Anti-Viral Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications
Abstract
1. Introduction
2. Oxidative Stress-Related Pathogenesis
3. The Keap1–Nrf2 Pathway
4. Keap1/Nrf2 Targeted Drug Design
5. Nrf2 Targeting by Curcumin as a Therapeutic Agent
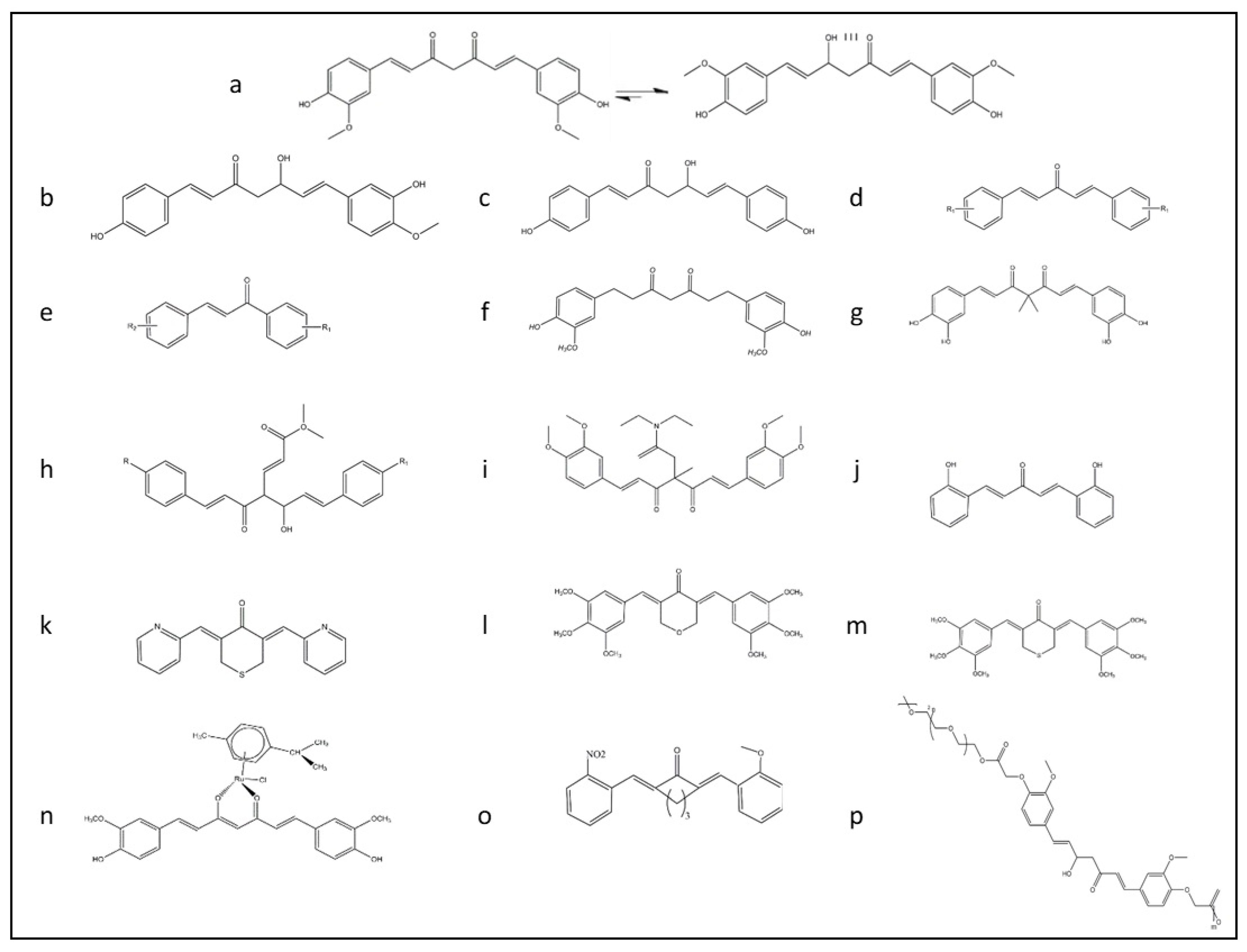
6. Formulation and Functional Improvement of Curcumin
7. Amelioration of the Candidate Diseases through Nrf2 Modulation by Curcumin
7.1. Kidney Diseases
7.2. Neurological Diseases
7.3. Liver Diseases
7.4. Diabetes
7.5. Cancer
8. Anti-Viral Effects of Curcumin
Curcumin Prevents Viral Infection Via Modulating Nrf2
9. Potential Mechanism of Curcumin on Pandemic Virus COVID-19
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arefin, S.; Buchanan, S.; Hobson, S.; Steinmetz, J.; Alsalhi, S.; Shiels, P.G.; Kublickiene, K.; Stenvinkel, P. Nrf2 in early vascular ageing: Calcification, senescence and therapy. Clin. Chim. Acta 2020, 505, 108–118. [Google Scholar] [CrossRef]
- Kurutas, E.B. The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: Current state. Nutr. J. 2015, 15, 1–22. [Google Scholar] [CrossRef]
- Wei, W.; Ma, N.; Fan, X.; Yu, Q.; Ci, X. The role of Nrf2 in acute kidney injury: Novel molecular mechanisms and therapeutic approaches. Free Radic. Biol. Med. 2020, 158, 1–12. [Google Scholar] [CrossRef]
- Kim, J.; Cha, Y.-N.; Surh, Y.-J. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2010, 690, 12–23. [Google Scholar] [CrossRef]
- Clarke, J.L.; Murray, J.B.; Park, B.K.; Copple, I.M. Roles of Nrf2 in drug and chemical toxicity. Curr. Opin. Toxicol. 2016, 1, 104–110. [Google Scholar] [CrossRef][Green Version]
- Dai, X.; Yan, X.; Wintergerst, K.A.; Cai, L.; Keller, B.B.; Tan, Y. Nrf2: Redox and metabolic regulator of stem cell state and function. Trends Mol. Med. 2020, 26, 185–200. [Google Scholar] [CrossRef]
- Ramezani, A.; Nahad, M.P.; Faghihloo, E. The role of Nrf2 transcription factor in viral infection. J. Cell. Biochem. 2018, 119, 6366–6382. [Google Scholar] [CrossRef]
- Li, Q.; Xing, S.; Chen, Y.; Liao, Q.; Li, Q.; Liu, Y.; He, S.; Feng, F.; Chen, Y.; Zhang, J. Reasonably activating Nrf2: A long-term, effective and controllable strategy for neurodegenerative diseases. Eur. J. Med. Chem. 2020, 185, 111862. [Google Scholar] [CrossRef]
- Saso, L.; Gürer-Orhan, H.; Stepanić, V. Modulators of oxidative stress: Chemical and pharmacological aspects. Antioxidants 2020, 9, 657. [Google Scholar] [CrossRef]
- Adefegha, S.A. Functional foods and nutraceuticals as dietary intervention in chronic diseases; Novel perspectives for health promotion and disease prevention. J. Diet. Suppl. 2018, 15, 977–1009. [Google Scholar] [CrossRef]
- Wang, L.; Li, W.; Cheng, D.; Guo, Y.; Wu, R.; Yin, R.; Li, S.; Kuo, H.-C.; Hudlikar, R.; Yang, H. Pharmacokinetics and pharmacodynamics of three oral formulations of curcumin in rats. J. Pharmacokinet. Pharmacodyn. 2020, 47, 131–144. [Google Scholar] [CrossRef]
- Kharat, M.; McClements, D.J. Recent advances in colloidal delivery systems for nutraceuticals: A case study—Delivery by Design of curcumin. J. Colloid Interface Sci. 2019, 557, 506–518. [Google Scholar] [CrossRef]
- Lee, W.-H.; Loo, C.-Y.; Bebawy, M.; Luk, F.; Mason, R.S.; Rohanizadeh, R. Curcumin and its derivatives: Their application in neuropharmacology and neuroscience in the 21st century. Curr. Neuropharmacol. 2013, 11, 338–378. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, N.; He, H.; Tang, X. Pharmaceutical strategies of improving oral systemic bioavailability of curcumin for clinical application. J. Control. Release 2019, 316, 359–380. [Google Scholar] [CrossRef]
- Shome, S.; Talukdar, A.D.; Choudhury, M.D.; Bhattacharya, M.K.; Upadhyaya, H. Curcumin as potential therapeutic natural product: A nanobiotechnological perspective. J. Pharm. Pharmacol. 2016, 68, 1481–1500. [Google Scholar] [CrossRef]
- Li, H.; Sureda, A.; Devkota, H.P.; Pittalà, V.; Barreca, D.; Silva, A.S.; Tewari, D.; Xu, S.; Nabavi, S.M. Curcumin, the golden spice in treating cardiovascular diseases. Biotechnol. Adv. 2020, 38, 107343. [Google Scholar] [CrossRef]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free radicals: Properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef]
- Lushchak, V.I. Free radicals, reactive oxygen species, oxidative stress and its classification. Chem. Biol. Interact. 2014, 224, 164–175. [Google Scholar] [CrossRef]
- Wu, X.; Cobbina, S.J.; Mao, G.; Xu, H.; Zhang, Z.; Yang, L. A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environ. Sci. Pollut. Res. 2016, 23, 8244–8259. [Google Scholar] [CrossRef]
- Ma, Q. Transcriptional responses to oxidative stress: Pathological and toxicological implications. Pharmacol. Ther. 2010, 125, 376–393. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS sources in physiological and pathological conditions. Oxidative Med. Cell. Longev. 2016, 2016. [Google Scholar] [CrossRef]
- Khan, A.U.; Wilson, T. Reactive oxygen species as cellular messengers. Chem. Biol. 1995, 2, 437–445. [Google Scholar] [CrossRef]
- Burgos-Morón, E.; Abad-Jiménez, Z.; Martínez de Marañón, A.; Iannantuoni, F.; Escribano-López, I.; López-Domènech, S.; Salom, C.; Jover, A.; Mora, V.; Roldan, I. Relationship between oxidative stress, ER stress, and inflammation in type 2 diabetes: The battle continues. J. Clin. Med. 2019, 8, 1385. [Google Scholar] [CrossRef]
