Oxidant-Mediated Protein Amino Acid Conversion
Abstract
1. The Rationale for the Concept of Oxidant-Mediated Protein Amino Acid Conversions
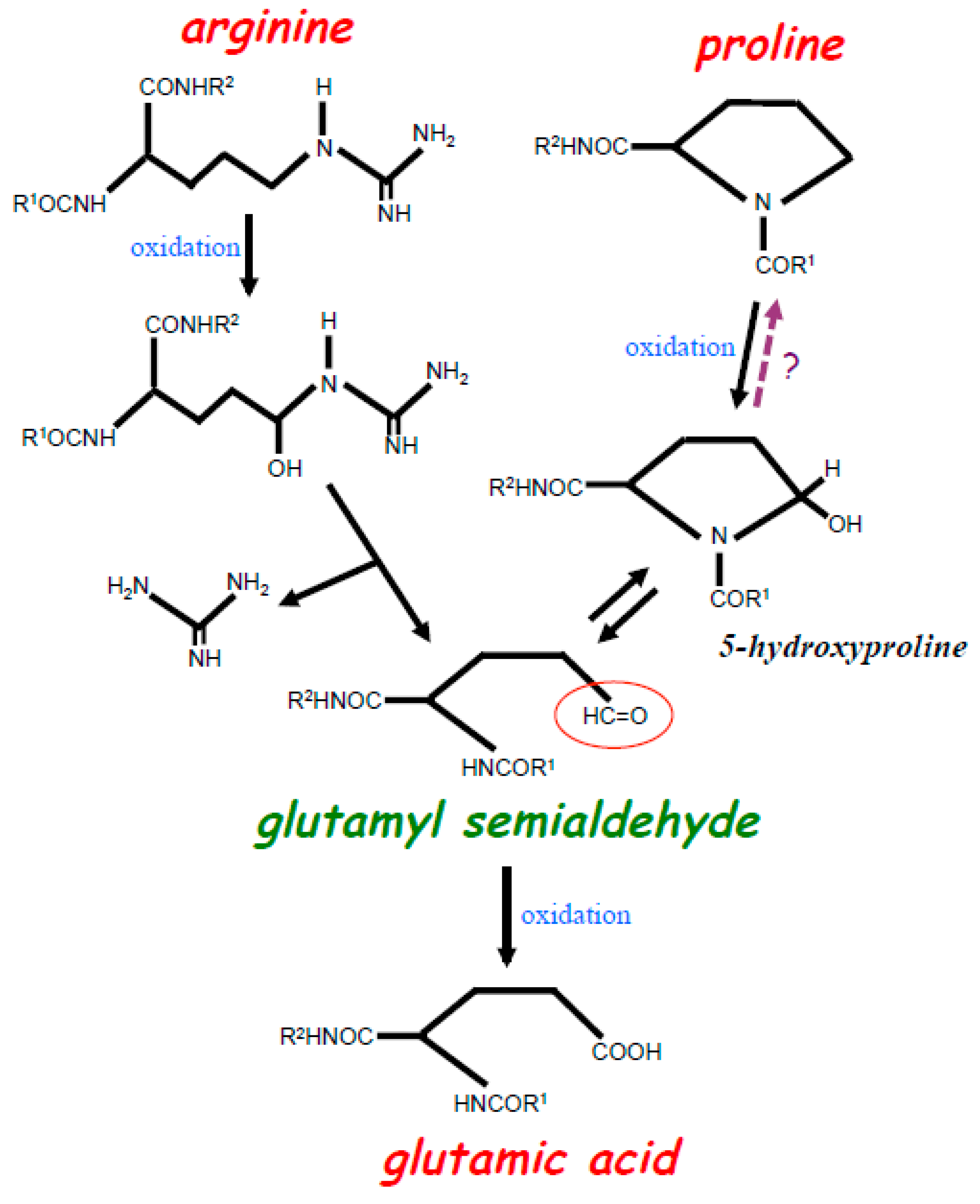
2. Experimental Evidence for the Occurrence of Protein Amino Acid Conversions in the Biological System
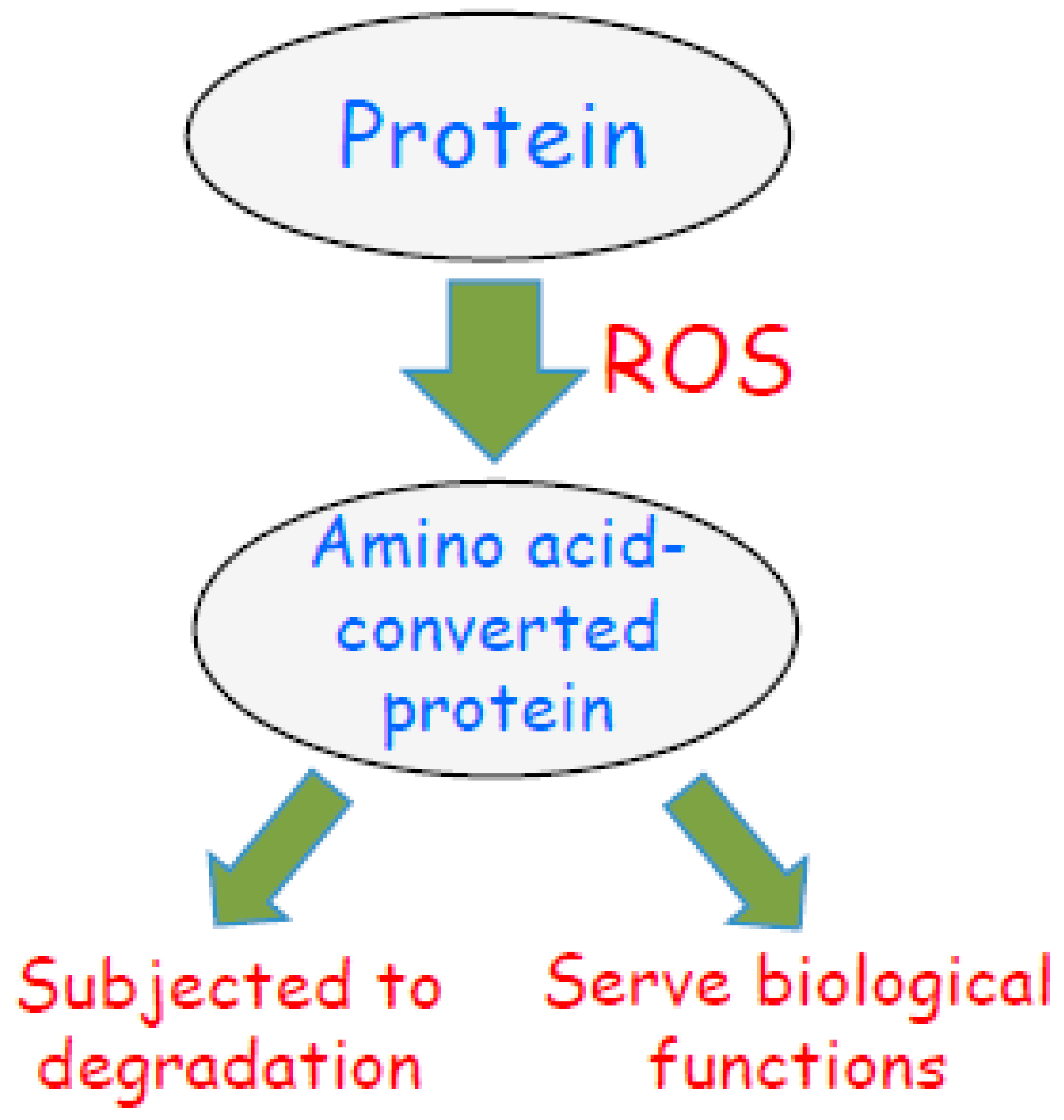
3. Role of Oxidant-Mediated Protein Amino Acid Conversion in Biology
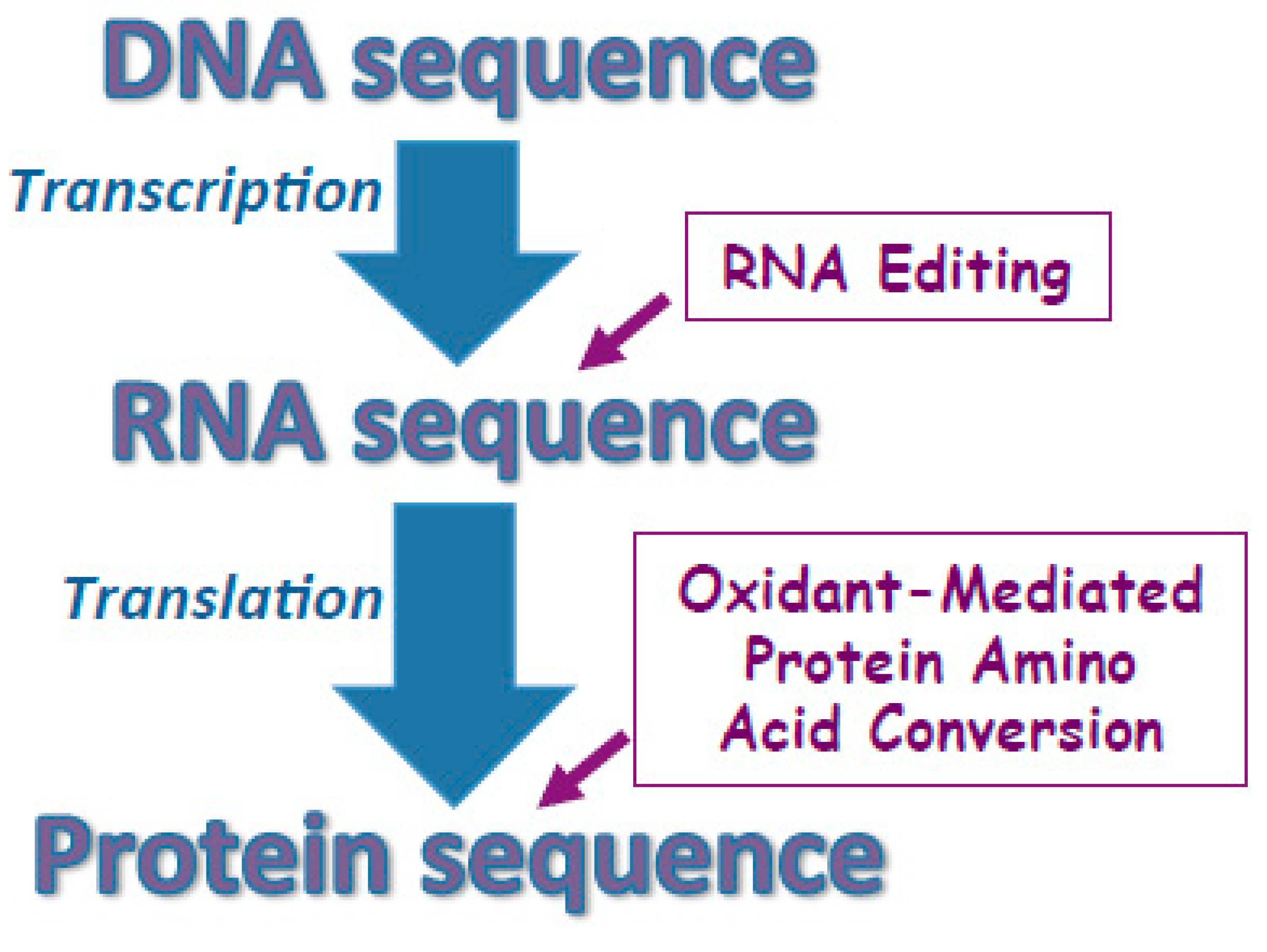
3.1. Challenging the Dogma of DNA Strictly Defining the Primary Structures of Proteins
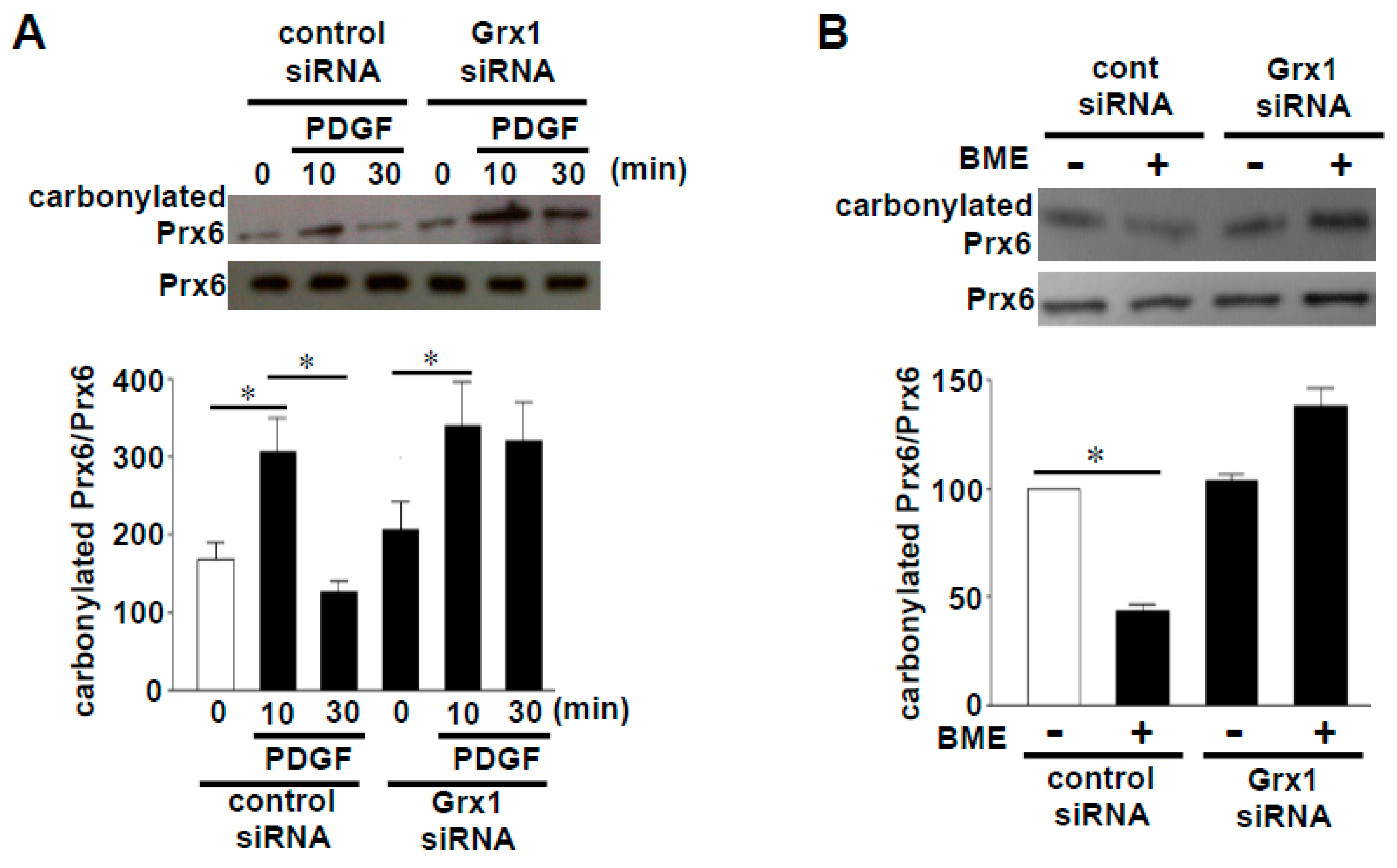
3.2. Challenging the Dogma of Proteostasis of Oxidized Proteins
4. Limitations and Future Directions
5. Conclusions
Funding
Conflicts of Interest
References
- Halliwell, B.; Gutteridge, J. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Levine, R.L. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic. Biol. Med. 2002, 32, 790–796. [Google Scholar] [CrossRef]
