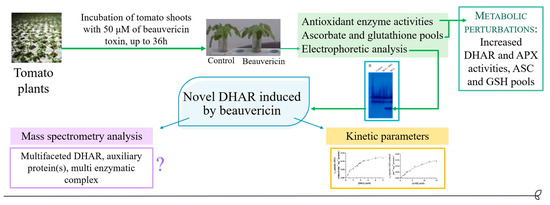
A Novel and Potentially Multifaceted Dehydroascorbate Reductase Increasing the Antioxidant Systems is Induced by Beauvericin in Tomato
Abstract
1. Introduction
2. Materials and Methods
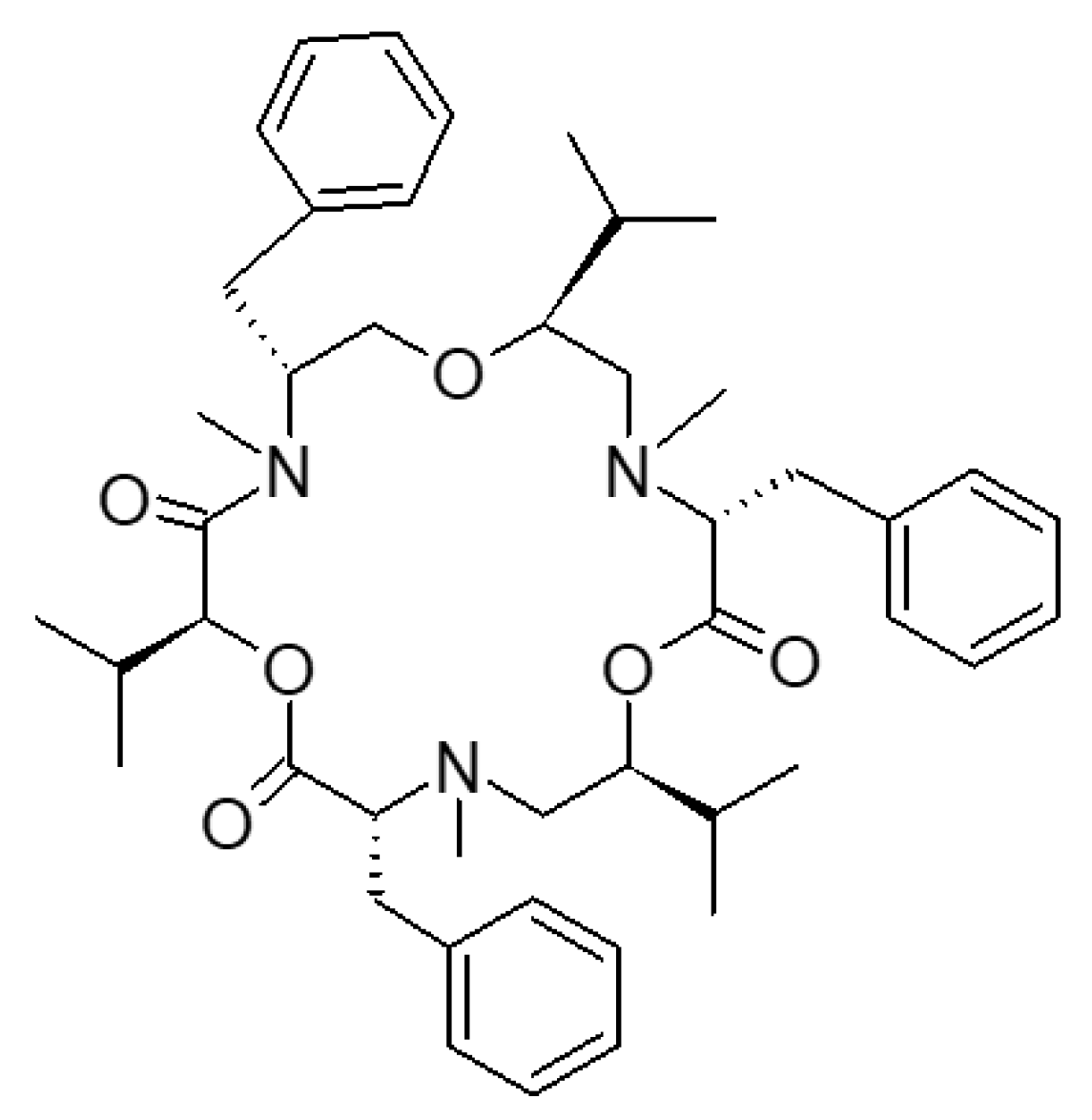
2.1. Chemicals
2.2. Plant Material
2.3. Determination of Ascorbate and Glutathione Pools
2.4. Proteins Extraction and Quantification
2.5. Enzyme Activity Measurements
2.6. Electrophoretic Analyses
2.6.1. Native-Polyacrylamide Gel Electrophoresis
2.6.2. Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis
2.7. Western Blot Analysis
2.8. Protein Identification by Mass Spectrometry
2.9. Kinetic Measurements
2.10. Statistical Analyses
3. Results
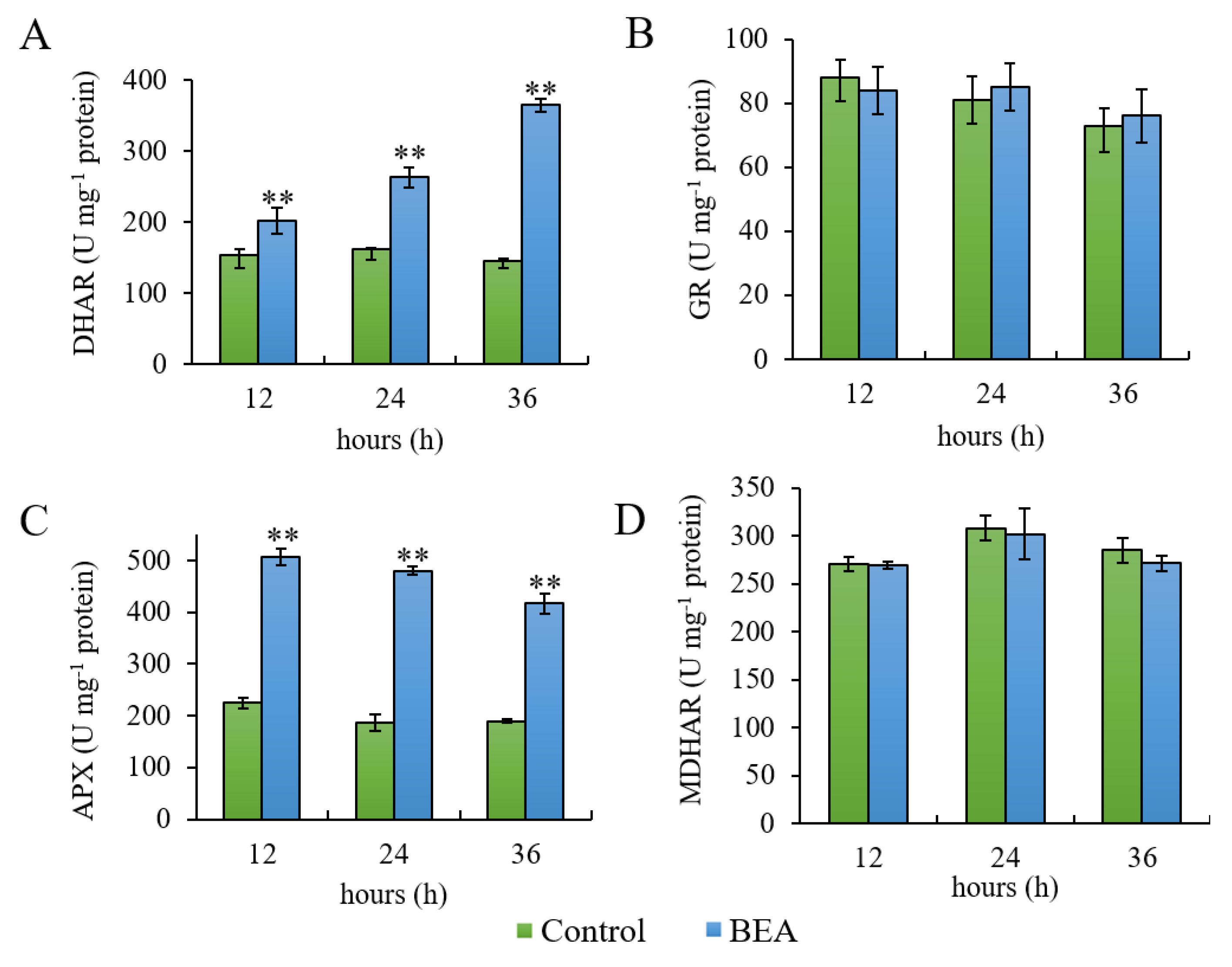
3.1. Determination of Ascorbate and Glutathione Pools
3.2. Enzyme Activity Measurements
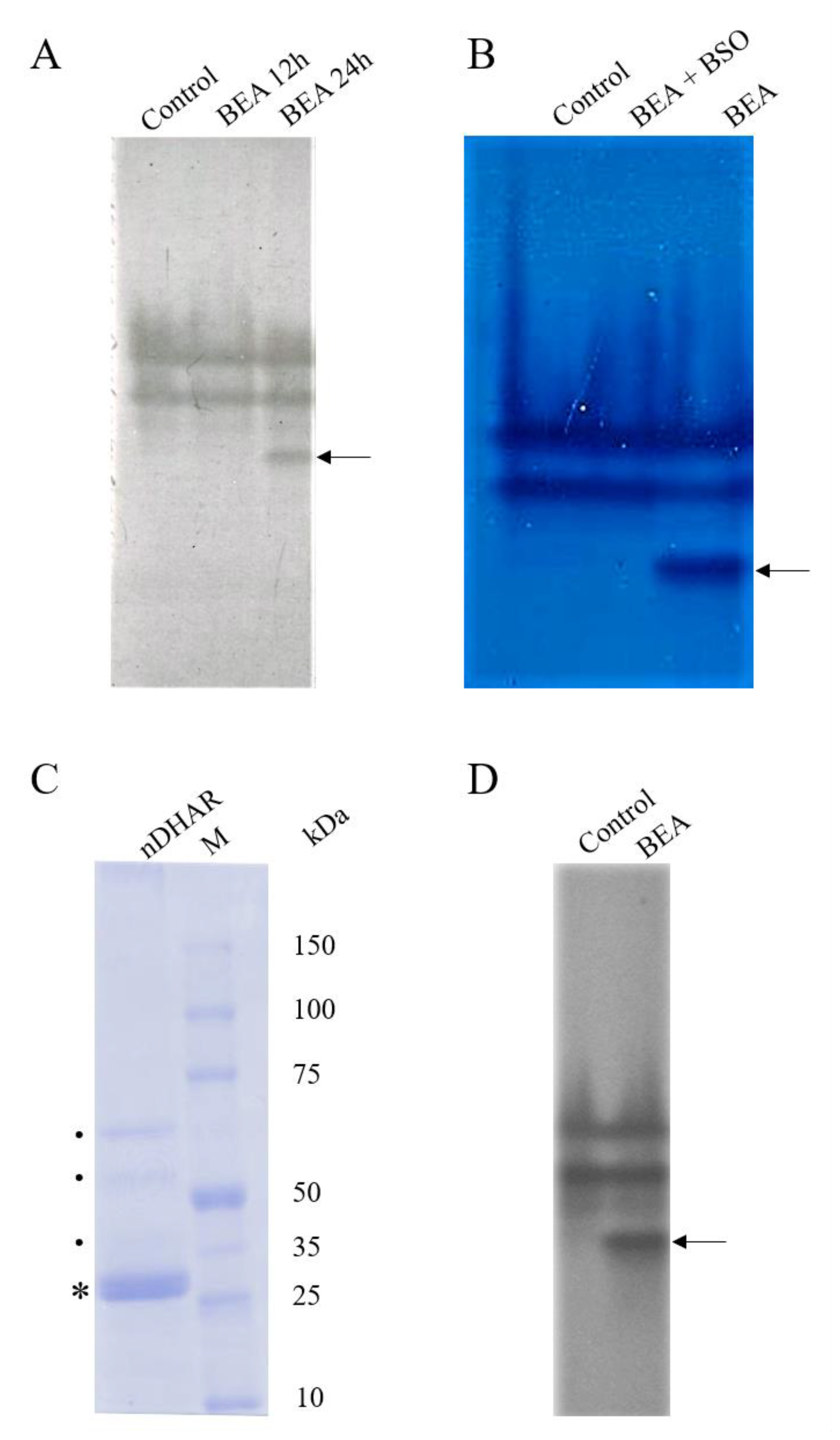
3.3. Electrophoretic Analyses
3.4. Western Blot Analysis
3.5. Protein Identification by Mass Spectrometry
3.6. Kinetic Measurements
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gruber-Dorninger, C.; Novak, B.; Nagl, V.; Berthiller, F. Emerging mycotoxins: Beyond traditionally determined food contaminants. J. Agric. Food Chem. 2017, 65, 7052–7070. [Google Scholar] [CrossRef]
- Jestoi, M. Emerging Fusarium-mycotoxins fusaproliferin, beauvericin, enniatins, and moniliformin—A review. Crit. Rev. Food Sci. Nutr. 2008, 48, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, V.C.; Mirabelli, V.; Cimmarusti, M.T.; Haidukowski, M.; Leslie, J.F.; Logrieco, A.F.; Caliandro, R.; Fanelli, F.; Mulè, G. Enniatin and beauvericin biosynthesis in Fusarium species: Production profiles and structural determinant prediction. Toxins 2017, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Contaminants in the Food Chain (CONTAM). Scientific Opinion on the risks to human and animal health related to the presence of beauvericin and enniatins in food and feed. EFSA J. 2014, 12, 1–174. [Google Scholar] [CrossRef]