- Kandlur, A.; Satyamoorthy, K.; Gangadharan, G. Oxidative stress in cognitive and epigenetic aging: A retrospective glance. Front. Mol. Neurosci. 2020, 13. [Google Scholar] [CrossRef]
- Deshmukh, P.; Unni, S.; Krishnappa, G.; Padmanabhan, B. The Keap1-Nrf2 pathway: Promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys. Rev. 2017, 9, 41–56. [Google Scholar] [CrossRef]
- Bellezza, I.; Giambanco, I.; Minelli, A.; Donato, R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta BBA Mol. Cell Res. 2018, 1865, 721–733. [Google Scholar] [CrossRef]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Ahmed, S.M.U.; Luo, L.; Namani, A.; Wang, X.J.; Tang, X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2017, 1863, 585–597. [Google Scholar] [CrossRef]
- Mohan, S.; Gupta, D. Crosstalk of toll-like receptors signaling and Nrf2 pathway for regulation of inflammation. Biomed. Pharmacother. 2018, 108, 1866–1878. [Google Scholar] [CrossRef]
- Madden, S.K.; Itzhaki, L.S. Structural and mechanistic insights into the Keap1-Nrf2 system as a route to drug discovery. Biochim. Biophys. Acta BBA Proteins Proteom. 2020, 1868, 140405. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Robledinos-Antón, N.; Fernández-Ginés, R.; Manda, G.; Cuadrado, A. Activators and inhibitors of NRF2: A review of their potential for clinical development. Oxidative Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Kong, A.-N. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 2009, 48, 91–104. [Google Scholar] [CrossRef]
- Cleasby, A.; Yon, J.; Day, P.J.; Richardson, C.; Tickle, I.J.; Williams, P.A.; Callahan, J.F.; Carr, R.; Concha, N.; Kerns, J.K. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS ONE 2014, 9, e98896. [Google Scholar] [CrossRef]
- Furukawa, M.; Xiong, Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 2005, 25, 162–171. [Google Scholar] [CrossRef]
- Richardson, B.; Jain, A.; Speltz, T.; Moore, T. Non-electrophilic modulators of the canonical Keap1/Nrf2 pathway. Bioorg. Med. Chem. Lett. 2015, 25, 2261–2268. [Google Scholar] [CrossRef]
- Canning, P.; Sorrell, F.J.; Bullock, A.N. Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med. 2015, 88, 101–107. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Kostov, R.V.; Canning, P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch. Biochem. Biophys. 2017, 617, 84–93. [Google Scholar] [CrossRef]
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef]
- Bai, F.; Zhang, B.; Hou, Y.; Yao, J.; Xu, Q.; Xu, J.; Fang, J. Xanthohumol analogues as potent Nrf2 activators against oxidative stress mediated damages of PC12 cells. ACS Chem. Neurosci. 2019, 10, 2956–2966. [Google Scholar] [CrossRef] [PubMed]
- Adelusi, T.I.; Du, L.; Hao, M.; Zhou, X.; Xuan, Q.; Apu, C.; Sun, Y.; Lu, Q.; Yin, X. Keap1/Nrf2/ARE signaling unfolds therapeutic targets for redox imbalanced-mediated diseases and diabetic nephropathy. Biomed. Pharmacother. 2020, 123, 109732. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, C.; Wu, Z.; Xing, C.; Miao, Z. Small molecules inhibiting Keap1–Nrf2 protein–protein interactions: A novel approach to activate Nrf2 function. MedChemComm 2017, 8, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Wen, S.; Li, Y.-X.; Gao, X.-X.; Zhang, X.; Jiang, Z.-Y. Recent progress in Keap1-Nrf2 protein-protein interaction inhibitors. Eur. J. Med. Chem. 2020, 202, 112532. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Massiah, M.A.; Bozak, R.E.; Hicks, R.J.; Talalay, P. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc. Natl. Acad. Sci. USA 2001, 98, 3404–3409. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Padmanabhan, B.; Kobayashi, A.; Shang, C.; Hirotsu, Y.; Yokoyama, S.; Yamamoto, M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007, 27, 7511–7521. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “Tethering” mechanism a two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006, 281, 24756–24768. [Google Scholar] [CrossRef]
- Davies, T.G.; Wixted, W.E.; Coyle, J.E.; Griffiths-Jones, C.; Hearn, K.; McMenamin, R.; Norton, D.; Rich, S.J.; Richardson, C.; Saxty, G. Monoacidic inhibitors of the Kelch-like ECH-associated protein 1: Nuclear factor erythroid 2-related factor 2 (KEAP1: NRF2) protein-protein interaction with high cell potency identified by fragment-based discovery. J. Med. Chem. 2016, 59, 3991–4006. [Google Scholar] [CrossRef]
- Qu, Z.; Sun, J.; Zhang, W.; Yu, J.; Zhuang, C. Transcription factor NRF2 as a promising therapeutic target for Alzheimer’s disease. Free Radic. Biol. Med. 2020, 159, 87–102. [Google Scholar] [CrossRef]
- Huang, Y.; Cao, S.; Zhang, Q.; Zhang, H.; Fan, Y.; Qiu, F.; Kang, N. Biological and pharmacological effects of hexahydrocurcumin, a metabolite of curcumin. Arch. Biochem. Biophys. 2018, 646, 31–37. [Google Scholar] [CrossRef]
