR/501Y.V3 | ORF1a—S1188L, K1795Q, del3675/3677 ORF1b—P314L, E1264D S—L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F ORF3a—S253P ORF8—S253P, E92K N—P80R, R203K, G204R |
| 4 | Delta | B.1.617.2 | 21A, 21I, 21J | G/478K.V1 | ORF1a—A1306S, P2046L, P2287S, V2930L, T3255I, T3646A ORF1b—P314L, G662S, P1000L, A1918V S—T19R, E156G, del157/158, L452R, T478K, D614G, P681R, D950N ORF3a—S26L M—I82T ORF7a—V82A, T120I ORF7b—T40I ORF8—S84L, del119/120 N—D63G, R203M, G215C, D377Y |
| 5 | Epsilon | B.1.427 B.1.429 | 21C | GH/452R.V1 | ORF1a—T265I ORF1b—P314L, D1183Y S—S13I, W152C, L452R, D614G ORF3a—Q57H, ORF8—S84L N—T205I |
| 6 | Eta | B.1.525 | 21D | G/484K.V3 | ORF1a—T2007I, del3675/3677 ORF1b—P314F S—Q52R, A67V, del69/70, del144/144, E484K, D614G, Q677H, F888L E—L21F M—I82F ORF6—del2/3 ORF8—S84L N—S2Y, del3/3, A12G, T205I |
| 7 | Lota | B.1.526 | 21F | GH/253G.V1 | ORF1a—T265I, L3201P, del3675/3677 ORF1b—P314L, Q1011H S—L5F, T95I, D253G, D614G ORF3a—P42L, Q57H ORF8—T11I, S84L |
| 8 | Kappa | B.1.617.1 | 21B | G/452R.V3 | ORF1a—T1567I, T3646A ORF1b—P314L, M1352I, K2310R S—L452R, E484Q, D614G, P681R, Q1071H ORF3a—S26L ORF7a—V82A ORF8—S84L N—R203M, D377Y |
| 9 | Mu | B.1.621 | 21H | GH | ORF1a—T1055A, T1538I, T3255I, Q3729R ORF1b—P314L, P1342S S—T95I, Y145N, R346K, E484K, N501Y, D614G, P681H, D950N ORF3a—Q57H, del256/257 ORF8—T11K, P38S, S67F, S84L N—T205I |
| 10 | Zeta | P.2 | 20B/S.484K | GR/484K.V2 | ORF1a—L3468V, L3930F ORF1b—P314L S—E484K, D614G, V1176F ORF8—S84L N—A119S, R203K, G204R, M234I |
| S. No. | Platform | Vaccine Candidate | Number |
|---|---|---|---|
| 1 | PS | Protein subunit | 52 |
| 2 | VVnr | Viral Vector (non-replicating) | 21 |
| 3 | DNA | DNA | 16 |
| 4 | IV | Inactivated Virus | 21 |
| 5 | RNA | RNA | 30 |
| 6 | VVr | Viral Vector (replicating) | 4 |
| 7 | VLP | Virus Like Particle | 6 |
| 8 | VVr + APC | VVr + Antigen Presenting Cell | 2 |
| 9 | LAV | Live Attenuated Virus | 2 |
| 10 | VVnr + APC | VVnr + Antigen Presenting Cell | 1 |
| 11 | BacAg-SpV | Bacterial antigen-spore expression vector | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tulimilli, S.V.; Dallavalasa, S.; Basavaraju, C.G.; Kumar Rao, V.; Chikkahonnaiah, P.; Madhunapantula, S.V.; Veeranna, R.P. Variants of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Vaccine Effectiveness. Vaccines 2022, 10, 1751. https://doi.org/10.3390/vaccines10101751
Tulimilli SV, Dallavalasa S, Basavaraju CG, Kumar Rao V, Chikkahonnaiah P, Madhunapantula SV, Veeranna RP. Variants of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Vaccine Effectiveness. Vaccines. 2022; 10(10):1751. https://doi.org/10.3390/vaccines10101751
Chicago/Turabian StyleTulimilli, SubbaRao V., Siva Dallavalasa, Chaithanya G. Basavaraju, Vinay Kumar Rao, Prashanth Chikkahonnaiah, SubbaRao V. Madhunapantula, and Ravindra P. Veeranna. 2022. "Variants of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Vaccine Effectiveness" Vaccines 10, no. 10: 1751. https://doi.org/10.3390/vaccines10101751
APA StyleTulimilli, S. V., Dallavalasa, S., Basavaraju, C. G., Kumar Rao, V., Chikkahonnaiah, P., Madhunapantula, S. V., & Veeranna, R. P. (2022). Variants of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Vaccine Effectiveness. Vaccines, 10(10), 1751. https://doi.org/10.3390/vaccines10101751








