Vaccine Breakthrough Infections by SARS-CoV-2 Variants after ChAdOx1 nCoV-19 Vaccination in Healthcare Workers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Confirmation of Breakthrough Infection
2.2. Humoral Immune Response
2.3. Whole Genome Sequencing
2.4. Sequencing Data Analysis
2.5. Phylogenetic and Mutation Analysis
2.6. Data Availability
2.7. Data Analysis
3. Results
3.1. Characteristics of Breakthrough Infections in HCWs
3.1.1. SARS-CoV-2 Infections in HCWs
3.1.2. Distribution of VBT Infections as Per Work Profile of HCW
3.1.3. Reinfection
3.1.4. Clinical Presentation of VBT Infections
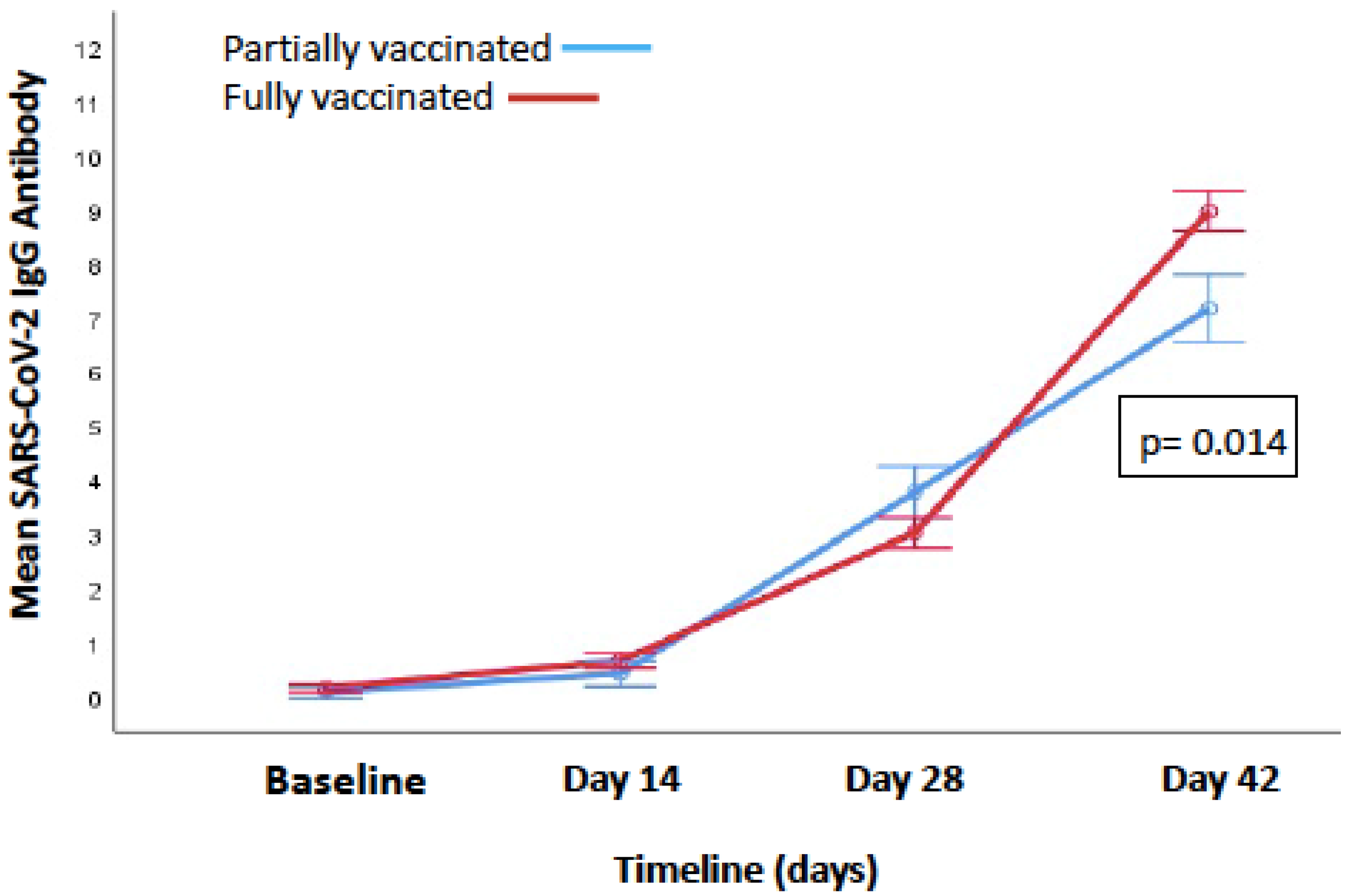
3.1.5. Change in IgG Antibody Levels from Baseline to 14 Days after the Second Dose
3.1.6. Antibody Response Post-Infection
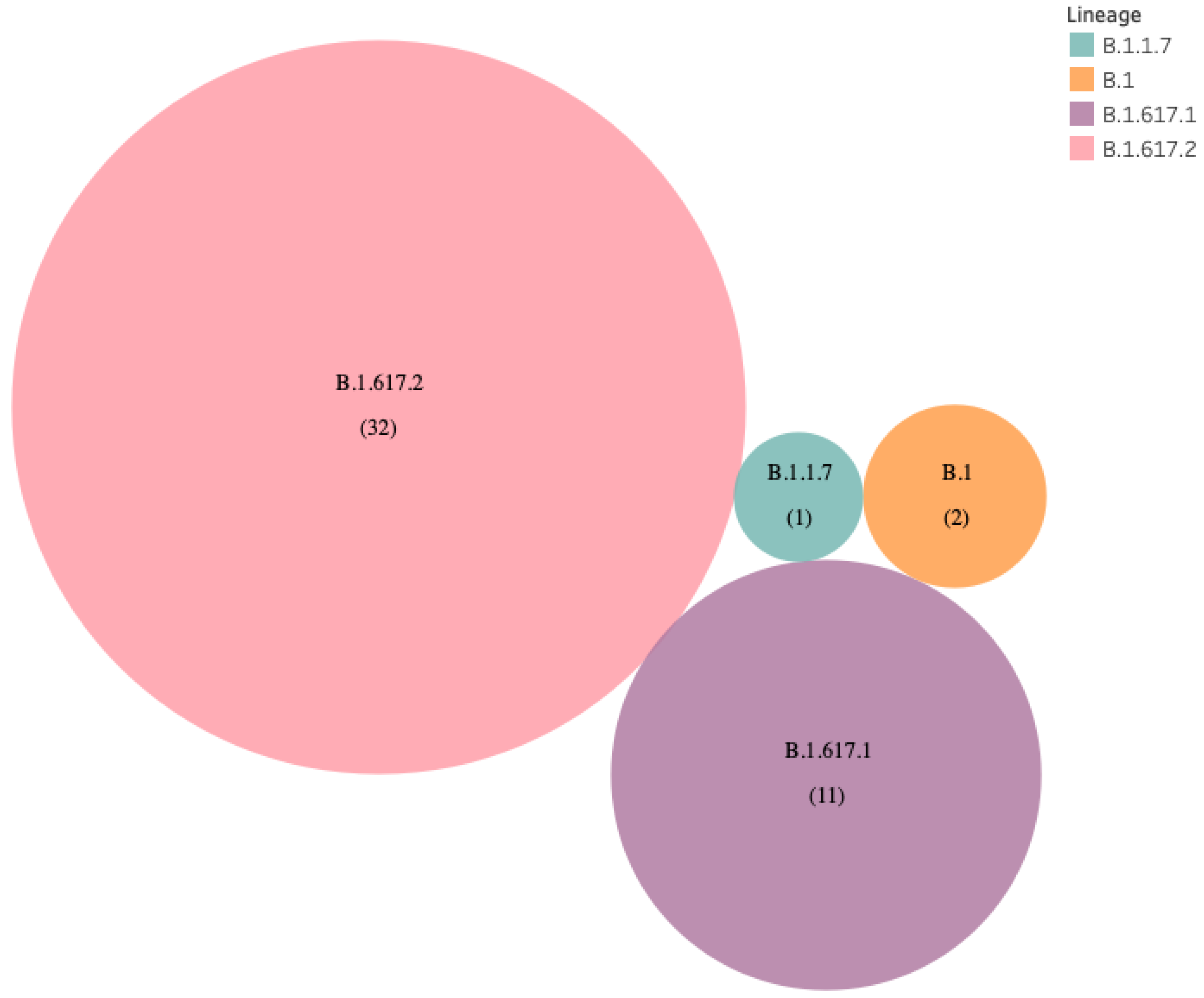
3.1.7. Genomic Sequencing of VBT Infections