- Najjar, F.M.; Taghavi, F.; Ghadari, R.; Sheibani, N.; Moosavi-Movahedi, A.A. Destructive effect of non-enzymatic glycation on catalase and remediation via curcumin. Arch. Biochem. Biophys. 2017, 630, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Jamwal, R. Bioavailable curcumin formulations: A review of pharmacokinetic studies in healthy volunteers. J. Integr. Med. 2018, 16, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Huminiecki, L.; Horbańczuk, J.; Atanasov, A.G. The functional genomic studies of curcumin. Semin. Cancer Biol. 2017, 46, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.; Sanderson, I.R.; MacDonald, T.T. Curcumin as a therapeutic agent: The evidence from in vitro, animal and human studies. Br. J. Nutr. 2010, 103, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Hatcher, H.; Planalp, R.; Cho, J.; Torti, F.; Torti, S. Curcumin: From ancient medicine to current clinical trials. Cell. Mol. Life Sci. 2008, 65, 1631–1652. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Acharya, A.; Ray, R.; Agrawal, R.; Raghuwanshi, R.; Jain, P. Cellular and molecular mechanisms of curcumin in prevention and treatment of disease. Crit. Rev. Food Sci. Nutr. 2020, 60, 887–939. [Google Scholar] [CrossRef]
- Negrette-Guzmán, M. Combinations of the antioxidants sulforaphane or curcumin and the conventional antineoplastics cisplatin or doxorubicin as prospects for anticancer chemotherapy. Eur. J. Pharmacol. 2019, 859, 172513. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Dashwood, R.H. Epigenetic regulation of NRF2/KEAP1 by phytochemicals. Antioxidants 2020, 9, 865. [Google Scholar] [CrossRef]
- Jiang, C.; Luo, P.; Li, X.; Liu, P.; Li, Y.; Xu, J. Nrf2/ARE is a key pathway for curcumin-mediated protection of TMJ chondrocytes from oxidative stress and inflammation. Cell Stress Chaperones 2020, 25, 1–12. [Google Scholar] [CrossRef]
- Li, X.; Chen, Y.; Mao, Y.; Dai, P.; Sun, X.; Zhang, X.; Cheng, H.; Wang, Y.; Banda, I.; Wu, G. Curcumin protects osteoblasts from oxidative stress-induced dysfunction via GSK3β-Nrf2 signaling pathway. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Abrahams, S.; Haylett, W.L.; Johnson, G.; Carr, J.A.; Bardien, S. Antioxidant effects of curcumin in models of neurodegeneration, aging, oxidative and nitrosative stress: A review. Neuroscience 2019, 406, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Lee, C. Therapeutic modulation of virus-induced oxidative stress via the Nrf2-dependent antioxidative pathway. Oxidative Med. Cell. Longev. 2018, 2018, 6208067. [Google Scholar] [CrossRef] [PubMed]
- Paunkov, A.; Chartoumpekis, D.V.; Ziros, P.G.; Sykiotis, G.P. A bibliometric review of the Keap1/Nrf2 pathway and its related antioxidant compounds. Antioxidants 2019, 8, 353. [Google Scholar] [CrossRef]
- Meng, F.-C.; Zhou, Y.-Q.; Ren, D.; Wang, R.; Wang, C.; Lin, L.-G.; Zhang, X.-Q.; Ye, W.-C.; Zhang, Q.-W. Turmeric: A review of its chemical composition, quality control, bioactivity, and pharmaceutical application. In Natural and Artificial Flavoring Agents and Food Dyes; Elsevier: Amsterdam, The Netherlands, 2018; pp. 299–350. [Google Scholar]
- Jiang, T.; Liao, W.; Charcosset, C. Recent advances in encapsulation of curcumin in nanoemulsions: A review of encapsulation technologies, bioaccessibility and applications. Food Res. Int. 2020, 132, 109035. [Google Scholar] [CrossRef] [PubMed]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A review of its’ effects on human health. Foods 2017, 6, 92. [Google Scholar] [CrossRef]
- Kimura, S.; Kiriyama, A.; Araki, K.; Yoshizumi, M.; Enomura, M.; Inoue, D.; Furubayashi, T.; Yutani, R.; Teraoka, R.; Tanaka, A. Novel strategy for improving the bioavailability of curcumin based on a new membrane transport mechanism that directly involves solid particles. Eur. J. Pharm. Biopharm. 2018, 122, 1–5. [Google Scholar] [CrossRef]
- Cheng, D.; Li, W.; Wang, L.; Lin, T.; Poiani, G.; Wassef, A.; Hudlikar, R.; Ondar, P.; Brunetti, L.; Kong, A.-N. Pharmacokinetics, pharmacodynamics, and PKPD modeling of curcumin in regulating antioxidant and epigenetic gene expression in healthy human volunteers. Mol. Pharm. 2019, 16, 1881–1889. [Google Scholar] [CrossRef]
- Mohammadian, M.; Waly, M.I.; Moghadam, M.; Emam-Djomeh, Z.; Salami, M.; Moosavi-Movahedi, A.A. Nanostructured food proteins as efficient systems for the encapsulation of bioactive compounds. Food Sci. Hum. Wellness 2020, 9, 199–213. [Google Scholar] [CrossRef]
- Deck, L.M.; Hunsaker, L.A.; Vander Jagt, T.A.; Whalen, L.J.; Royer, R.E.; Vander Jagt, D.L. Activation of anti-oxidant Nrf2 signaling by enone analogues of curcumin. Eur. J. Med. Chem. 2018, 143, 854–865. [Google Scholar] [CrossRef]
- Shin, J.W.; Chun, K.-S.; Kim, D.-H.; Kim, S.-J.; Kim, S.H.; Cho, N.-C.; Na, H.-K.; Surh, Y.-J. Curcumin induces stabilization of Nrf2 protein through Keap1 cysteine modification. Biochem. Pharmacol. 2020, 173, 113820. [Google Scholar] [CrossRef]
- Tu, Z.-S.; Wang, Q.; Sun, D.-D.; Dai, F.; Zhou, B. Design, synthesis, and evaluation of curcumin derivatives as Nrf2 activators and cytoprotectors against oxidative death. Eur. J. Med. Chem. 2017, 134, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, R.M.C.; Pruccoli, L.; Bisi, A.; Gobbi, S.; Rampa, A.; Martinez, A.; Pérez, C.; Martinez-Gonzalez, L.; Paglione, M.; Di Schiavi, E. Novel curcumin-diethyl fumarate hybrid as a dualistic GSK-3β inhibitor/Nrf2 inducer for the treatment of Parkinson’s disease. ACS Chem. Neurosci. 2020, 11, 2728–2740. [Google Scholar] [CrossRef] [PubMed]
