Differential Sex-Dependent Regulation of the Alveolar Macrophage miRNome of SP-A2 and co-ex (SP-A1/SP-A2) and Sex Differences Attenuation after 18 h of Ozone Exposure
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Filtered Air (FA) and Ozone (O3) Exposure
2.3. RNA Preparation, Library Construction, and Sequencing
2.4. miRNA Data Analysis
2.5. Ingenuity Pathway Analysis (IPA)
2.6. Gene Expression Analysis
2.7. Statistical Analysis
3. Results
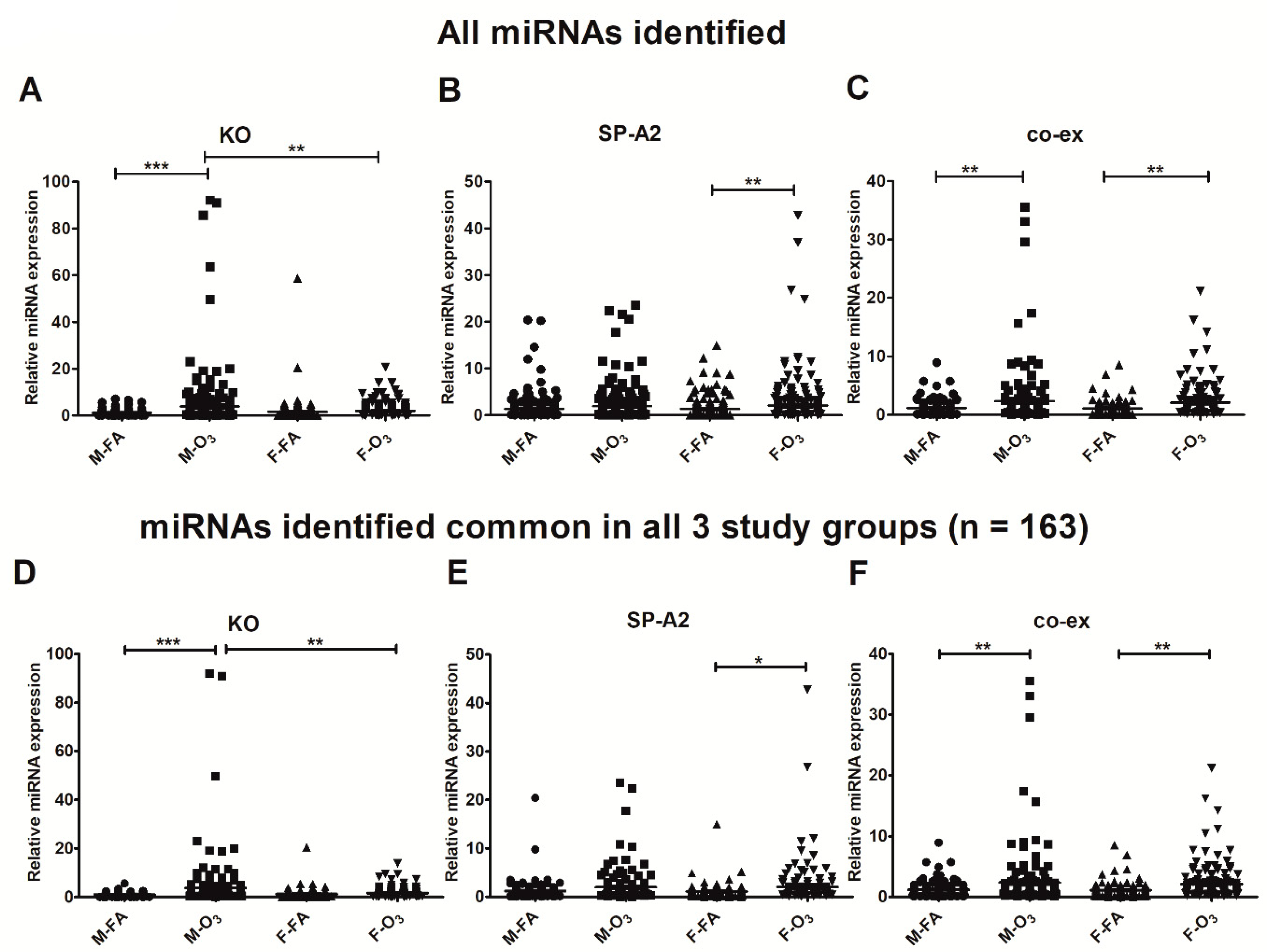
3.1. Effect of SP-A2 (1A0), SP-A1/SP-A2 (6A2/1A0, co-ex), and KO on the Expression of AM miRNome
Sex Differences
3.2. miRNAs, the Levels of which Changed ≥2-Fold in Response to FA, O3, and Sex
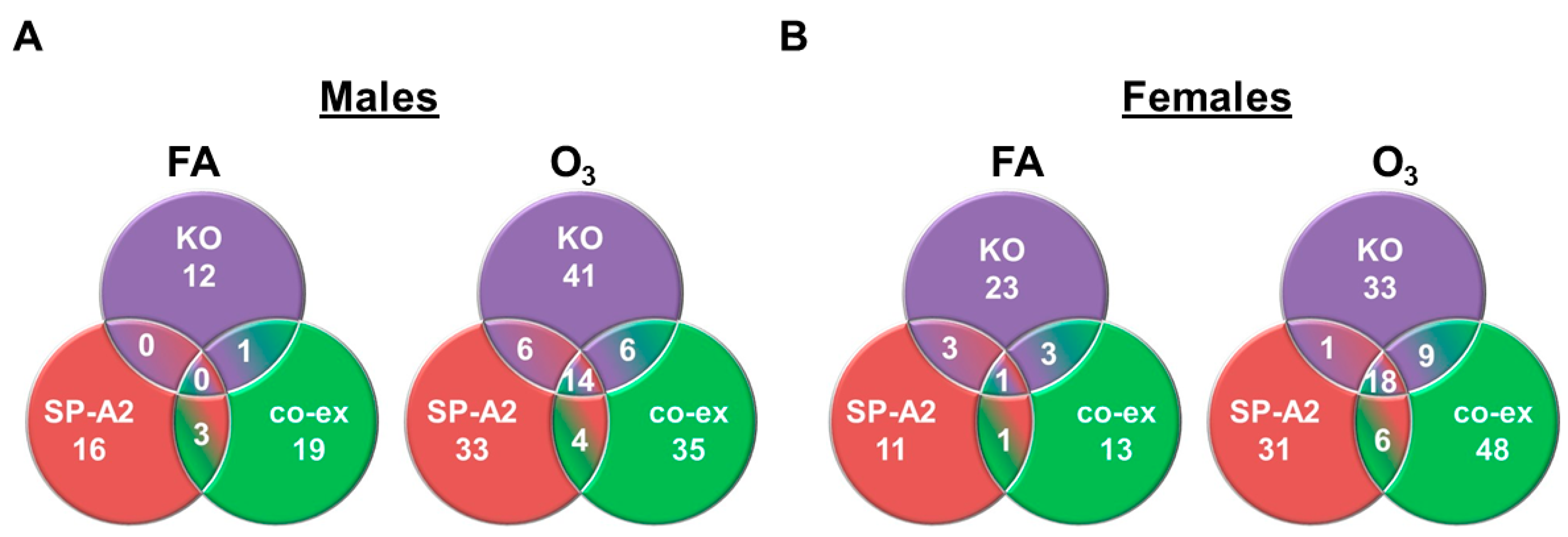
3.3. Shared miRNAs among the Three Studied Groups in Response to FA or O3 in Males and Females
3.4. Ingenuity Pathway Analysis (IPA) Pathways
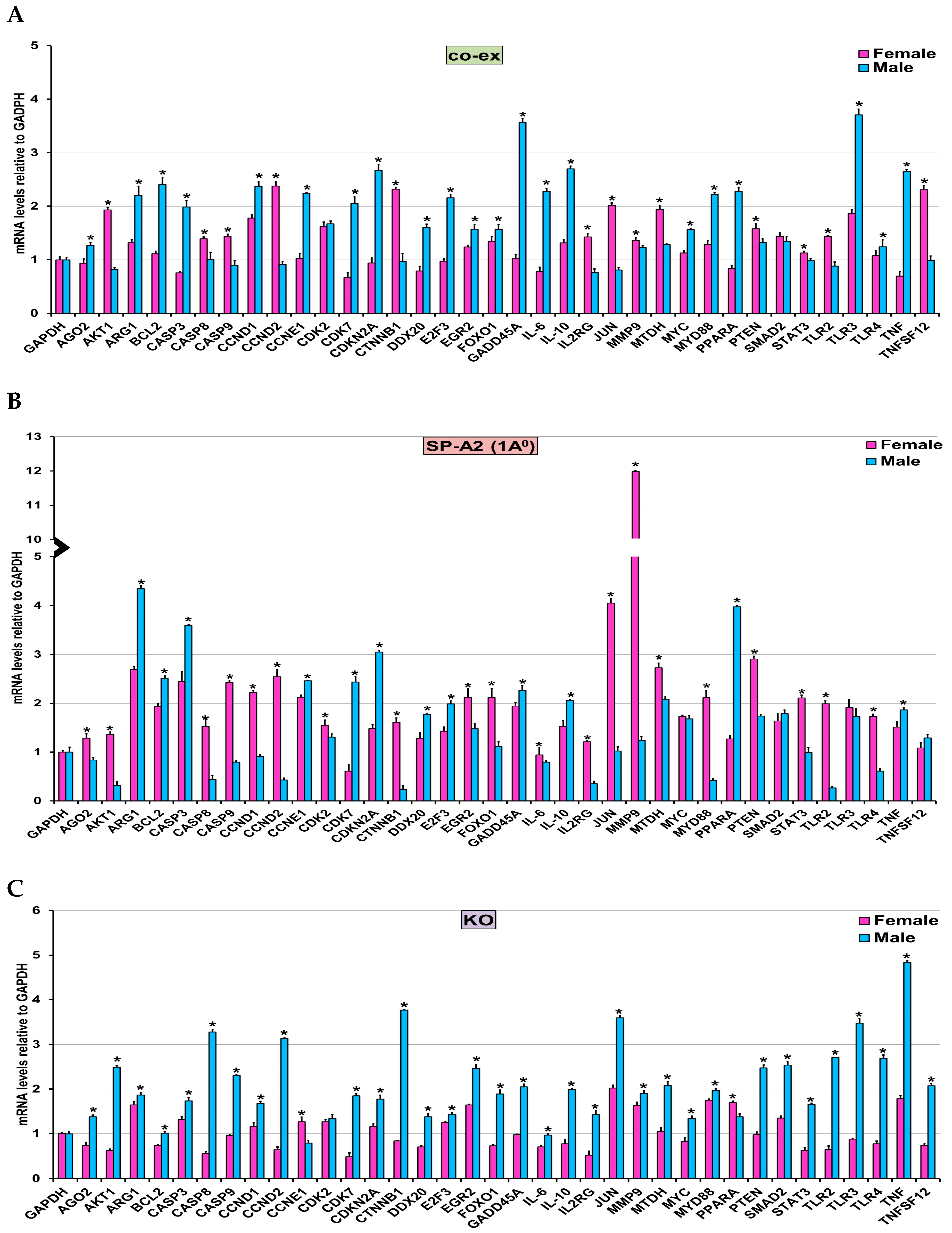
3.5. Validation of miRNA Target Genes
3.6. In Response to O3 Exposure
3.6.1. SP-A Genotype-Independent miRNAs (i.e., Found in Common among the Three Groups (SP-A2, co-ex, KO))
3.6.2. SP-A Genotype-Dependent miRNAs (i.e., Not Found in Common among the Three Groups)
4. Discussion
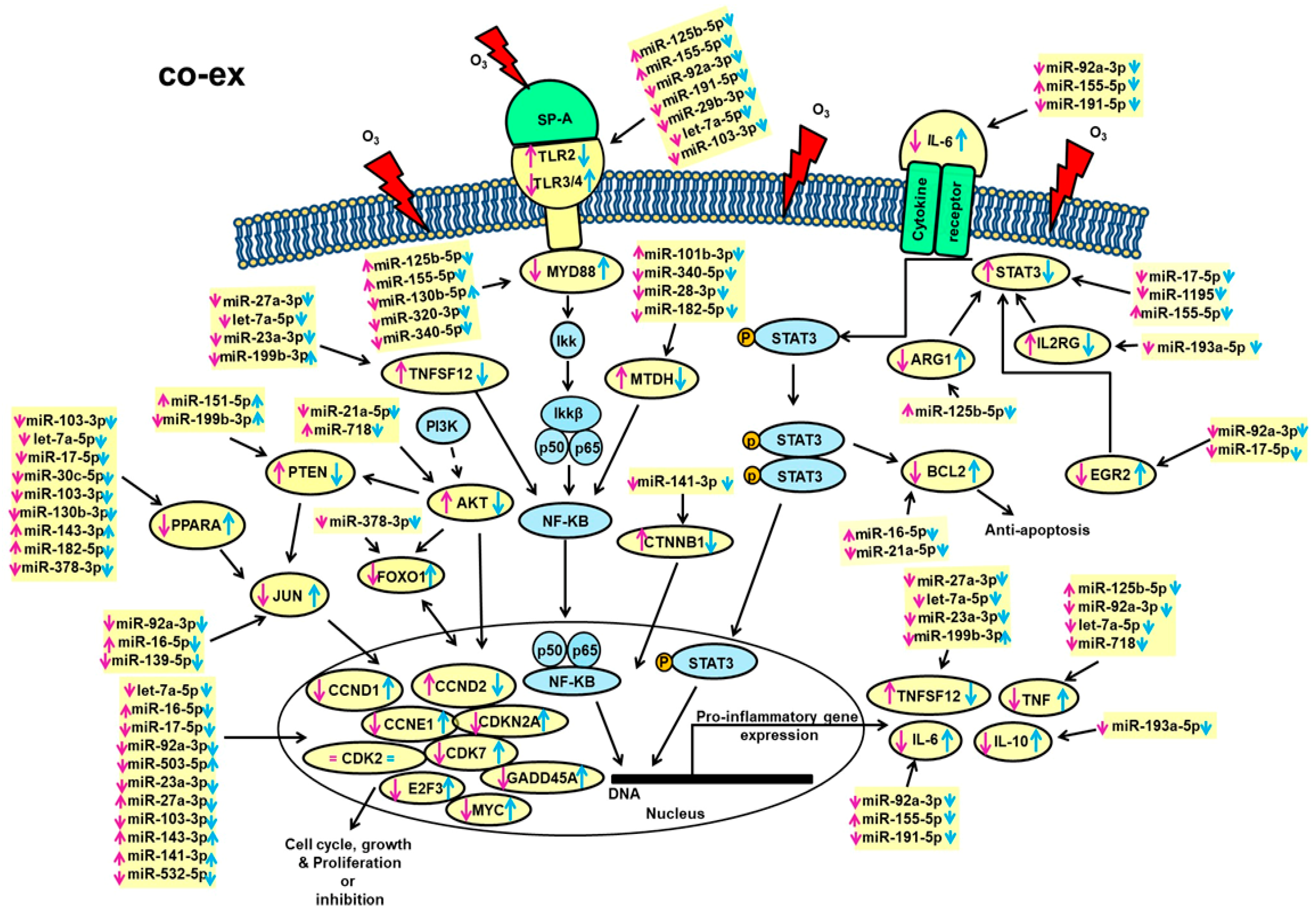
4.1. Anti-Apoptosis, Cell Cycle, Growth, and Proliferation
4.2. Proinflammatory Responses
5. Overall Comments
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
Ethics Statement
Abbreviations
References
- Brunekreef, B. The continuing challenge of air pollution. Eur. Respir. J. 2010, 36, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Al-Hegelan, M.; Tighe, R.M.; Castillo, C.; Hollingsworth, J.W. Ambient ozone and pulmonary innate immunity. Immunol. Res. 2011, 49, 173–191. [Google Scholar] [CrossRef]
- Bromberg, P.A. Mechanisms of the acute effects of inhaled ozone in humans. Biochimica Biophysica Acta 2016, 1860, 2771–2781. [Google Scholar] [CrossRef]
- Jerrett, M.; Burnett, R.T.; Pope, C.A., 3rd; Ito, K.; Thurston, G.; Krewski, D.; Shi, Y.; Calle, E.; Thun, M. Long-term ozone exposure and mortality. N. Engl. J. Med. 2009, 360, 1085–1095. [Google Scholar] [CrossRef]
- Kelly, F.J.; Fussell, J.C. Air pollution and airway disease. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2011, 41, 1059–1071. [Google Scholar] [CrossRef]
- Voter, K.Z.; Whitin, J.C.; Torres, A.; Morrow, P.E.; Cox, C.; Tsai, Y.; Utell, M.J.; Frampton, M.W. Ozone exposure and the production of reactive oxygen species by bronchoalveolar cells in humans. Inhal. Toxicol. 2001, 13, 465–483. [Google Scholar] [CrossRef]
- Almqvist, C.; Worm, M.; Leynaert, B. Impact of gender on asthma in childhood and adolescence: A GA2LEN review. Allergy 2008, 63, 47–57. [Google Scholar] [CrossRef] [PubMed]
- De Torres, J.P.; Casanova, C.; Celli, B.R. Sex, forced expiratory volume in 1 s decline, body weight change and C-reactive protein in COPD patients. The Eur. Respir. J. 2009, 34, 776. [Google Scholar] [CrossRef]
- Falagas, M.E.; Mourtzoukou, E.G.; Vardakas, K.Z. Sex differences in the incidence and severity of respiratory tract infections. Respir. Med. 2007, 101, 1845–1863. [Google Scholar] [CrossRef] [PubMed]
- Varkey, A.B. Chronic obstructive pulmonary disease in women: Exploring gender differences. Curr. Opin. Pulm. Med. 2004, 10, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Crouch, E.; Wright, J.R. Surfactant proteins a and d and pulmonary host defense. Annu. Rev. Physiol. 2001, 63, 521–554. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.R. Pulmonary surfactant: A front line of lung host defense. J. Clin. Investig. 2003, 111, 1453–1455. [Google Scholar] [CrossRef] [PubMed]
- Crouch, E.C. Collectins and pulmonary host defense. Am. J. Respir. Cell Mol. Biol. 1998, 19, 177–201. [Google Scholar] [CrossRef] [PubMed]
- Phelps, D.S. Surfactant regulation of host defense function in the lung: A question of balance. Pediatric Pathol. Mol. Med. 2001, 20, 269–292. [Google Scholar] [CrossRef]
- Wright, J.R. Immunomodulatory funtions of surfactant. Physilogical Rev. 1997, 77, 932–962. [Google Scholar]
- Wright, J.R. Immunoregulatory functions of surfactant proteins. Nat. Rev. Immunol. 2005, 5, 58–68. [Google Scholar] [CrossRef]
- Floros, J.; Phelps, D.S. Pulmonary surfactant protein A; structure, expression, and its role in innate host defense. In Update of Intensive Care Medicine; Nakos, Lekka, M., Eds.; University of Ioannina: Ioannina, Greece, 2002; pp. 87–102. [Google Scholar]
- Hollingsworth, J.W.; Kleeberger, S.R.; Foster, W.M. Ozone and pulmonary innate immunity. Proc. Am. Thorac. Soc. 2007, 4, 240–246. [Google Scholar] [CrossRef]