- Mallebrera, B.; Prosperini, A.; Font, G.; Ruiz, M.J. In vitro mechanisms of Beauvericin toxicity: A review. Food Chem. Toxicol. 2018, 111, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Sood, S.; Sandhu, S.S.; Mukherjee, T.K. Pharmacological and Therapeutic Potential of Beauvericin: A Short Review. J. Proteom. Bioinform. 2017, 10, 18–23. [Google Scholar] [CrossRef]
- Tong, Y.; Liu, M.; Zhang, Y.; Liu, X.; Huang, R.; Song, F.; Dai, H.; Ren, B.; Sun, N.; Pei, G.; et al. Beauvericin counteracted multi-drug resistant Candida albicans by blocking ABC transporters. Synth. Syst. Biotechnol. 2016, 1, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ruan, C.; Bai, X.; Zhang, M.; Zhu, S.; Jiang, Y. Isolation and identification of the antimicrobial agent beauvericin from the endophytic Fusarium oxysporum 5-19 with NMR and ESI-MS/MS. Biomed. Res. Int. 2016, 2016, 1–5. [Google Scholar] [CrossRef]
- Wu, Q.; Patocka, J.; Nepovimova, E.; Kuca, K. A review on the synthesis and bioactivity aspects of beauvericin, a fusarium mycotoxin. Front. Pharmacol. 2018, 9, 1338–1350. [Google Scholar] [CrossRef]
- Paciolla, C.; Dipierro, N.; Mule, G.; Logrieco, A.; Dipierro, S. The mycotoxins beauvericin and T-2 induce cell death and alteration to the ascorbate metabolism in tomato protoplasts. Physiol. Mol. Plant. Pathol. 2004, 65, 49–56. [Google Scholar] [CrossRef]
- Šrobárová, A.; da Silva, J.A.T.; Kogan, G.; Ritieni, A.; Santini, A. Beauvericin decreases cell viability of wheat. Chem. Biodivers. 2009, 6, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Casini, A.F.; Nishikimi, M. Molecular cloning and functional expression of rat liver glutathione-dependent dehydroascorbate reductase. J. Biol. Chem. 1998, 273, 28708–28712. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Sano, S. Molecular and functional characterization of monodehydro-ascorbate and dehydroascorbate reductases. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M.A., Munné-Bosch, S., Burritt, D.J., Diaz-Vivancos, P., Fujita, M., Lorence, A., Eds.; Springer: Cham, Switzerland, 2017; Volume 1, pp. 129–156. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef]