- Su, I.-J.; Chang, H.-Y.; Wang, H.-C.; Tsai, K.-J. A curcumin analog exhibits multiple biologic effects on the pathogenesis of Alzheimer’s disease and improves behavior, inflammation, and β-amyloid accumulation in a mouse model. Int. J. Mol. Sci. 2020, 21, 5459. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Jiang, T.; Long, M.; Chen, J.; Ren, D.-M.; Wong, P.K.; Chapman, E.; Zhou, B.; Zhang, D.D. A curcumin derivative that inhibits vinyl carbamate-induced lung carcinogenesis via activation of the Nrf2 protective response. Antioxid. Redox Signal. 2015, 23, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Pung, D.; Su, Z.-Y.; Guo, Y.; Zhang, C.; Yang, A.Y.; Zheng, X.; Du, Z.-Y.; Zhang, K.; Kong, A.-N. Epigenetics reactivation of Nrf2 in prostate TRAMP C1 cells by curcumin analogue FN1. Chem. Res. Toxicol. 2016, 29, 694–703. [Google Scholar] [CrossRef]
- Li, W.; Su, Z.-Y.; Guo, Y.; Zhang, C.; Wu, R.; Gao, L.; Zheng, X.; Du, Z.-Y.; Zhang, K.; Kong, A.-N. Curcumin derivative epigenetically reactivates Nrf2 antioxidative stress signaling in mouse prostate cancer TRAMP C1 cells. Chem. Res. Toxicol. 2018, 31, 88–96. [Google Scholar] [CrossRef]
- Garufi, A.; Baldari, S.; Pettinari, R.; Montani, M.S.G.; D’Orazi, V.; Pistritto, G.; Crispini, A.; Giorno, E.; Toietta, G.; Marchetti, F. A ruthenium (II)-curcumin compound modulates NRF2 expression balancing the cancer cell death/survival outcome according to p53 status. J. Exp. Clin. Cancer Res. 2020, 39, 1–15. [Google Scholar] [CrossRef]
- Xiang, L.; Zhang, Q.; Chi, C.; Wu, G.; Lin, Z.; Li, J.; Gu, Q.; Chen, G. Curcumin analog A13 alleviates oxidative stress by activating Nrf2/ARE pathway and ameliorates fibrosis in the myocardium of high-fat-diet and streptozotocin-induced diabetic rats. Diabetol. Metab. Syndr. 2020, 12, 1–8. [Google Scholar] [CrossRef]
- Mohajeri, M.; Sadeghizadeh, M.; Najafi, F.; Javan, M. Polymerized nano-curcumin attenuates neurological symptoms in EAE model of multiple sclerosis through down regulation of inflammatory and oxidative processes and enhancing neuroprotection and myelin repair. Neuropharmacology 2015, 99, 156–167. [Google Scholar] [CrossRef]
- Pandey, M.K.; Kumar, S.; Thimmulappa, R.K.; Parmar, V.S.; Biswal, S.; Watterson, A.C. Design, synthesis and evaluation of novel PEGylated curcumin analogs as potent Nrf2 activators in human bronchial epithelial cells. Eur. J. Pharm. Sci. 2011, 43, 16–24. [Google Scholar] [CrossRef]
- Sandhir, R.; Yadav, A.; Mehrotra, A.; Sunkaria, A.; Singh, A.; Sharma, S. Curcumin nanoparticles attenuate neurochemical and neurobehavioral deficits in experimental model of Huntington’s disease. Neuromol. Med. 2014, 16, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Askarizadeh, A.; Barreto, G.E.; Henney, N.C.; Majeed, M.; Sahebkar, A. Neuroprotection by curcumin: A review on brain delivery strategies. Int. J. Pharm. 2020, 585, 119476. [Google Scholar] [CrossRef] [PubMed]
- Nabila, N.; Suada, N.K.; Denis, D.; Yohan, B.; Adi, A.C.; Veterini, A.S.; Anindya, A.L.; Sasmono, R.T.; Rachmawati, H. Antiviral action of curcumin encapsulated in nanoemulsion against four serotypes of dengue virus. Pharm. Nanotechnol. 2020, 8, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Marbawati, D.; Umniyati, S.R. Effects of curcumin and pentagamavunon-0 against dengue-2 virus Infection in vero cells; an in vitro study. Procedia Environ. Sci. 2015, 23, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.X.; Li, C.M.; Huang, C.Z. Curcumin modified silver nanoparticles for highly efficient inhibition of respiratory syncytial virus infection. Nanoscale 2016, 8, 3040–3048. [Google Scholar] [CrossRef] [PubMed]
- Lagoa, R.; Silva, J.; Rodrigues, J.R.; Bishayee, A. Advances in phytochemical delivery systems for improved anticancer activity. Biotechnol. Adv. 2020, 38, 107382. [Google Scholar] [CrossRef] [PubMed]
- Gupte, P.A.; Giramkar, S.A.; Harke, S.M.; Kulkarni, S.K.; Deshmukh, A.P.; Hingorani, L.L.; Mahajan, M.P.; Bhalerao, S.S. Evaluation of the efficacy and safety of Capsule Longvida® optimized curcumin (solid lipid curcumin particles) in knee osteoarthritis: A pilot clinical study. J. Inflamm. Res. 2019, 12, 145–152. [Google Scholar] [CrossRef]
- Nahar, P.P.; Slitt, A.L.; Seeram, N.P. Anti-inflammatory effects of novel standardized solid lipid curcumin formulations. J. Med. Food 2015, 18, 786–792. [Google Scholar] [CrossRef]
- Sudheeran, S.P.; Jacob, D.; Mulakal, J.N.; Nair, G.G.; Maliakel, A.; Maliakel, B.; Kuttan, R.; Krishnakumar, I. Safety, tolerance, and enhanced efficacy of a bioavailable formulation of curcumin with fenugreek dietary fiber on occupational stress: A randomized, double-blind, placebo-controlled pilot study. J. Clin. Psychopharmacol. 2016, 36, 236–243. [Google Scholar] [CrossRef]
- Campbell, M.S.; Berrones, A.J.; Krishnakumar, I.; Charnigo, R.J.; Westgate, P.M.; Fleenor, B.S. Responsiveness to curcumin intervention is associated with reduced aortic stiffness in young, obese men with higher initial stiffness. J. Funct. Foods 2017, 29, 154–160. [Google Scholar] [CrossRef]
- Tortik, N.; Spaeth, A.; Plaetzer, K. Photodynamic decontamination of foodstuff from Staphylococcus aureus based on novel formulations of curcumin. Photochem. Photobiol. Sci. 2014, 13, 1402–1409. [Google Scholar] [CrossRef] [PubMed]