- Su, W.Y.; Gordon, T. Alterations in surfactant protein A after acute exposure to ozone. J. Appl. Physiol. (Bethesda, Md: 1985) 1996, 80, 1560–1567. [Google Scholar] [CrossRef]
- Floros, J.; Phelps, D.S. Pulmonary surfactant. In Anesthesia: Biologic Foundations; Yaksh, T.L., Lynch, C., III, Zapol, W.M., Maze, M., Biebuyck, J.F., Saidman, L.J., Eds.; Lippincott-Raven Publishers: Philadelphia, PA, USA, 1997; pp. 1259–1280. [Google Scholar]
- Batenburg, J.J.; Haagsman, H.P. The lipids of pulmonary surfactant: Dynamics and interactions with proteins. Prog. Lipid Res. 1998, 37, 235–276. [Google Scholar] [CrossRef]
- Bates, S.R.; Dodia, C.; Tao, J.Q.; Fisher, A.B. Surfactant protein-A plays an important role in lung surfactant clearance: Evidence using the surfactant protein-A gene-targeted mouse. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008, 294, L325–L333. [Google Scholar] [CrossRef]
- Johansson, J.; Curstedt, T. Molecular structures and interactions of pulmonary surfactant components. Eur. J. Biochem. 1997, 244, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.J.; Greene, K.; Voelker, D.R. Surfactant protein A and surfactant protein D in health and disease. Am. J. Physiol. 1998, 275, L1–L13. [Google Scholar] [CrossRef] [PubMed]
- Mikerov, A.N.; Gan, X.; Umstead, T.M.; Miller, L.; Chinchilli, V.M.; Phelps, D.S.; Floros, J. Sex differences in the impact of ozone on survival and alveolar macrophage function of mice after Klebsiella pneumoniae infection. Respir. Res. 2008, 9, 24. [Google Scholar] [CrossRef]
- Mikerov, A.N.; Haque, R.; Gan, X.; Guo, X.; Phelps, D.S.; Floros, J. Ablation of SP-A has a negative impact on the susceptibility of mice to Klebsiella pneumoniae infection after ozone exposure: Sex differences. Respir. Res. 2008, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Mikerov, A.N.; Hu, S.; Durrani, F.; Gan, X.; Wang, G.; Umstead, T.M.; Phelps, D.S.; Floros, J. Impact of sex and ozone exposure on the course of pneumonia in wild type and SP-A (-/-) mice. Microb. Pathog. 2012, 52, 239–249. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mikerov, A.N.; Phelps, D.S.; Gan, X.; Umstead, T.M.; Haque, R.; Wang, G.; Floros, J. Effect of ozone exposure and infection on bronchoalveolar lavage: Sex differences in response patterns. Toxicol. Lett. 2014, 230, 333–344. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haque, R.; Umstead, T.M.; Freeman, W.M.; Floros, J.; Phelps, D.S. The impact of surfactant protein-A on ozone-induced changes in the mouse bronchoalveolar lavage proteome. Proteome Sci. 2009, 7, 12. [Google Scholar] [CrossRef]
- Haque, R.; Umstead, T.M.; Ponnuru, P.; Guo, X.; Hawgood, S.; Phelps, D.S.; Floros, J. Role of surfactant protein-A (SP-A) in lung injury in response to acute ozone exposure of SP-A deficient mice. Toxicol. Appl. Pharmacol. 2007, 220, 72–82. [Google Scholar] [CrossRef]
- Floros, J.; Hoover, R.R. Genetics of the hydrophilic surfactant proteins A and D. Biochimica Biophysica Acta 1998, 1408, 312–322. [Google Scholar] [CrossRef]
- Hoover, R.R.; Floros, J. Organization of the human SP-A and SP-D loci at 10q22-q23. Physical and radiation hybrid mapping reveal gene order and orientation. Am. J. Respir. Cell Mol. Biol. 1998, 18, 353–362. [Google Scholar] [CrossRef]
- DiAngelo, S.; Lin, Z.; Wang, G.; Phillips, S.; Ramet, M.; Luo, J.; Floros, J. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis. Markers 1999, 15, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Karinch, A.M.; Floros, J. 5′ splicing and allelic variants of the human pulmonary surfactant protein A genes. Am. J. Respir. Cell Mol. Biol. 1995, 12, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Verdugo, I.; Wang, G.; Floros, J.; Casals, C. Structural analysis and lipid-binding properties of recombinant human surfactant protein a derived from one or both genes. Biochemistry 2002, 41, 14041–14053. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wang, G.; Phelps, D.S.; Al-Mondhiry, H.; Floros, J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: Effect of ozone-induced SP-A oxidation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 286, L546–L553. [Google Scholar] [CrossRef] [PubMed]