- Noshi, M.; Yamada, H.; Hatanaka, R.; Tanabe, N.; Tamoi, M.; Shigeoka, S. Arabidopsis dehydroascorbate reductase 1 and 2 modulate redox states of ascorbate-glutathione cycle in the cytosol in response to photooxidative stress. Biosci. Biotechnol. Biochem. 2017, 81, 523–533. [Google Scholar] [CrossRef]
- Huang, D. Dietary antioxidants and health promotion. Antioxidants 2018, 7, 9. [Google Scholar] [CrossRef]
- Paciolla, C.; Fortunato, S.; Dipierro, N.; Paradiso, A.; De Leonardis, S.; Mastropasqua, L.; de Pinto, M.C. Vitamin C in Plants: From Functions to Biofortification. Antioxidants 2019, 8, 519. [Google Scholar] [CrossRef]
- Vicente, O.; Boscaiu, M. Flavonoids: Antioxidant compounds for plant defence… and for a healthy human diet. Not. Bot. Horti Agrobot. Cluj Napoca 2018, 46, 14–21. [Google Scholar] [CrossRef]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369. [Google Scholar] [CrossRef]
- Loi, M.; Paciolla, C.; Logrieco, A.F.; Mulè, G. Plant Bioactive Compounds in Pre-and Postharvest Management for Aflatoxins Reduction. Front. Microbiol. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Hussain, S.; Rao, M.J.; Anjum, M.A.; Ejaz, S.; Zakir, I.; Ali, M.A.; Ahmad, N.; Ahmad, S. Oxidative stress and antioxidant defense in plants under drought conditions. In Plant Abiotic Stress Tolerance, 1st ed.; Hasanuzzaman, M., Rehman, K., Nahar, K., Alharby, H.F., Eds.; Springer: Cham, Switzerland, 2019; Volume 1, pp. 207–219. [Google Scholar]
- Hasanuzzaman, M.; Hossain, M.A.; da Silva, J.A.T.; Fujita, M. Plant Response and Tolerance to Abiotic Oxidative Stress: Antioxidant Defense Is a Key Factor. In Crop Stress and its Management: Perspectives and Strategies; Venkateswarlu, B., Shanker, A., Shanker, C., Maheswari, M., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 261–315. [Google Scholar] [CrossRef]