- Razi, M.A.; Wakabayashi, R.; Tahara, Y.; Goto, M.; Kamiya, N. Genipin-stabilized caseinate-chitosan nanoparticles for enhanced stability and anti-cancer activity of curcumin. Colloids Surf. B Biointerfaces 2018, 164, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, M.; Salami, M.; Mohammadian, M.; Emam-Djomeh, Z.; Jahanbani, R.; Moosavi-Movahedi, A.A. Physicochemical and bio-functional properties of walnut proteins as affected by trypsin-mediated hydrolysis. Food Biosci. 2020, 36, 100611. [Google Scholar] [CrossRef]
- Moghadam, M.; Salami, M.; Mohammadian, M.; Delphi, L.; Sepehri, H.; Emam-Djomeh, Z.; Moosavi-Movahedi, A.A. Walnut protein–curcumin complexes: Fabrication, structural characterization, antioxidant properties, and in vitro anticancer activity. J. Food Meas. Charact. 2020, 14, 876–885. [Google Scholar] [CrossRef]
- Jose, A.; Labala, S.; Venuganti, V.V.K. Co-delivery of curcumin and STAT3 siRNA using deformable cationic liposomes to treat skin cancer. J. Drug Target. 2017, 25, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.; Tagami, T.; Kishi, T.; Ozeki, T. Curcumin marinosomes as promising nano-drug delivery system for lung cancer. Int. J. Pharm. 2018, 540, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Aqil, F.; Munagala, R.; Jeyabalan, J.; Agrawal, A.K.; Gupta, R. Exosomes for the enhanced tissue bioavailability and efficacy of curcumin. AAPS J. 2017, 19, 1691–1702. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, M.; Moghadam, M.; Salami, M.; Emam-Djomeh, Z.; Alavi, F.; Momen, S.; Moosavi-Movahedi, A.A. Whey protein aggregates formed by non-toxic chemical cross-linking as novel carriers for curcumin delivery: Fabrication and characterization. J. Drug Deliv. Sci. Technol. 2020, 56, 101531. [Google Scholar] [CrossRef]
- Esmaili, M.; Ghaffari, S.M.; Moosavi-Movahedi, Z.; Atri, M.S.; Sharifizadeh, A.; Farhadi, M.; Yousefi, R.; Chobert, J.-M.; Haertlé, T.; Moosavi-Movahedi, A.A. Beta casein-micelle as a nano vehicle for solubility enhancement of curcumin; food industry application. LWT Food Sci. Technol. 2011, 44, 2166–2172. [Google Scholar] [CrossRef]
- Alavi, F.; Emam-Djomeh, Z.; Yarmand, M.S.; Salami, M.; Momen, S.; Moosavi-Movahedi, A.A. Cold gelation of curcumin loaded whey protein aggregates mixed with k-carrageenan: Impact of gel microstructure on the gastrointestinal fate of curcumin. Food Hydrocoll. 2018, 85, 267–280. [Google Scholar] [CrossRef]
- Mohammadian, M.; Salami, M.; Alavi, F.; Momen, S.; Emam-Djomeh, Z.; Moosavi-Movahedi, A.A. Fabrication and characterization of curcumin-loaded complex coacervates made of gum Arabic and whey protein nanofibrils. Food Biophys. 2019, 14, 425–436. [Google Scholar] [CrossRef]
- Al Fayi, M.; Otifi, H.; Alshyarba, M.; Dera, A.A.; Rajagopalan, P. Thymoquinone and curcumin combination protects cisplatin-induced kidney injury, nephrotoxicity by attenuating NFκB, KIM-1 and ameliorating Nrf2/HO-1 signalling. J. Drug Target. 2020, 28, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, J.; Chirino, Y.I.; Molina-Jijón, E.; Andérica-Romero, A.C.; Tapia, E.; Pedraza-Chaverrí, J. Renoprotective effect of the antioxidant curcumin: Recent findings. Redox Biol. 2013, 1, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Shelton, L.M.; Park, B.K.; Copple, I.M. Role of Nrf2 in protection against acute kidney injury. Kidney Int. 2013, 84, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Shopit, A.; Niu, M.; Wang, H.; Tang, Z.; Li, X.; Tesfaldet, T.; Ai, J.; Ahmad, N.; Al-Azab, M.; Tang, Z. Protection of diabetes-induced kidney injury by phosphocreatine via the regulation of ERK/Nrf2/HO-1 signaling pathway. Life Sci. 2020, 242, 117248. [Google Scholar] [CrossRef]
- Lee, J.-S.; Surh, Y.-J. Nrf2 as a novel molecular target for chemoprevention. Cancer Lett. 2005, 224, 171–184. [Google Scholar] [CrossRef]
- Balogun, E.; Hoque, M.; Gong, P.; Killeen, E.; Green, C.J.; Foresti, R.; Alam, J.; Motterlini, R. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem. J. 2003, 371, 887–895. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Liu, H.; Yuan, J.; Yin, Y.; Wang, T.; Cheng, B.; Sun, S.; Guo, Z. Curcumin ameliorates glyoxylate-induced calcium oxalate deposition and renal injuries in mice. Phytomedicine 2019, 61, 152861. [Google Scholar] [CrossRef]
- Choi, B.-H.; Kang, K.-S.; Kwak, M.-K. Effect of redox modulating NRF2 activators on chronic kidney disease. Molecules 2014, 19, 12727–12759. [Google Scholar] [CrossRef]
- Zhang, X.; Liang, D.; Guo, L.; Liang, W.; Jiang, Y.; Li, H.; Zhao, Y.; Lu, S.; Chi, Z.H. Curcumin protects renal tubular epithelial cells from high glucose-induced epithelial-to-mesenchymal transition through Nrf2-mediated upregulation of heme oxygenase-1. Mol. Med. Rep. 2015, 12, 1347–1355. [Google Scholar] [CrossRef]
- Wu, J.; Pan, X.; Fu, H.; Zheng, Y.; Dai, Y.; Yin, Y.; Chen, Q.; Hao, Q.; Bao, D.; Hou, D. Effect of curcumin on glycerol-induced acute kidney injury in rats. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Michaličková, D.; Hrnčíř, T.; Canová, N.K.; Slanař, O. Targeting Keap1/Nrf2/ARE signaling pathway in multiple sclerosis. Eur. J. Pharmacol. 2020, 873, 172973. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, M.J.; Borcar, A.; Proctor, J.L.; Greco, T.; Rosenthal, R.E.; Fiskum, G. Transcriptional activation of antioxidant gene expression by Nrf2 protects against mitochondrial dysfunction and neuronal death associated with acute and chronic neurodegeneration. Exp. Neurol. 2020, 328, 113247. [Google Scholar] [CrossRef]