- Mikerov, A.N.; Umstead, T.M.; Huang, W.; Liu, W.; Phelps, D.S.; Floros, J. SP-A1 and SP-A2 variants differentially enhance association of Pseudomonas aeruginosa with rat alveolar macrophages. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005, 288, L150–L158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mikerov, A.N.; Wang, G.; Umstead, T.M.; Zacharatos, M.; Thomas, N.J.; Phelps, D.S.; Floros, J. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect. Immun. 2007, 75, 1403–1412. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Noutsios, G.T.; Thorenoor, N.; Zhang, X.; Phelps, D.S.; Umstead, T.M.; Durrani, F.; Floros, J. SP-A2 contributes to miRNA-mediated sex differences in response to oxidative stress: Pro-inflammatory, anti-apoptotic, and anti-oxidant pathways are involved. Biol. Sex Differ. 2017, 8, 37. [Google Scholar] [CrossRef]
- Oberley, R.E.; Snyder, J.M. Recombinant human SP-A1 and SP-A2 proteins have different carbohydrate-binding characteristics. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003, 284, L871–L881. [Google Scholar] [CrossRef]
- Selman, M.; Lin, H.M.; Montano, M.; Jenkins, A.L.; Estrada, A.; Lin, Z.; Wang, G.; DiAngelo, S.L.; Guo, X.; Umstead, T.M.; et al. Surfactant protein A and B genetic variants predispose to idiopathic pulmonary fibrosis. Hum. Genet. 2003, 113, 542–550. [Google Scholar] [CrossRef]
- Thorenoor, N.; Umstead, T.M.; Zhang, X.; Phelps, D.S.; Floros, J. Survival of surfactant protein-A1 and SP-A2 transgenic mice after klebsiella pneumoniae infection, exhibits Sex-, Gene-, and Variant specific differences; treatment with surfactant protein improves survival. Front. Immunol. 2018, 9, 2404. [Google Scholar] [CrossRef]
- Thorenoor, N.; Zhang, X.; Umstead, T.M.; Scott Halstead, E.; Phelps, D.S.; Floros, J. Differential effects of innate immune variants of surfactant protein-A1 (SFTPA1) and SP-A2 (SFTPA2) in airway function after Klebsiella pneumoniae infection and sex differences. Respir. Res. 2018, 19, 23. [Google Scholar] [CrossRef]
- Wang, G.; Bates-Kenney, S.R.; Tao, J.Q.; Phelps, D.S.; Floros, J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry 2004, 43, 4227–4239. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Myers, C.; Mikerov, A.; Floros, J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry 2007, 46, 8425–8435. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Phelps, D.S.; Umstead, T.M.; Floros, J. Human SP-A protein variants derived from one or both genes stimulate TNF-alpha production in the THP-1 cell line. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000, 278, L946–L954. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Taneva, S.; Keough, K.M.; Floros, J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochimica Biophysica Acta 2007, 1768, 2060–2069. [Google Scholar] [CrossRef][Green Version]
- Wang, G.; Umstead, T.M.; Phelps, D.S.; Al-Mondhiry, H.; Floros, J. The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ. Health Perspect. 2002, 110, 79–84. [Google Scholar] [CrossRef]
- Karinch, A.M.; Deiter, G.; Ballard, P.L.; Floros, J. Regulation of expression of human SP-A1 and SP-A2 genes in fetal lung explant culture. Biochimica Biophysica Acta 1998, 1398, 192–202. [Google Scholar] [CrossRef]
- Kumar, A.; Snyder, J. Differential regulation of SP-A1 and SP-A2 genes by cAMP, glucocorticoids, and insulin. Am. J. Physiol. Lung Cell. Mol. Physiol. 1998, 274, L177–L185. [Google Scholar] [CrossRef]
- Scavo, L.M.; Ertsey, R.; Gao, B.Q. Human surfactant proteins A1 and A2 are differentially regulated during development and by soluble factors. Am. J. Physiol. 1998, 275, L653–L669. [Google Scholar] [CrossRef]
- Silveyra, P.; DiAngelo, S.L.; Floros, J. An 11-nt sequence polymorphism at the 3′UTR of human SFTPA1 and SFTPA2 gene variants differentially affect gene expression levels and miRNA regulation in cell culture. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 307, L106–L119. [Google Scholar] [CrossRef]
- Silveyra, P.; Raval, M.; Simmons, B.; Diangelo, S.; Wang, G.; Floros, J. The untranslated exon B of human surfactant protein A2 mRNAs is an enhancer for transcription and translation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011, 301, L795–L803. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tagaram, H.R.; Wang, G.; Umstead, T.M.; Mikerov, A.N.; Thomas, N.J.; Graff, G.R.; Hess, J.C.; Thomassen, M.J.; Kavuru, M.S.; Phelps, D.S.; et al. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L1052–L1063. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Guo, X.; Floros, J. Human SP-A 3′-UTR variants mediate differential gene expression in basal levels and in response to dexamethasone. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003, 284, L738–L748. [Google Scholar] [CrossRef]