3.1.8. Comparison of Ct Values
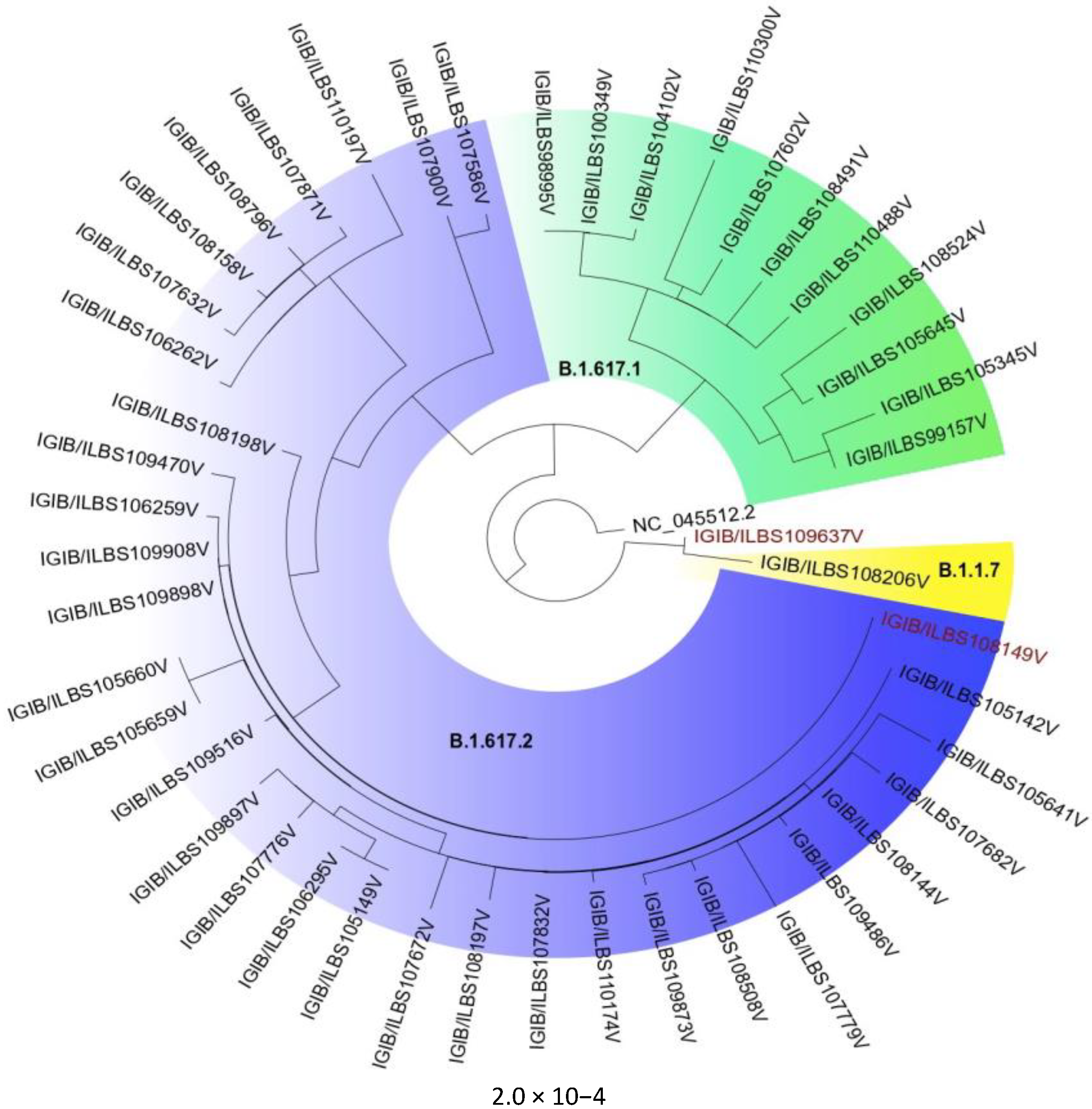
3.1.9. Phylogenetic and Mutation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frederiksen, L.S.F.; Zhang, Y.; Foged, C.; Thakur, A. The Long Road Toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies. Front. Immunol. 2020, 11, 1817. [Google Scholar] [CrossRef]
- Ministry of Health and Family Welfare. Cumulative COVID Vaccination Coverage Report 29 May 2021. Available online: https://www.mohfw.gov.in/pdf/CumulativeCovidVaccinationCoverageReport29thMay2021.pdf (accessed on 30 May 2021).
- Mallapaty, S. India’s massive COVID surge puzzles scientists. Nat. Cell Biol. 2021, 592, 667–668. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Samal, J.; Kumar, V.; Sharma, J.; Agrawal, U.; Ehtesham, N.; Sundar, D.; Rahman, S.; Hira, S.; Hasnain, S. Structure-Function