- Suzuki, Y.J.; Carini, M.; Butterfield, D.A. Protein carbonylation. Antioxid. Redox Signal. 2010, 12, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Amici, A.; Levine, R.L.; Tsai, L.; Stadtman, E.R. Conversion of amino acid residues in proteins and amino acid homopolymers to carbonyl derivatives by metal-catalyzed oxidation reactions. J. Biol. Chem. 1989, 264, 3341–3346. [Google Scholar] [PubMed]
- Berlett, B.S.; Stadtman, E.R. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997, 272, 20313–20316. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Cheema, A.K.; Zhang, L.; Suzuki, Y.J. Protein carbonylation as a novel mechanism in redox signaling. Circ. Res. 2008, 102, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Marcocci, L.; Liu, L.; Suzuki, Y.J. Cell signaling by protein carbonylation and decarbonylation. Antioxid. Redox Signal. 2010, 12, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Bansal, G.; Marcocci, L.; Suzuki, Y.J. Proposed role of primary protein carbonylation in cell signaling. Redox Rep. 2012, 17, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Marcocci, L.; Das, D.; Wang, X.; Luo, H.; Zungu-Edmondson, M.; Suzuki, Y.J. Mechanism of protein decarbonylation. Free Radic. Biol. Med. 2013, 65, 1126–1133. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. Peroxiredoxin 6: A bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antioxid. Redox Signal. 2011, 15, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch Biochem. Biophys. 2017, 617, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, M.; Yu, Z.X.; Ferrans, V.J.; Irani, K.; Finkel, T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 1995, 270, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.J.; Forman, H.J.; Sevanian, A. Oxidants as stimulators of signal transduction. Free Radic. Biol. Med. 1997, 22, 269–285. [Google Scholar] [CrossRef]