- Tsotakos, N.; Silveyra, P.; Lin, Z.; Thomas, N.; Vaid, M.; Floros, J. Regulation of translation by upstream translation initiation codons of surfactant protein A1 splice variants. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L58–L75. [Google Scholar] [CrossRef]
- Noutsios, G.T.; Silveyra, P.; Bhatti, F.; Floros, J. Exon B of human surfactant protein A2 mRNA, alone or within its surrounding sequences, interacts with 14-3-3; role of cis-elements and secondary structure. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 304, L722–L735. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Guo, X.; Floros, J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005, 289, L497–L508. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Guo, X.; Silveyra, P.; Kimball, S.R.; Floros, J. Cap-independent translation of human SP-A 5′-UTR variants: A double-loop structure and cis-element contribution. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 296, L635–L647. [Google Scholar] [CrossRef]
- Wang, G.; Umstead, T.M.; Hu, S.; Mikerov, A.N.; Phelps, D.S.; Floros, J. Differential Effects of Human SP-A1 and SP-A2 on the BAL Proteome and Signaling Pathways in Response to Klebsiella pneumoniae and Ozone Exposure. Front. Immunol. 2019, 10, 561. [Google Scholar] [CrossRef]
- Phelps, D.S.; Umstead, T.M.; Floros, J. Sex differences in the acute in vivo effects of different human SP-A variants on the mouse alveolar macrophage proteome. J. Proteom. 2014, 108, 427–444. [Google Scholar] [CrossRef]
- Phelps, D.S.; Umstead, T.M.; Silveyra, P.; Hu, S.; Wang, G.; Floros, J. Differences in the alveolar macrophage proteome in transgenic mice expressing human SP-A1 and SP-A2. J. Proteom. Genom. Res. 2013, 1, 2–26. [Google Scholar] [CrossRef]
- Tsotakos, N.; Phelps, D.S.; Yengo, C.M.; Chinchilli, V.M.; Floros, J. Single-cell analysis reveals differential regulation of the alveolar macrophage actin cytoskeleton by surfactant proteins A1 and A2: Implications of sex and aging. Biol. Sex Differ. 2016, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Thorenoor, N.; Kawasawa, Y.I.; Gandhi, C.K.; Floros, J. Sex-specific regulation of gene expression networks by Surfactant protein A (SP-A) variants in alveolar macrophages in response to Klebsiella pneumoniae. Front. Immunol. Mol. Innate Immun. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rodriguez, E.; Pascual, A.; Arroyo, R.; Floros, J.; Perez-Gil, J. Human pulmonary surfactant protein sp-a1 provides maximal efficiency of lung interfacial films. Biophys. J. 2016, 111, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Ezzie, M.E.; Crawford, M.; Cho, J.H.; Orellana, R.; Zhang, S.; Gelinas, R.; Batte, K.; Yu, L.; Nuovo, G.; Galas, D.; et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax 2012, 67, 122–131. [Google Scholar] [CrossRef]
- Sayed, D.; Abdellatif, M. MicroRNAs in development and disease. Physiol. Rev. 2011, 91, 827–887. [Google Scholar] [CrossRef]
- Sessa, R.; Hata, A. Role of microRNAs in lung development and pulmonary diseases. Pulm. Circ. 2013, 3, 315–328. [Google Scholar] [CrossRef]
- Feketea, G.; Bocsan, C.I.; Popescu, C.; Gaman, M.; Stanciu, L.A.; Zdrenghea, M.T. A review of macrophage micrornas’ role in human asthma. Cells 2019, 8, 420. [Google Scholar] [CrossRef]
- Hoefel, G.; Tay, H.; Foster, P. MicroRNAs in lung diseases. Chest 2019, 156, 991–1000. [Google Scholar] [CrossRef]
- Miao, C.; Xiong, Y.; Zhang, G.; Chang, J. MicroRNAs in idiopathic pulmonary fibrosis, new research progress and their pathophysiological implication. Exp. Lung Res. 2018, 44, 178–190. [Google Scholar] [CrossRef]
- Wang, J.X.; Gao, J.; Ding, S.L.; Wang, K.; Jiao, J.Q.; Wang, Y.; Sun, T.; Zhou, L.Y.; Long, B.; Zhang, X.J.; et al. Oxidative modification of mir-184 enables it to target bcl-xl and bcl-w. Mol. Cell 2015, 59, 50–61. [Google Scholar] [CrossRef]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.S.; Zhang, Z.; McManus, M.T.; Harfe, B.D.; Sun, X. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 2208–2213. [Google Scholar] [CrossRef]
- Greene, C.M.; Gaughan, K.P. microRNAs in asthma: Potential therapeutic targets. Curr. Opin. Pulm. Med. 2013, 19, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Levänen, B.; Bhakta, N.R.; Torregrosa Paredes, P.; Barbeau, R.; Hiltbrunner, S.; Pollack, J.L.; Sköld, C.M.; Svartengren, M.; Grunewald, J.; Gabrielsson, S.; et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol. 2013, 131, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Molina-Pinelo, S.; Pastor, M.D.; Suarez, R.; Romero-Romero, B.; González De la Peña, M.; Salinas, A.; García-Carbonero, R.; De Miguel, M.J.; Rodríguez-Panadero, F.; Carnero, A.; et al. MicroRNA clusters: Dysregulation in lung adenocarcinoma and COPD. Eur. Respir. J. 2014, 43, 1740–1749. [Google Scholar] [CrossRef] [PubMed]