- Abou El-ezz, D.; Maher, A.; Sallam, N.; El-brairy, A.; Kenawy, S. Trans-cinnamaldehyde modulates hippocampal Nrf2 factor and inhibits amyloid beta aggregation in LPS-induced neuroinflammation mouse model. Neurochem. Res. 2018, 43, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Suwanwela, N.C.; Patumraj, S. Curcumin by down-regulating NF-kB and elevating Nrf2, reduces brain edema and neurological dysfunction after cerebral I/R. Microvasc. Res. 2016, 106, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Wang, H.; Fang, J.; Zhu, Y.; Zhou, J.; Wang, X.; Zhou, Y.; Zhou, M. Curcumin provides neuroprotection in model of traumatic brain injury via the Nrf2-ARE signaling pathway. Brain Res. Bull. 2018, 140, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, M.S.; Banik, S.; Akter, M.; Rahman, M.M.; Sikder, M.T.; Hosokawa, T.; Saito, T.; Kurasaki, M. Curcumin alleviates arsenic-induced toxicity in PC12 cells via modulating autophagy/apoptosis. Ecotoxicol. Environ. Saf. 2020, 200, 110756. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, B.; Dhiman, M.; Mittal, S.; Mantha, A.K. Curcumin revitalizes Amyloid beta (25–35)-induced and organophosphate pesticides pestered neurotoxicity in SH-SY5Y and IMR-32 cells via activation of APE1 and Nrf2. Metab. Brain Dis. 2017, 32, 2045–2061. [Google Scholar] [CrossRef]
- Xie, Y.-L.; Chu, J.-G.; Jian, X.-M.; Dong, J.-Z.; Wang, L.-P.; Li, G.-X.; Yang, N.-B. Curcumin attenuates lipopolysaccharide/d-galactosamine-induced acute liver injury by activating Nrf2 nuclear translocation and inhibiting NF-kB activation. Biomed. Pharmacother. 2017, 91, 70–77. [Google Scholar] [CrossRef]
- Liu, Z.; Dou, W.; Zheng, Y.; Wen, Q.; Qin, M.; Wang, X.; Tang, H.; Zhang, R.; Lv, D.; Wang, J. Curcumin upregulates Nrf2 nuclear translocation and protects rat hepatic stellate cells against oxidative stress. Mol. Med. Rep. 2016, 13, 1717–1724. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, S.; Xiang, B.; Li, L.; Lin, Y. Curcumin attenuates oxaliplatin-induced liver injury and oxidative stress by activating the Nrf2 pathway. Drug Des. Dev. Ther. 2020, 14, 73. [Google Scholar] [CrossRef]
- Lu, C.; Jiang, Y.; Zhang, F.; Shao, J.; Wu, L.; Wu, X.; Lian, N.; Chen, L.; Jin, H.; Chen, Q. Tetramethylpyrazine prevents ethanol-induced hepatocyte injury via activation of nuclear factor erythroid 2-related factor 2. Life Sci. 2015, 141, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chang, X.; Zhan, H.; Zhang, Q.; Li, C.; Gao, Q.; Yang, M.; Luo, Z.; Li, S.; Sun, Y. Curcumin and Baicalin ameliorate ethanol-induced liver oxidative damage via the Nrf2/HO-1 pathway. J. Food Biochem. 2020, 44, e13425. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Osorio, A.S.; Gonzalez-Reyes, S.; Pedraza-Chaverri, J. Natural Nrf2 activators in diabetes. Clin. Chim. Acta 2015, 448, 182–192. [Google Scholar] [CrossRef] [PubMed]
- He, H.-J.; Wang, G.-Y.; Gao, Y.; Ling, W.-H.; Yu, Z.-W.; Jin, T.-R. Curcumin attenuates Nrf2 signaling defect, oxidative stress in muscle and glucose intolerance in high fat diet-fed mice. World J. Diabetes 2012, 3, 94–104. [Google Scholar] [CrossRef]
- Pugazhenthi, S.; Akhov, L.; Selvaraj, G.; Wang, M.; Alam, J. Regulation of heme oxygenase-1 expression by demethoxy curcuminoids through Nrf2 by a PI3-kinase/Akt-mediated pathway in mouse β-cells. Am. J. Physiol. Endocrinol. Metab. 2007, 293, 645–655. [Google Scholar] [CrossRef]
- Mani, B.K.; Osborne-Lawrence, S.; Metzger, N.; Zigman, J.M. Lowering oxidative stress in ghrelin cells stimulates ghrelin secretion. Am. J. Physiol. Endocrinol. Metab. 2020, 319, 330–337. [Google Scholar] [CrossRef]
- Vasileva, L.V.; Savova, M.S.; Amirova, K.M.; Dinkova-Kostova, A.T.; Georgiev, M.I. Obesity and NRF2-mediated cytoprotection: Where is the missing link? Pharmacol. Res. 2020, 156, 104760. [Google Scholar] [CrossRef]
- Jiang, Z.-Y.; Lu, M.-C.; You, Q.-D. Nuclear factor erythroid 2-related factor 2 (Nrf2) inhibition: An emerging strategy in cancer therapy. J. Med. Chem. 2018, 62, 3840–3856. [Google Scholar] [CrossRef]
- Xue, D.; Zhou, X.; Qiu, J. Emerging role of NRF2 in ROS-mediated tumor chemoresistance. Biomed. Pharmacother. Biomed. Pharmacother. 2020, 131, 110676. [Google Scholar] [CrossRef]
- Paciello, F.; Fetoni, A.R.; Mezzogori, D.; Rolesi, R.; Di Pino, A.; Paludetti, G.; Grassi, C.; Troiani, D. The dual role of curcumin and ferulic acid in counteracting chemoresistance and cisplatin-induced ototoxicity. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, M.R.; Chapman, E.; Zhang, D.D. NRF2 and the hallmarks of cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-Y.; Shien, J.-H.; Tiley, L.; Chiou, S.-S.; Wang, S.-Y.; Chang, T.-J.; Lee, Y.-J.; Chan, K.-W.; Hsu, W.-L. Curcumin inhibits influenza virus infection and haemagglutination activity. Food Chem. 2010, 119, 1346–1351. [Google Scholar] [CrossRef]
- Mazumder, A.; Raghavan, K.; Weinstein, J.; Kohn, K.W.; Pommier, Y. Inhibition of human immunodeficiency virus type-1 integrase by curcumin. Biochem. Pharmacol. 1995, 49, 1165–1170. [Google Scholar] [CrossRef]