- Chen, Z.; Young, T.E.; Ling, J.; Chang, S.C.; Gallie, D.R. Increasing vitamin C content of plants through enhanced ascorbate recycling. PNAS 2003, 100, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Holler, S.; Ueda, Y.; Wu, L.; Wang, Y.; Hajirezaei, M.R.; Ghaffari, M.R.; von Wiren, N.; Frei, M. Ascorbate biosynthesis and its involvement in stress tolerance and plant development in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wang, S.; Eltayeb, A.E.; Uddin, M.I.; Yamamoto, Y.; Tsuji, W.; Takeuchi, Y.; Tanaka, K. Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 2010, 231, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Paciolla, C.; Ippolito, M.P.; Logrieco, A.; Dipierro, N.; Mule, G.; Dipierro, S. A different trend of antioxidant defence responses makes tomato plants less susceptible to beauvericin than to T-2 mycotoxin phytotoxicity. Physiol. Mol. Plant. Path. 2008, 72, 3–9. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Macknight, R.C.; Laing, W.A.; Bulley, S.M.; Broad, R.C.; Johnson, A.A.; Hellens, R.P. Increasing ascorbate levels in crops to enhance human nutrition and plant abiotic stress tolerance. Curr. Opin. Biotechnol. 2017, 44, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Kouri, K.; Lemmens, M.; Lemmens-Gruber, R. Beauvericin-induced channels in ventricular myocytes and liposomes. Biochim. Biophys. Acta 2003, 1609, 203–210. [Google Scholar] [CrossRef]
- Demidchik, V.; Shabala, S. Mechanisms of cytosolic calcium elevation in plants: The role of ion channels, calcium extrusion systems and NADPH oxidase-mediated ‘ROS-Ca2+ Hub’. Funct. Plant. Biol. 2018, 45, 9–27. [Google Scholar] [CrossRef]
- Medvedev, S.S. Principles of calcium signal generation and transduction in plant cells. Russ. J. Plant. Physiol. 2018, 65, 771–783. [Google Scholar] [CrossRef]
- Bilska, K.; Wojciechowska, N.; Alipour, S.; Kalemba, E.M. Ascorbic Acid—The Little-Known Antioxidant in Woody Plants. Antioxidants 2019, 8, 645. [Google Scholar] [CrossRef]
- Rahantaniaina, M.S.; Li, S.; Chatel-Innocenti, G.; Tuzet, A.; Mhamdi, A.; Vanacker, H.; Noctor, G. Glutathione oxidation in response to intracellular H2O2: Key but overlapping roles for dehydroascorbate reductases. Plant. Signal. Behav. 