- Omran, A.; Elimam, D.; Yin, F. MicroRNAs: New insights into chronic childhood diseases. BioMed Res. Int. 2013, 2013, 291826. [Google Scholar] [CrossRef]
- Van Pottelberge, G.R.; Mestdagh, P.; Bracke, K.R.; Thas, O.; van Durme, Y.M.; Joos, G.F.; Vandesompele, J.; Brusselle, G.G. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care. Med. 2011, 183, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Pinkerton, K.E.; Harbaugh, M.; Han, M.K.; Jourdan Le Saux, C.; Van Winkle, L.S.; Martin, W.J., 2nd; Kosgei, R.J.; Carter, E.J.; Sitkin, N.; Smiley-Jewell, S.M.; et al. Women and lung disease. Sex differences and global health disparities. Am. J. Respir. Crit. Care. Med. 2015, 192, 11–16. [Google Scholar] [CrossRef]
- Townsend, E.A.; Miller, V.M.; Prakash, Y.S. Sex differences and sex steroids in lung health and disease. Endocr. Rev. 2012, 33, 1–47. [Google Scholar] [CrossRef]
- Dai, R.; Ahmed, S.A. Sexual dimorphism of miRNA expression: A new perspective in understanding the sex bias of autoimmune diseases. Ther. Clin. Risk Manag. 2014, 10, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Noutsios, G.T.; Thorenoor, N.; Zhang, X.; Phelps, D.S.; Umstead, T.M.; Durrani, F.; Floros, J. Major effect of oxidative stress on the male, but not female, sp-a1 type ii cell mirnome. Front. Immunol. 2019, 10, 1514. [Google Scholar] [CrossRef] [PubMed]
- Thorenoor, N.; Kawasawa, Y.I.; Gandhi, C.K.; Zhang, X.; Floros, J. Differential impact of co-expressed sp-a1/sp-a2 protein on am mirnome; sex differences. Front. Immunol. 2019, 10, 1960. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Guo, X.; Diangelo, S.; Thomas, N.J.; Floros, J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: Formation of tubular myelin in vivo requires both gene products. J. Biol. Chem. 2010, 285, 11998–12010. [Google Scholar] [CrossRef]
- Hatch, G.E.; Slade, R.; Harris, L.P.; McDonnell, W.F.; Devlin, R.B.; Koren, H.S.; Costa, D.L.; McKee, J. Ozone dose and effect in humans and rats. A comparison using oxygen-18 labeling and bronchoalveolar lavage. Am. J. Respir. Crit. Care. Med. 1994, 150, 676–683. [Google Scholar] [CrossRef]
- Phelps, D.S.; Umstead, T.M.; Floros, J. Sex differences in the response of the alveolar macrophage proteome to treatment with exogenous surfactant protein-A. Proteome Sci. 2012, 10, 44. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013, 14, 219. [Google Scholar] [CrossRef]
- Abbas, I.; Verdin, A.; Escande, F.; Saint-Georges, F.; Cazier, F.; Mulliez, P.; Courcot, D.; Shirali, P.; Gosset, P.; Garcon, G. In vitro short-term exposure to air pollution PM2.5-0.3 induced cell cycle alterations and genetic instability in a human lung cell coculture model. Environ. Res. 2016, 147, 146–158. [Google Scholar] [CrossRef]
- Abu-Elmagd, M.; Alghamdi, M.A.; Shamy, M.; Khoder, M.I.; Costa, M.; Assidi, M.; Kadam, R.; Alsehli, H.; Gari, M.; Pushparaj, P.N.; et al. Evaluation of the effects of airborne particulate matter on bone marrow-mesenchymal stem cells (bm-mscs): Cellular, molecular and systems biological approaches. Int. J. Environ. Res. Public Health 2017, 14, 440. [Google Scholar] [CrossRef]
- Li, G.; Liao, Y.; Hu, J.; Lu, L.; Zhang, Y.; Li, B.; An, T. Activation of NF-kappaB pathways mediating the inflammation and pulmonary diseases associated with atmospheric methylamine exposure. Environ. Pollut. (Barking, Essex: 1987) 2019, 252, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Ouyang, H.; Abdalla, B.A.; Xu, H.; Nie, Q.; Zhang, X. MiR-16 controls myoblast proliferation and apoptosis through directly suppressing Bcl2 and FOXO1 activities. Biochim. Biophys. Acta. Gene Regul. Mech. 2017, 1860, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wan, R.; Hu, G.; Yang, L.; Xiong, J.; Wang, F.; Shen, J.; He, S.; Guo, X.; Ni, J.; et al. MiR-15b and miR-16 induce the apoptosis of rat activated pancreatic stellate cells by targeting Bcl-2 in vitro. Pancreatology 2012, 12, 91–99. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, Z.; Du, X.; Davies, H.; Huo, X.; Fang, M. Mir-16 and fluoxetine both reverse autophagic and apoptotic change in chronic unpredictable mild stress model rats. Front. Neurosci. 2017, 11, 428. [Google Scholar] [CrossRef]
- Attar, M.; Arefian, E.; Nabiuni, M.; Adegani, F.J.; Bakhtiari, S.H.; Karimi, Z.; Barzegar, M.; Soleimani, M. MicroRNA 17-92 expressed by a transposone-based vector changes expression level of cell-cycle-related genes. Cell Biol. Int. 2012, 36, 1005–1012. [Google Scholar] [CrossRef]