- Sui, Z.; Salto, R.; Li, J.; Craik, C.; de Montellano, P.R.O. Inhibition of the HIV-1 and HIV-2 proteases by curcumin and curcumin boron complexes. Bioorg. Med. Chem. 1993, 1, 415–422. [Google Scholar] [CrossRef]
- Ali, A.; Banerjea, A.C. Curcumin inhibits HIV-1 by promoting Tat protein degradation. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Rechtman, M.M.; Har-Noy, O.; Bar-Yishay, I.; Fishman, S.; Adamovich, Y.; Shaul, Y.; Halpern, Z.; Shlomai, A. Curcumin inhibits hepatitis B virus via down-regulation of the metabolic coactivator PGC-1α. FEBS Lett. 2010, 584, 2485–2490. [Google Scholar] [CrossRef]
- Paulraj, F.; Abas, F.; Lajis, N.H.; Othman, I.; Hassan, S.S.; Naidu, R. The curcumin analogue 1, 5-bis (2-hydroxyphenyl)-1, 4-pentadiene-3-one induces apoptosis and downregulates E6 and E7 oncogene expression in HPV16 and HPV18-infected cervical cancer cells. Molecules 2015, 20, 11830–11860. [Google Scholar] [CrossRef]
- Ferreira, V.H.; Nazli, A.; Dizzell, S.E.; Mueller, K.; Kaushic, C. The anti-inflammatory activity of curcumin protects the genital mucosal epithelial barrier from disruption and blocks replication of HIV-1 and HSV-2. PLoS ONE 2015, 10, e0124903. [Google Scholar] [CrossRef]
- Mathew, D.; Hsu, W.-L. Antiviral potential of curcumin. J. Funct. Foods 2018, 40, 692–699. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Yao, C.-A. Potential role of Nrf2 activators with dual antiviral and anti-inflammatory properties in the management of viral pneumonia. Infect. Drug Resist. 2020, 13, 1735–1741. [Google Scholar] [CrossRef] [PubMed]
- Praditya, D.; Kirchhoff, L.; Brüning, J.; Rachmawati, H.; Steinmann, J.; Steinmann, E. Anti-infective properties of the golden spice curcumin. Front. Microbiol. 2019, 10, 912–928. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-Y.; Chen, D.-Y.; Wen, H.-W.; Ou, J.-L.; Chiou, S.-S.; Chen, J.-M.; Wong, M.-L.; Hsu, W.-L. Inhibition of Enveloped Viruses Infectivity by Curcumin. PLoS ONE 2013, 8, e62482. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Doroghazi, J.; Roemer, M.E.; Triezenberg, S.J. Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity. Virology 2008, 373, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Gong, L.; Wang, Z.; Han, F.; Liu, H.; Lu, X.; Liu, L. Curcumin inhibits human cytomegalovirus by downregulating heat shock protein 90. Mol. Med. Rep. 2015, 12, 4789–4793. [Google Scholar] [CrossRef] [PubMed]
- Tomita, M.; Matsuda, T.; Kawakami, H.; Uchihara, J.-N.; Okudaira, T.; Masuda, M.; Ohshiro, K.; Mori, N. Curcumin targets Akt cell survival signaling pathway in HTLV-I-infected T-cell lines (Retraction of vol 97, pg 322, 2006). Cancer Sci. 2011, 102, 498–U214. [Google Scholar] [CrossRef]
- Poursina, Z.; Mohammadi, A.; Yazdi, S.Z.; Humpson, I.; Vakili, V.; Boostani, R.; Rafatpanah, H. Curcumin increased the expression of c-FLIP in HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) patients. J. Cell. Biochem. 2019, 120, 15740–15745. [Google Scholar] [CrossRef]
- Chen, M.-H.; Lee, M.-Y.; Chuang, J.-J.; Li, Y.-Z.; Ning, S.-T.; Chen, J.-C.; Liu, Y.-W. Curcumin inhibits HCV replication by induction of heme oxygenase-1 and suppression of AKT. Int. J. Mol. Med. 2012, 30, 1021–1028. [Google Scholar] [CrossRef]
- Dutta, K.; Ghosh, D.; Basu, A. Curcumin protects neuronal cells from Japanese encephalitis virus-mediated cell death and also inhibits infective viral particle formation by dysregulation of ubiquitin–proteasome system. J. Neuroimmune Pharmacol. 2009, 4, 328–337. [Google Scholar] [CrossRef]
- Si, X.; Wang, Y.; Wong, J.; Zhang, J.; McManus, B.M.; Luo, H. Dysregulation of the ubiquitin-proteasome system by curcumin suppresses coxsackievirus B3 replication. J. Virol. 2007, 81, 3142–3150. [Google Scholar] [CrossRef]
- Padilla-s, L.; Rodríguez, A.; Gonzales, M.M.; Gallego-g, J.C.; Castaño-o, J.C. Inhibitory effects of curcumin on dengue virus type 2-infected cells in vitro. Arch. Virol. 2014, 159, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Lin, L.; Chen, Y.; Wu, S.; Si, X.; Wu, H.; Zhai, X.; Wang, Y.; Tong, L.; Pan, B. Curcumin inhibits the replication of enterovirus 71 in vitro. Acta Pharm. Sin. B 2014, 4, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Colpitts, C.C.; Schang, L.M.; Rachmawati, H.; Frentzen, A.; Pfaender, S.; Behrendt, P.; Brown, R.J.; Bankwitz, D.; Steinmann, J.; Ott, M. Turmeric curcumin inhibits entry of all hepatitis C virus genotypes into human liver cells. Gut 2014, 63, 1137–1149. [Google Scholar]
- Edwards, M.R.; Johnson, B.; Mire, C.E.; Xu, W.; Shabman, R.S.; Speller, L.N.; Leung, D.W.; Geisbert, T.W.; Amarasinghe, G.K.; Basler, C.F. The Marburg Virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep. 2014, 6, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Gu, L.; Su, Y.; Wang, Q.; Zhao, Y.; Chen, X.; Deng, H.; Li, W.; Wang, G.; Li, K. Inhibition of curcumin on influenza A virus infection and influenzal pneumonia via oxidative stress, TLR2/4, p38/JNK MAPK and NF-κB pathways. Int. Immunopharmacol. 2018, 54, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Komaravelli, N.; Ansar, M.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein-ring finger protein 4 dependent pathway. Free Radic. Biol. Med. 2017, 113, 494–504. [Google Scholar] [CrossRef]
- Liu, Z.; Ying, Y. The inhibitory effect of curcumin on virus-induced cytokine storm and its potential use in the associated severe pneumonia. Front. Cell Dev. Biol. 2020, 8, 479. [Google Scholar] [CrossRef]