- Suzuki, Y.J.; Hao, J.J. Results supporting the concept of the oxidant-mediated protein amino acid conversion, a naturally occurring protein engineering process, in human cells. Version 2. F1000Res. 2017, 6, 594. [Google Scholar] [CrossRef] [PubMed]
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, C.L.; Adoue, V.; Majewski, J. RNA editing of protein sequences: A rare event in human transcriptomes. RNA 2012, 18, 1586–1596. [Google Scholar] [CrossRef] [PubMed]
- Nyström, T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005, 24, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Grune, T.; Reinheckel, T.; Davies, K.J. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997, 11, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L. Proteolysis induced by metal-catalyzed oxidation. Revis. Biol. Celular. 1989, 21, 347–360. [Google Scholar] [PubMed]
- Weil-Malherbe, H.; Krebs, H.A. Metabolism of amino-acids: The conversion of proline into glutamic acid in kidney. Biochem J. 1935, 29, 2077–2081. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.B.; Strecker, H.J. The interconversion of glutamic acid and proline. IV. The oxidation of proline by rat liver mitochondria. J. Biol. Chem. 1962, 237, 1876–1882. [Google Scholar] [PubMed]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, Y.J. Oxidant-Mediated Protein Amino Acid Conversion. Antioxidants 2019, 8, 50. https://doi.org/10.3390/antiox8020050
Suzuki YJ. Oxidant-Mediated Protein Amino Acid Conversion. Antioxidants. 2019; 8(2):50. https://doi.org/10.3390/antiox8020050
Chicago/Turabian StyleSuzuki, Yuichiro J. 2019. "Oxidant-Mediated Protein Amino Acid Conversion" Antioxidants 8, no. 2: 50. https://doi.org/10.3390/antiox8020050
APA StyleSuzuki, Y. J. (2019). Oxidant-Mediated Protein Amino Acid Conversion. Antioxidants, 8(2), 50. https://doi.org/10.3390/antiox8020050