Analyses of New SARS-COVID-2 Variants B.1.1.7, B.1.351 and B.1.1.28.1: Clinical, Diagnostic, Therapeutic and Public Health Implications. Viruses 2021, 13, 439. [Google Scholar] [CrossRef]
- COVID-19 Vaccine Breakthrough Infections Reported to CDC—United States, 1 January–30 April. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 792. [CrossRef]
- Tyagi, K.; Ghosh, A.; Nair, D.; Dutta, K.; Bhandari, P.S.; Ansari, I.A.; Misra, A. Breakthrough COVID19 Infections after Vaccinations in Healthcare and Other Workers in a Chronic Care Medical Facility in New Delhi, India. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 1007–1008. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAM tools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chernomor, O.; Von Haeseler, A.; Minh, B.Q. Data from: Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree v1.3. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. 2010. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 31 May 2021).
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Joyner, M.J.; Senefeld, J.W.; Klassen, S.A.; John, R.M.; Johnson, P.W.; Theel, E.S.; Wiggins, C.C.; Bruno, K.A.; Klompas, A.M.; Lesser, E.R.; et al. Effect of convalescent plasma on mortality among hospitalized patients with COVID-19: Initial three-month experience. MedRxiv 2020. [Google Scholar] [CrossRef]
- Ministry of Health and Family Welfare, Government of National Capital Territory of Delhi. Delhi Health Bulletin Dated 31 May 2021. Available online: http://health.delhigovt.nic.in/wps/wcm/connect/57048e0042e1cbd6bd69bf28c2355f02/DHB31MA.pdf?MOD=AJPERES&lmod=-783403617&CACHEID=57048e0042e1cbd6bd69bf28c2355f02.pdf (accessed on 31 May 2021).