- Shu, J.; Xia, Z.; Li, L.; Liang, E.T.; Slipek, N.; Shen, D.; Foo, J.; Subramanian, S.; Steer, C.J. Dose-dependent differential mRNA target selection and regulation by let-7a-7f and miR-17-92 cluster microRNAs. RNA Biol. 2012, 9, 1275–1287. [Google Scholar] [CrossRef]
- Wang, F.; Mao, A.; Tang, J.; Zhang, Q.; Yan, J.; Wang, Y.; Di, C.; Gan, L.; Sun, C.; Zhang, H. microRNA-16-5p enhances radiosensitivity through modulating Cyclin D1/E1-pRb-E2F1 pathway in prostate cancer cells. J. Cell. Physiol. 2018. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, C.; Wang, M.; Li, Z.; Casimiro, M.C.; Liu, M.; Wu, K.; Whittle, J.; Ju, X.; Hyslop, T.; et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell Biol. 2008, 182, 509–517. [Google Scholar] [CrossRef]
- Kim, E.K.; Choi, E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta 2010, 1802, 396–405. [Google Scholar] [CrossRef]
- Seger, R.; Krebs, E.G. The mapk signaling cascade. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1995, 9, 726–735. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, W.Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. Res. 2015, 35, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Essers, M.A.; Weijzen, S.; de Vries-Smits, A.M.; Saarloos, I.; de Ruiter, N.D.; Bos, J.L.; Burgering, B.M. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004, 23, 4802–4812. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.N.; Lin, Z.; Gandhi, C.K.; Amatya, S.; Wang, Y.; Lin, L.Z.; Floros, J. Sex and sp-a2 dependent nad(h) redox alterations in mouse alveolar macrophages in response to ozone exposure: Potential implications for covid-19. Antioxidants 2020, 915, in press. [Google Scholar] [CrossRef]
- Migita, K.; Iwanaga, N.; Izumi, Y.; Kawahara, C.; Kumagai, K.; Nakamura, T.; Koga, T.; Kawakami, A. TNF-alpha-induced miR-155 regulates IL-6 signaling in rheumatoid synovial fibroblasts. BMC Res. Notes 2017, 10, 403. [Google Scholar] [CrossRef]
- Vigorito, E.; Kohlhaas, S.; Lu, D.; Leyland, R. miR-155: An ancient regulator of the immune system. Immunol. Rev. 2013, 253, 146–157. [Google Scholar] [CrossRef]
- Ceppi, M.; Pereira, P.M.; Dunand-Sauthier, I.; Barras, E.; Reith, W.; Santos, M.A.; Pierre, P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2735–2740. [Google Scholar] [CrossRef]
- Hunter, C.A.; Jones, S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015, 16, 448–457. [Google Scholar] [CrossRef]
- Yu, M.; Zheng, X.; Witschi, H.; Pinkerton, K.E. The role of interleukin-6 in pulmonary inflammation and injury induced by exposure to environmental air pollutants. Toxicol. Sci. Off. J. Soc. Toxicol. 2002, 68, 488–497. [Google Scholar] [CrossRef]
- Carpagnano, G.E.; Kharitonov, S.A.; Wells, A.U.; Pantelidis, P.; Du Bois, R.M.; Barnes, P.J. Increased vitronectin and endothelin-1 in the breath condensate of patients with fibrosing lung disease. Respir. Int. Rev. Thorac. Dis. 2003, 70, 154–160. [Google Scholar] [CrossRef]
- Rincon, M.; Irvin, C.G. Role of IL-6 in asthma and other inflammatory pulmonary diseases. Int. J. Biol. Sci. 2012, 8, 1281–1290. [Google Scholar] [CrossRef] [PubMed]
- Wedzicha, J.A.; Seemungal, T.A.; MacCallum, P.K.; Paul, E.A.; Donaldson, G.C.; Bhowmik, A.; Jeffries, D.J.; Meade, T.W. Acute exacerbations of chronic obstructive pulmonary disease are accompanied by elevations of plasma fibrinogen and serum IL-6 levels. Thromb. Haemost. 2000, 84, 210–215. [Google Scholar] [PubMed]
- Cataldo, D.; Munaut, C.; Noel, A.; Frankenne, F.; Bartsch, P.; Foidart, J.M.; Louis, R. MMP-2- and MMP-9-linked gelatinolytic activity in the sputum from patients with asthma and chronic obstructive pulmonary disease. Int. Arch. Allergy Immunol. 2000, 123, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Ishizaki, M.; Kudoh, S.; Kitaichi, M.; Yamanaka, N. Localization of matrix metalloproteinases-1, -2, and -9 and tissue inhibitor of metalloproteinase-2 in interstitial lung diseases. Lab. Investig. J. Tech. Methods Pathol. 1998, 78, 687–698. [Google Scholar]
- Lanchou, J.; Corbel, M.; Tanguy, M.; Germain, N.; Boichot, E.; Theret, N.; Clement, B.; Lagente, V.; Malledant, Y. Imbalance between matrix metalloproteinases (MMP-9 and MMP-2) and tissue inhibitors of metalloproteinases (TIMP-1 and TIMP-2) in acute respiratory distress syndrome patients. Crit. Care Med. 2003, 31, 536–542. [Google Scholar] [CrossRef]