2017, 12, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Deponte, M. The incomplete glutathione puzzle: Just guessing at numbers and figures? Antioxid. Redox Signal. 2017, 27, 1130–1161. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Li, H.; Ouyang, B.; Zhang, J.; Ye, Z. Cloning and mapping of genes involved in tomato ascorbic acid biosynthesis and metabolism. Plant. Sci. 2006, 170, 120–127. [Google Scholar] [CrossRef]
- De Gara, L.; Paciolla, C.; Tommasi, F.; Liso, R.; Arrigoni, O. «In vivo» Inhibition of Galactono-γ-Lactone Conversion to Ascorbate by Lycorine. J. Plant. Physiol. 1994, 144, 649–653. [Google Scholar] [CrossRef]
- De Gara, L.; Paciolla, C.; De Tullio, C.M.; Motto, M.; Arrigoni, O. Ascorbate-dependent hydrogen peroxide detoxification and ascorbate regeneration during germination of a highly productive maize hybrid: Evidence of an improved detoxification mechanism against reactive oxygen species. Physiol. Plant. 2000, 109, 7–13. [Google Scholar] [CrossRef]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic acid-a potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant. Sci. 2017, 8, 613–630. [Google Scholar] [CrossRef]
- Dixon, D.P.; Edwards, R. Glutathione transferases. Arab. Book 2010, 8, 1–15. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Lin, S.-T.; Ko, T.P.; Wu, S.M.; Lin, T.H.; Chang, Y.C.; Huang, F.A.; Lee, T.M. Enzymatic characterization and crystal structure analysis of Chlamydomonas reinhardtii dehydroascorbate reductase and their implications for oxidative stress. Plant. Physiol. Biochem. 2017, 120, 144–155. [Google Scholar] [CrossRef]
- Vethanayagam, J.G.; Green, E.H.; Rose, R.C.; Bode, A.M. Glutathione-dependent ascorbate recycling activity of rat serum albumin. Free Radic. Biol. Med. 1999, 26, 1591–1598. [Google Scholar] [CrossRef]
- Lu, Y.L.; Chia, C.Y.; Liu, Y.W.; Hou, W.C. Biological activities and applications of dioscorins, the major tuber storage proteins of yam. J. Tradit. Complement. Med. 2012, 2, 41–46. [Google Scholar] [CrossRef]