- Victor, P.J.; Mathews, K.P.; Paul, H.; Mammen, J.J.; Murugesan, M. Protective Effect of COVID-19 Vaccine Among Health Care Workers During the Second Wave of the Pandemic in India. Mayo Clin. Proc. 2021, 96, 2493–2494. [Google Scholar] [CrossRef] [PubMed]
- Hall, V.J.; Foulkes, S.; Saei, A.; Andrews, N.; Oguti, B.; Charlett, A.; Wellington, E.; Stowe, J.; Gillson, N.; Atti, A.; et al. COVID-19 vaccine coverage in health-care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): A prospective, multicentre, cohort study. Lancet 2021, 397, 1725–1735. [Google Scholar] [CrossRef]
- Keehner, J.; Horton, L.E.; Pfeffer, M.A.; Longhurst, C.A.; Schooley, R.T.; Currier, J.S.; Abeles, S.R.; Torriani, F.J. SARS-CoV-2 infection after vaccination in health care workers in California. N. Engl. J. Med. 2021, 384, 1774–1775. [Google Scholar] [CrossRef]
- Benenson, S.; Oster, Y.; Cohen, M.J.; Nir-Paz, R. BNT162b2 mRNA Covid-19 Vaccine Effectiveness among Health Care Workers. N. Engl. J. Med. 2021, 384, 1775–1777. [Google Scholar] [CrossRef]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and immunogenicity of the ChAdOx1 nCOVID-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Philomina, J.B.; Jolly, B.; John, N.; Bhoyar, R.C.; Majeed, N.; Senthivel, V.; Cp, F.; Rophina, M.; Vasudevan, B.; Imran, M.; et al. Genomic survey of SARS-CoV-2 vaccine breakthrough infections in healthcare workers from Kerala, India. J. Infect. 2021, 83, 237–279. [Google Scholar] [CrossRef]
- Singh, U.B.; Rophina, M.; Chaudhry, R.; Senthive, V.; Bala, K.; Bhoyar, R.C.; Jolly, B.; Jamshed, N.; Imran, M.; Gupta, R.; et al. Variants of Concern responsible for SARS-COVID-2 vaccine breakthrough infections from India. OSF Prepr. 2021. [Google Scholar] [CrossRef]
- Rophina, M.; Pandhare, K.; Shamnath, A.; Imran, M.; Jolly, B.; Scaria, V. Comprehensive Resource for Immune Escape Variants and Their Annotations. Available online: http://clingen.igib.res.in/esc/ (accessed on 31 May 2021).
- Dhar, M.S.; Marwal, R.; Radhakrishnan, V.; Ponnusamy, K.; Jolly, B.; Bhoyar, R.C.; Fatihi, S.; Datta, M.; Singh, P.; Sharma, U.; et al. Genomic characterization and Epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. medRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.-M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-COVID-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Weissman, D.; Alameh, M.-G.; de Silva, T.; Collini, P.; Hornsby, H.; Brown, R.; LaBranche, C.C.; Edwards, R.J.; Sutherland, L.; Santra, S.; et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe 2021, 29, 23–31. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; Denis, K.S.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Balazs, A.B. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021, 184, 2372–2383. [Google Scholar] [CrossRef] [PubMed]
- Tchesnokova, E.S.V.; Kulakesara, H.; Larson, L.; Bowers, V.; Rechkina, E.; Kisiela, D.; Sledneva, Y.; Choudhury, D.; Maslova, I.; Deng, K.; et al. Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-COVID-2 variants. bioRxiv 2021. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. SARS-COVID-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9, 1542. [Google Scholar] [CrossRef] [PubMed]