- Babaei, F.; Nassiri-Asl, M.; Hosseinzadeh, H. Curcumin (a constituent of turmeric): New treatment option against COVID-19. Food Sci. Nutr. 2020, 8, 5215–5227. [Google Scholar] [CrossRef]
- Zhu, L.; Ding, X.; Zhang, D.; Yuan, C.; Wang, J.; Ndegwa, E.; Zhu, G. Curcumin inhibits bovine herpesvirus type 1 entry into MDBK cells. Acta Virol. 2015, 59, 221–227. [Google Scholar] [CrossRef]
- Yang, M.; Lee, G.; Si, J.; Lee, S.-J.; You, H.J.; Ko, G. Curcumin shows antiviral properties against norovirus. Molecules 2016, 21, 1401. [Google Scholar] [CrossRef]
- von Rhein, C.; Weidner, T.; Henß, L.; Martin, J.; Weber, C.; Sliva, K.; Schnierle, B.S. Curcumin and Boswellia serrata gum resin extract inhibit chikungunya and vesicular stomatitis virus infections in vitro. Antivir. Res. 2016, 125, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Cesaro, T.; Carrau, L.; Vallet, T.; Vignuzzi, M. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antivir. Res. 2017, 142, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Obata, K.; Kojima, T.; Masaki, T.; Okabayashi, T.; Yokota, S.; Hirakawa, S.; Nomura, K.; Takasawa, A.; Murata, M.; Tanaka, S. Curcumin prevents replication of respiratory syncytial virus and the epithelial responses to it in human nasal epithelial cells. PLoS ONE 2013, 8, e70225. [Google Scholar] [CrossRef] [PubMed]
- Olagnier, D.; Farahani, E.; Thyrsted, J.; Blay-Cadanet, J.; Herengt, A.; Idorn, M.; Hait, A.; Hernaez, B.; Knudsen, A.; Iversen, M.B.; et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020, 11, 4938. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Pajares, M.; Benito, C.; Jiménez-Villegas, J.; Escoll, M.; Fernández-Ginés, R.; Yagüe, A.J.G.; Lastra, D.; Manda, G.; Rojo, A.I. Can activation of NRF2 be a strategy against COVID-19? Trends Pharmacol. Sci. 2020, 41, 598–610. [Google Scholar] [CrossRef]
- McCord, J.M.; Hybertson, B.M.; Cota-Gomez, A.; Geraci, K.P.; Gao, B. Nrf2 activator PB125® as a potential therapeutic agent against COVID-19. Antioxidants 2020, 9, 518. [Google Scholar] [CrossRef]
- Mendonca, P.; Soliman, K.F. Flavonoids Activation of the Transcription Factor Nrf2 as a Hypothesis Approach for the Prevention and Modulation of SARS-CoV-2 Infection Severity. Antioxidants 2020, 9, 659. [Google Scholar] [CrossRef]
- Wagener, F.A.; Pickkers, P.; Peterson, S.J.; Immenschuh, S.; Abraham, N.G. Targeting the heme-heme oxygenase system to prevent severe complications following COVID-19 infections. Antioxidants 2020, 9, 540. [Google Scholar] [CrossRef]
- Kandeel, M.; Al-Nazawi, M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020, 251, 117627. [Google Scholar] [CrossRef]
- Shanmugarajan, D.; Prabitha, P.; Kumar, B.P.; Suresh, B. Curcumin to inhibit binding of spike glycoprotein to ACE2 receptors: Computational modelling, simulations, and ADMET studies to explore curcuminoids against novel SARS-CoV-2 targets. RSC Adv. 2020, 10, 31385–31399. [Google Scholar] [CrossRef]

| Activity | Target Virus | Mechanism of Action | References |
|---|---|---|---|
| A specific mode of action | |||
| Anti-HIV | Human immunodeficiency virus |
| [135] [136] [137] |
| Anti-HBV | Hepatitis B virus |
| [138] |
| Anti-HPV | Human papilloma virus |
| [139] |
| Anti- HTLV-1 | Human T-cell leukemia virus type 1 |
| [147] [148] |
| Anti-HSV-1 | Herpes simplex virus 1 |
| [140] [145] |
| Anti-HCMV | Human cytomegalovirus |
| [146] |
| Anti-HCV | Hepatitis C virus |
| [154] [149] |
| Anti-CVB3 | Coxsackie virus B3 |
| [151] [152] [153] [150] |
| Anti-DNV2 | Dengue virus type 2 | ||
| Anti-EV71 | Enterovirus 71 | ||
| Anti-JEV | Japanese encephalitis Virus | ||
| Anti-EV71 | Enterovirus 71 |
| [153] |
| Anti-MARV | Marburg virus |
| [155] |
| Anti-IAV | Influenza A virus | [156] | |
| Anti-RSV | Respiratory syncytial virus | [157] | |
| Anti-SARS-CoV | SARS- coronavirus | [158] | |
| Anti-MERS-CoV | MERS- coronavirus | [158] | |
| Anti-SARS-CoV2 | SARS- coronavirus 2 | [159] | |
| Non-specific mode of action | |||
| Anti-HCV | Hepatitis C virus |
| [154] [160] [161] |
| Anti-BoHV-1 | Bovine herpesvirus type 1 | ||
| Anti-HuNoV | Human norovirus | ||
| Anti-CHIKV | Chikungunya virus |
| [162] [163] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahban, M.; Habibi-Rezaei, M.; Mazaheri, M.; Saso, L.; Moosavi-Movahedi, A.A. Anti-Viral Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications. Antioxidants 2020, 9, 1228. https://doi.org/10.3390/antiox9121228
Rahban M, Habibi-Rezaei M, Mazaheri M, Saso L, Moosavi-Movahedi AA. Anti-Viral Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications. Antioxidants. 2020; 9(12):1228. https://doi.org/10.3390/antiox9121228
Chicago/Turabian StyleRahban, Mahdie, Mehran Habibi-Rezaei, Mansoureh Mazaheri, Luciano Saso, and Ali A. Moosavi-Movahedi. 2020. "Anti-Viral Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications" Antioxidants 9, no. 12: 1228. https://doi.org/10.3390/antiox9121228
APA StyleRahban, M., Habibi-Rezaei, M., Mazaheri, M., Saso, L., & Moosavi-Movahedi, A. A. (2020). Anti-Viral Potential and Modulation of Nrf2 by Curcumin: Pharmacological Implications. Antioxidants, 9(12), 1228. https://doi.org/10.3390/antiox9121228