- Heyneke, E.; Fernie, A.R. Metabolic regulation of photosynthesis. Biochem. Soc. Trans. 2018, 46, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Woodrow, I.E.; Murphy, D.J.; Latzko, E. Regulation of stromal sedoheptulose 1, 7-bisphosphatase activity by pH and Mg2+ concentration. J. Biol. Chem. 1984, 259, 3791–3795. [Google Scholar] [PubMed]
- Gütle, D.D.; Roret, T.; Müller, S.J.; Couturier, J.; Lemaire, S.D.; Hecker, A.; Dhalleine, T.; Buchanan, B.B.; Reski, R.; Einsle, O.; et al. Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. PNAS 2016, 113, 6779–6784. [Google Scholar] [CrossRef]
- Portis, A.R., Jr.; Chon, C.J.; Mosbach, A.; Heldt, H.W. Fructose-and sedoheptulose bisphosphatase. The sites of a possible control of CO2 fixation by light-dependent changes of the stromal Mg2+ concentration. Biochim. Biophys. Acta Bioenerg. 1977, 461, 313–325. [Google Scholar] [CrossRef]
- Ding, F.; Wang, M.; Zhang, S.; Ai, X. Changes in SBPase activity influence photosynthetic capacity, growth, and tolerance to chilling stress in transgenic tomato plants. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Ding, F.; Wang, M.; Zhang, S. Overexpression of a Calvin cycle enzyme SBPase improves tolerance to chilling-induced oxidative stress in tomato plants. Sci. Hortic. 2017, 214, 27–33. [Google Scholar] [CrossRef]
- Vian, A.; Henry-Vian, C.; Davies, E. Rapid and systemic accumulation of chloroplast mRNA-binding protein transcripts after flame stimulus in tomato. Plant Physiol. 1999, 121, 517–524. [Google Scholar] [CrossRef]
- Ruggieri, G.M.; Triassi, A.; Alvarez, C.E.; Gola, A.; Wiggenhauser, J.; Budde, C.O.; Lara, M.V.; Drincovich, M.F.; Müller, G.L. Overexpression of glycine-rich RNA-binding protein in tomato renders fruits with higher protein content after cold storage. Biol. Plant. 2018, 62, 501–510. [Google Scholar] [CrossRef]
- Stanković, B.; Vian, A.; Henry-Vian, C.; Davies, E. Molecular cloning and characterization of a tomato cDNA encoding a systemically wound-inducible bZIP DNA-binding protein. Planta 2000, 212, 60–66. [Google Scholar] [CrossRef]
- Kolton, M.; Keren, I.; Shevtsov, S.; Shaya, F.; Peled-Zehavi, H.; Danon, A.; Ostersetzer-Biran, O. Plastidic redox switches: Ferredoxins as novel RNA-binding proteins. Endocytobiosis Cell Res. 2011, 21, 1–18. [Google Scholar]