| Category of HCW | Vaccinated N = 1639 | Partially Vaccinated N = 293 | Fully Vaccinated N = 1346 | Non-Vaccinated N = 219 | Total N = 1858 | Level of Significance p Value |
|---|---|---|---|---|---|---|
| Doctors n/N (%) | 30/129 (23.26) | 8/29 (27.59) | 22/100 (22) | 5/18 (27.78) | 35/147 (23.81) | 0.754 |
| Nursing staff n/N (%) | 76/314 (24.20) | 21/87 (24.14) | 55/227 (24.23) | 27/110 (24.55) | 103/424 (24.29) | 0.997 |
| Technicians n/N (%) | 16//215 (7.44) | 5/60 (8.33) | 11/155 (7.10) | 4/20 (20) | 20/235 (8.51) | 0.150 |
| Non-Medical n/N (%) | 14/321 (4.36) | 3/27 (11.11) | 11/294 (3.74) | 6/36 (16.67) | 20/357 (5.60) | 0.003 * |
| GDA n/N (%) | 20/660 (3.03) | 3/90 (3.33) | 17/570 (2.98) | 5/35 (14.29) | 25/695 (3.60) | 0.002 * |
| Total n/N (%) | 156/1639 (9.52) | 40/293 (13.65) | 116/1346 (8.62) | 47/219 (21.46) | 203/1858 (10.93) | non-vaccinated vs. vaccinated <0.001 Partially vaccinated vs. fully vaccinated p = 0.008 |
| p value | <0.001 # | <0.001 # | 0.621 | <0.001 # |
| Infection Status | Non-Vaccinated (n = 219) | Partially Vaccinated (n = 293) | RR a Non-Vaccinated vs. Partially Vaccinated | Fully Vaccinated (n = 1346) | RR b Non-Vaccinated vs. Fully Vaccinated | RR c Partially Vaccinated vs. Fully Vaccinated |
|---|---|---|---|---|---|---|
| Developed infection N (95% CI) | 47 (21.46%) | 40 (13.65%) | 1.57 (1.07–2.31) p = 0.02 | 116 (8.61%) | 2.49 (1.83–3.39) p ≤ 0.001 | 1.58 (1.13–2.22) p = 0.01 |
| Mild infection | 24 (10.9%) | 32 (10.9%) | 1.0 (0.6–1.65) P = 0.99 | 113 (8.39%) | 1.30 (0.86–1.98) p = 0.21 | 1.30 (0.9–1.8) p = 0.17 |
| Moderate illness requiring Hospitalization | 20 (9.13%) | 8 (2.73%) | 3.35 (1.5–7.45) p = 0.002 | 3 (0.22%) | 40.97 * (12.2–136.7) p < 0.001 | 12.25 * (3.2–45.9) p < 0.001 |
| ICU admission | 3 (1.36%) | 0 (0) | - | 0 (0) | - | - |
| Death | 1 (0.46%) | 0 (0) | 0 (0) | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kale, P.; Gupta, E.; Bihari, C.; Patel, N.; Rooge, S.; Pandey, A.; Bajpai, M.; Khillan, V.; Chattopadhyay, P.; Devi, P.; et al. Vaccine Breakthrough Infections by SARS-CoV-2 Variants after ChAdOx1 nCoV-19 Vaccination in Healthcare Workers. Vaccines 2022, 10, 54. https://doi.org/10.3390/vaccines10010054
Kale P, Gupta E, Bihari C, Patel N, Rooge S, Pandey A, Bajpai M, Khillan V, Chattopadhyay P, Devi P, et al. Vaccine Breakthrough Infections by SARS-CoV-2 Variants after ChAdOx1 nCoV-19 Vaccination in Healthcare Workers. Vaccines. 2022; 10(1):54. https://doi.org/10.3390/vaccines10010054
Chicago/Turabian StyleKale, Pratibha, Ekta Gupta, Chhagan Bihari, Niharika Patel, Sheetalnath Rooge, Amit Pandey, Meenu Bajpai, Vikas Khillan, Partha Chattopadhyay, Priti Devi, and et al. 2022. "Vaccine Breakthrough Infections by SARS-CoV-2 Variants after ChAdOx1 nCoV-19 Vaccination in Healthcare Workers" Vaccines 10, no. 1: 54. https://doi.org/10.3390/vaccines10010054
APA StyleKale, P., Gupta, E., Bihari, C., Patel, N., Rooge, S., Pandey, A., Bajpai, M., Khillan, V., Chattopadhyay, P., Devi, P., Maurya, R., Jha, N., Mehta, P., Kumar, M., Sharma, P., Saifi, S., Swaminathan, A., Alam, S., Uppili, B., ... Sarin, S. K. (2022). Vaccine Breakthrough Infections by SARS-CoV-2 Variants after ChAdOx1 nCoV-19 Vaccination in Healthcare Workers. Vaccines, 10(1), 54. https://doi.org/10.3390/vaccines10010054