- Russell, R.E.; Culpitt, S.V.; DeMatos, C.; Donnelly, L.; Smith, M.; Wiggins, J.; Barnes, P.J. Release and activity of matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase-1 by alveolar macrophages from patients with chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2002, 26, 602–609. [Google Scholar] [CrossRef]
- Mishra, V.; DiAngelo, S.L.; Silveyra, P. Sex-specific IL-6-associated signaling activation in ozone-induced lung inflammation. Biol. Sex Differ. 2016, 7, 16. [Google Scholar] [CrossRef]
- Atkinson, J.J.; Senior, R.M. Matrix metalloproteinase-9 in lung remodeling. Am. J. Respir. Cell Mol. Biol. 2003, 28, 12–24. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Patel, K.D. Matrix metalloproteinase-2 (MMP-2) and MMP-9 in pulmonary pathology. Exp. Lung Res. 2005, 31, 599–621. [Google Scholar] [CrossRef]
- Kenyon, N.J.; van der Vliet, A.; Schock, B.C.; Okamoto, T.; McGrew, G.M.; Last, J.A. Susceptibility to ozone-induced acute lung injury in iNOS-deficient mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002, 282, L540–L545. [Google Scholar] [CrossRef][Green Version]
- Geraghty, P.; Wyman, A.E.; Garcia-Arcos, I.; Dabo, A.J.; Gadhvi, S.; Foronjy, R. STAT3 modulates cigarette smoke-induced inflammation and protease expression. Front. Physiol. 2013, 4, 267. [Google Scholar] [CrossRef] [PubMed]
- El Kasmi, K.C.; Holst, J.; Coffre, M.; Mielke, L.; de Pauw, A.; Lhocine, N.; Smith, A.M.; Rutschman, R.; Kaushal, D.; Shen, Y.; et al. General nature of the STAT3-activated anti-inflammatory response. J. Immunol. (Baltimore, Md: 1950) 2006, 177, 7880–7888. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ward, P.A. STAT3 and suppressor of cytokine signaling 3: Potential targets in lung inflammatory responses. Expert Opin. Ther. Targets 2007, 11, 869–880. [Google Scholar] [CrossRef]
- Ruwanpura, S.M.; McLeod, L.; Miller, A.; Jones, J.; Vlahos, R.; Ramm, G.; Longano, A.; Bardin, P.G.; Bozinovski, S.; Anderson, G.P.; et al. Deregulated Stat3 signaling dissociates pulmonary inflammation from emphysema in gp130 mutant mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012, 302, L627–L639. [Google Scholar] [CrossRef]
- Saleh, A.; Shan, L.; Halayko, A.J.; Kung, S.; Gounni, A.S. Critical role for STAT3 in IL-17A-mediated CCL11 expression in human airway smooth muscle cells. J. Immunol. (Baltimore, Md: 1950) 2009, 182, 3357–3365. [Google Scholar] [CrossRef] [PubMed]
- Caetano, M.S.; Hassane, M.; Van, H.T.; Bugarin, E.; Cumpian, A.M.; McDowell, C.L.; Cavazos, C.G.; Zhang, H.; Deng, S.; Diao, L.; et al. Sex specific function of epithelial STAT3 signaling in pathogenesis of K-ras mutant lung cancer. Nat. Commun. 2018, 9, 4589. [Google Scholar] [CrossRef] [PubMed]
- Carballo, M.; Conde, M.; El Bekay, R.; Martin-Nieto, J.; Camacho, M.J.; Monteseirin, J.; Conde, J.; Bedoya, F.J.; Sobrino, F. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J. Biol. Chem. 1999, 274, 17580–17586. [Google Scholar] [CrossRef]
- Durrani, F.; Phelps, D.S.; Weisz, J.; Silveyra, P.; Hu, S.; Mikerov, A.N.; Floros, J. Gonadal hormones and oxidative stress interaction differentially affects survival of male and female mice after lung Klebsiella pneumoniae infection. Exp. Lung Res. 2012, 38, 165–172. [Google Scholar] [CrossRef]
- Phelps, D.S.; Umstead, T.M.; Quintero, O.A.; Yengo, C.M.; Floros, J. In vivo rescue of alveolar macrophages from SP-A knockout mice with exogenous SP-A nearly restores a wild type intracellular proteome; actin involvement. Proteome Sci. 2011, 9, 67. [Google Scholar] [CrossRef]
- Ali, M.; Umstead, T.M.; Haque, R.; Mikerov, A.N.; Freeman, W.M.; Floros, J.; Phelps, D.S. Differences in the BAL proteome after Klebsiella pneumoniae infection in wild type and SP-A-/- mice. Proteome Sci. 2010, 8, 34. [Google Scholar] [CrossRef]
- Phelps, D.S.; Chinchilli, V.M.; Weisz, J.; Shearer, D.; Zhang, X.; Floros, J. Using toponomics to characterize phenotypic diversity in alveolar macrophages from male mice treated with exogenous SP-A1. Biomark. Res. 2020, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Voelker, D.R.; Lugogo, N.L.; Wang, G.; Floros, J.; Ingram, J.L.; Chu, H.W.; Church, T.D.; Kandasamy, P.; Fertel, D.; et al. Surfactant protein A is defective in abrogating inflammation in asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011, 301, L598–L606. [Google Scholar] [CrossRef] [PubMed]
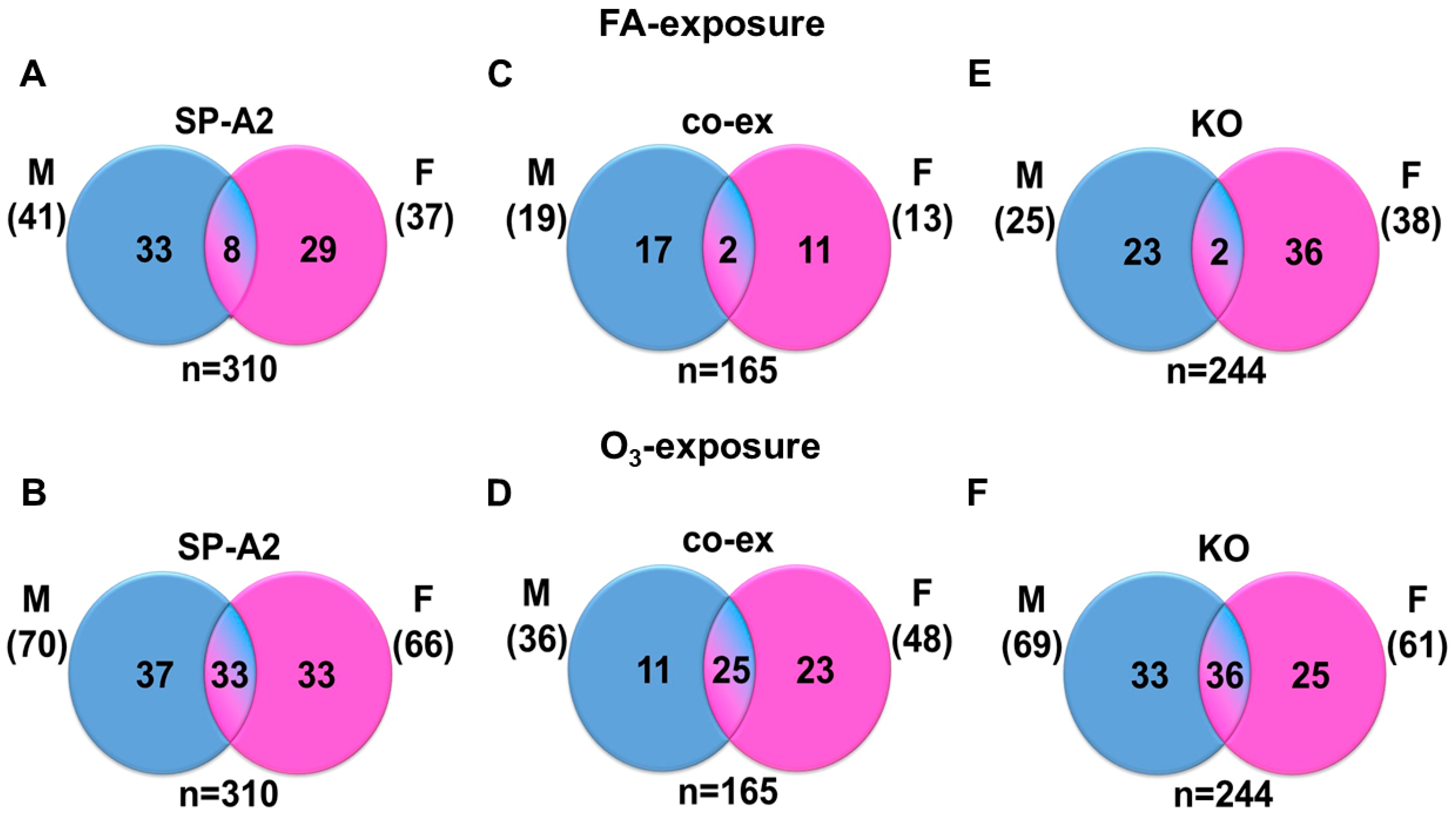
| Gene Variant and Number of miRNAs Identified | Male | Female | ||
|---|---|---|---|---|
| FA vs. O3 | FA vs. O3 | |||
| ≥2-fold (Increase) | ≥2-fold (Decrease) | ≥2-fold (Increase) | ≥2-fold (Decrease) | |
| SP-A2 (1A0) (n = 310) | 41 * | 70 * | 37 * | 66 * |
| co-ex (n = 165) | 19 * | 36 * | 13 * | 48 * |