- Marondedze, C.; Ludivine, T.; Serrano, N.L.; Lilley, K.S.; Gehring, C. The RNA-binding protein repertoire of Arabidopsis thaliana. Sci. Rep. 2016, 6, 29766. [Google Scholar] [CrossRef] [PubMed]



| Assigned Protein | Accession Number | Protein Coverage | Sequence | m/z |
|---|---|---|---|---|
| Chloroplast sedoheptulose-1,7-bisphosphatase | C5IU71 | 15.7% | HEFLLLDEGK | 1669.80 |
| YTGGMVPDNQIIVK | 1633.82 | |||
| YTGGMVPDNQIIVK (ox) | 1633.82 | |||
| FEETLYGSSR | 1188.57 | |||
| TTYVLALK | 908.49 | |||
| MFSPGNLR | 921.45 | |||
| GIFTNVTSPTAK | 1236.67 | |||
| chloroplast mRNA-binding protein | Q9XEJ6 | 14.9% | AVTLDGMAR | 948.47 |
| IFNCVSDR | 952.44 | |||
| FSEITGAGGR | 993.49 | |||
| NMHFYAEPR | 1163.52 | |||
| DCEEWFFDR | 1254.48 | |||
| ILEGEVFDAVLDNNGK | 1731.87 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loi, M.; De Leonardis, S.; Mulè, G.; Logrieco, A.F.; Paciolla, C. A Novel and Potentially Multifaceted Dehydroascorbate Reductase Increasing the Antioxidant Systems is Induced by Beauvericin in Tomato. Antioxidants 2020, 9, 435. https://doi.org/10.3390/antiox9050435
Loi M, De Leonardis S, Mulè G, Logrieco AF, Paciolla C. A Novel and Potentially Multifaceted Dehydroascorbate Reductase Increasing the Antioxidant Systems is Induced by Beauvericin in Tomato. Antioxidants. 2020; 9(5):435. https://doi.org/10.3390/antiox9050435
Chicago/Turabian StyleLoi, Martina, Silvana De Leonardis, Giuseppina Mulè, Antonio F. Logrieco, and Costantino Paciolla. 2020. "A Novel and Potentially Multifaceted Dehydroascorbate Reductase Increasing the Antioxidant Systems is Induced by Beauvericin in Tomato" Antioxidants 9, no. 5: 435. https://doi.org/10.3390/antiox9050435
APA StyleLoi, M., De Leonardis, S., Mulè, G., Logrieco, A. F., & Paciolla, C. (2020). A Novel and Potentially Multifaceted Dehydroascorbate Reductase Increasing the Antioxidant Systems is Induced by Beauvericin in Tomato. Antioxidants, 9(5), 435. https://doi.org/10.3390/antiox9050435