| KO (n = 244) | 25 * | 69 * | 38 * | 61 * |
| miRNA ID | SP-A2 (1A0) | co-ex (SP-A1 (6A2)/SP-A2 (1A0) | KO | Target Molecule | |||
|---|---|---|---|---|---|---|---|
| Fold Change in Males | Fold Change in Females | Fold Change in Males | Fold Change in Females | Fold Change in Males | Fold Change in Females | ||
| let-7a-5p | 1.395 | 0.942 | 0.912 | 1.356 | 0.498 | 1.020 | AGO2, CCND1, CCND2, CCNE1, CDKN2A, CDK7, E2F3, MMP9, PPARA, TLR4, TNF, TNFSF12 |
| miR-16-5p | 1.621 | 0.787 | 1.045 | 2.301 † | 0.620 | 0.929 | CCND1, CCND2, CCNE1, CDK7, TNFSF12, E2F3, BCL2, JUN |
| miR-17-5p | 1.994 † | 0.734 | 0.960 | 0.953 | 0.212 † | 0.199 † | CCND1, CCND2, CCNE1, CDK7, STAT3, EGR2, E2F3, MYC, PPARA, TNFSF12 |
| miR-21a-5p | 0.649 | 0.553 | 1.338 | 0.563 | 0.966 | 0.994 | BCL2, AKT |
| miR-23a-3p | 1.111 | 0.842 | 0.808 | 0.895 | 0.782 | 1.225 | E2F3, TNFSF12 |
| miR-25-5p | 0.692 | 1.692 | 0.557 | 0.807 | 1.219 | 0.235 † | SMAD2 |
| miR-27a-3p | 1.253 | 1.085 | 0.960 | 2.045 † | 1.163 | 1.206 | E2F3, TNFSF12 |
| miR-28-3p | 1.216 | 0.587 | 0.203 † | 0.826 | 0.177 † | 0.843 | MTDH |
| miR-29b-3p | 0.546 | 1.402 | 1.540 | 1.258 | 2.539 † | 0.707 | AGO2, TLR3 |
| miR-30c-5p | 1.293 | 0.870 | 0.976 | 0.987 | 1.617 | 0.864 | AGO2, DDX20, PPARA |
| miR-101b-3p | 0.456 † | 1.146 | 1.153 | 2.339 † | 1.001 | 1.147 | MTDH |
| miR-103-3p | 1.027 | 0.869 | 1.219 | 1.108 | 0.680 | 1.005 | E2F3, PPARA, AGO2, TLR4 |
| miR-125b-5p | 1.556 | 6.921 † | 1.643 | 5.025 † | 3.585 † | 2.030 † | TLR2, TNF, ARG1, MYD88 |
| miR-130b-3p | 4.656 † | 3.584 † | 0.128 † | 0.505 | 1.207 | 0.554 | PPARA |
| miR-130b-5p | 2.613 † | 1.204 | 2.263 † | 0.420 † | 0.582 | 0.236 † | MYD88 |
| miR-139-5p | 5.637 † | 2.121 † | 0.961 | 1.422 | 3.378 † | 1.527 | AGO2, JUN |
| miR-141-3p | 0.888 | 1.362 | 2.015 † | 6.530 † | 6.623 † | 5.760 † | CTNNB1, GADD45A |
| miR-143-3p | 3.613 † | 4.148 † | 4.202 † | 16.191 † | 9.130 † | 5.773 † | E2F3, PPARA |
| miR-151-5p | 1.345 | 2.847 † | 35.602 † | 7.713 † | 2.508 † | 1.670 | PTEN, AGO2 |
| miR-155-5p | 6.792 † | 0.192 † | 1.077 | 2.088 † | 0.421 † | 2.479 † | IL-6, TLR2, MYD88, STAT3 |
| miR-181a-5p | 0.788 | 0.970 | 0.578 | 0.585 | 0.875 | 1.040 | SMAD2 |
| miR-182-5p | 1.622 | 1.216 | 0.375 † | 3.279 † | 1.984 | 1.235 | PPARA, MTDH |
| miR-191-5p | 1.311 | 1.093 | 1.029 | 0.960 | 0.707 | 1.017 | IL-6, TLR3 |
| miR-193a-5p | 0.677 | 0.842 | 0.758 | 0.701 | 1.277 | 0.796 | IL-10, IL2RG |
| miR-199b-3p | 5.455 † | 2.659 † | 1.829 | 0.543 | 7.971 † | 3.341 † | PTEN, TNFSF12 |
| miR-320-3p | 0.802 | 1.498 | 0.994 | 0.942 | 1.072 | 0.649 | MYD88 |
| miR-320b | 1.114 | 0.324 † | 0.700 | 0.876 | 0.333 † | 0.773 | MMP9, SMAD2 |
| miR-340-5p | 0.843 | 0.748 | 1.607 | 0.646 | 2.031 † | 0.438 † | MTDH, MYD88 |
| miR-378-3p | 1.246 | 0.631 | 0.764 | 1.004 | 0.778 | 0.808 | PPARA, FOXO1, CASP9 |
| miR-455-3p | 3.142 † | 5.087 † | 2.554 † | 3.777 † | 4.915 † | 3.509 † | TNFSF12 |
| miR-503-5p | 0.934 | 0.984 | 2.333 † | 0.919 | 0.498 | 1.468 | CDK2 |
| miR-532-5p | 1.235 | 0.632 | 0.424 † | 0.922 | 0.566 | 0.795 | MYC |
| miR-92a-3p | 1.338 | 2.008 † | 0.903 | 0.888 | 1.131 | 0.380 † | CCND1, CCNE1, CDK7, IL-6, TLR2, TLR3, EGR2, JUN, E2F3, TNF, SMAD2 |
| miR-718 | 1.053 | 0.932 | 0.819 | 2.754 † | 1.214 | 1.187 | TNF, AKT |
| miR-1195 | 1.392 | 0.431 † | 0.375 † | 0.699 | 0.655 | 0.767 | STAT3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thorenoor, N.; Phelps, D.S.; Floros, J. Differential Sex-Dependent Regulation of the Alveolar Macrophage miRNome of SP-A2 and co-ex (SP-A1/SP-A2) and Sex Differences Attenuation after 18 h of Ozone Exposure. Antioxidants 2020, 9, 1190. https://doi.org/10.3390/antiox9121190
Thorenoor N, Phelps DS, Floros J. Differential Sex-Dependent Regulation of the Alveolar Macrophage miRNome of SP-A2 and co-ex (SP-A1/SP-A2) and Sex Differences Attenuation after 18 h of Ozone Exposure. Antioxidants. 2020; 9(12):1190. https://doi.org/10.3390/antiox9121190
Chicago/Turabian StyleThorenoor, Nithyananda, David S. Phelps, and Joanna Floros. 2020. "Differential Sex-Dependent Regulation of the Alveolar Macrophage miRNome of SP-A2 and co-ex (SP-A1/SP-A2) and Sex Differences Attenuation after 18 h of Ozone Exposure" Antioxidants 9, no. 12: 1190. https://doi.org/10.3390/antiox9121190
APA StyleThorenoor, N., Phelps, D. S., & Floros, J. (2020). Differential Sex-Dependent Regulation of the Alveolar Macrophage miRNome of SP-A2 and co-ex (SP-A1/SP-A2) and Sex Differences Attenuation after 18 h of Ozone Exposure. Antioxidants, 9(12), 1190. https://doi.org/10.3390/antiox9121190






