Abstract
Chronic rhinosinusitis without nasal polyps (CRSsNP) is a chronic inflammatory disease that lacks a clear pathogenesis/pathophysiology. While large studies focused on elucidating the pathophysiology of CRS with NPs (CRSwNP), this study aimed to use a systemic evaluation approach to identify the redox gene expression profile, its association with oxidative damage in CRSsNP, and the differences between CRSsNP and -wNP. The expression of 84 redox genes was analyzed using real-time PCR array in control and CRSsNP nasal mucosae. Changes in the mRNA and protein levels of these redox differentially expressed genes (DEGs) were verified using a customized real-time PCR array, RT-PCR, and Western blotting in an additional 18 patients. 4-Hydroxynonenal (lipid peroxidation) and 3-nitrotyrosine (protein nitrosylation) expression, representing oxidative stress (OxS) and nitrosative stress (NsS) status, were examined using immunohistochemistry. We found 27 DEGs (24 upregulated and 3 downregulated) in CRSsNP. AKR1C2, GCLM, GPX2, NOS2, and NQO1 were upregulated and LPO was downregulated more than 4-fold. These changes led to a substantial increase in OxS in CRSsNP nasal mucosa. In a comparison of the currently identified 27 DEGs with the 23 previously reported CRSwNP genes, there were 16 unique redox DEGs expressed between CRSsNP and -wNP. A String protein interaction network analysis revealed that CRSsNP possessed “an adaptive antioxidant defense signature”, while CRSwNP showed “a pro-inflammatory and -oxidant pathway”. Collectively, we systemically performed transcriptomic analysis to profile OxS-related genes in CRSsNP and highlighted the unique redox gene sets and pathway differences between CRSsNP and -wNP.
1. Introduction
Oxidative stress (OxS) is a phenomenon caused by an imbalance between the production and accumulation of reactive oxygen/nitrogen species (ROS/RNS) in cells and tissues and the ability of a biological system to detoxify these reactive products [1,2]. Superoxide radicals (O2•−), hydrogen peroxide (H2O2), hydroxyl radicals (•OH), and singlet oxygen (1O2) are commonly defined as ROS [3], while nitric oxide (NO•), peroxynitrite (ONOO−), S-nitrosothiols (RSNOs), and others are RNS [4]. The accumulation of ROS/RNS-induced damage, including damage to cellular molecules such as DNA, proteins, and lipids, is responsible for the development of diseases, especially chronic inflammatory diseases [1,5]. In contrast, the antioxidant defense system of cells, mainly consisting of enzymes such as superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPx), protects them from ROS/RNS-induced damage through antioxidant effects [6].
Chronic rhinosinusitis (CRS) can be subdivided into two major categories, CRS with nasal polyps (CRSwNP) and without nasal polyps (CRSsNP), based on the phenotypes of whether nasal polyps (NPs) are present or absent [7,8]. CRSsNP is characterized by mainly Th1-driven inflammation with high levels of IFN-γ and active TGFβ1 signaling with fibrosis, basement membrane thickening, goblet cell hyperplasia, subepithelial edema, and mononuclear cell infiltration [9]. The mechanism of action of remodeling in CRSsNP is still unclear. Therefore, a better understanding of CRSsNP pathogenesis/pathophysiology is needed to advance the current diagnosis and treatment of these patients.
While extensive studies have focused on OxS in CRSwNP [10,11,12,13,14], less attention has been paid to CRSsNP. A significant difference has been reported in the number of microplastics, which has been shown to cause oxidative damage and neurotoxicity between the CRSsNP and the control group [15]. The expression in the apical portion of the nasal epithelium of dual oxidases, which are responsible for H2O2 generation in the airway epithelium, is increased in CRSwP and CRSsP [16]. However, when comparing the control patients to their secondhand smoke (SHS)-exposed counterparts, SHS exposure was associated with statistically significantly higher levels of ROS-positive cells. The SHS exposure does not affect ROS levels in CRSsNP and CRSwNP patients [17]. No correlation was observed between any glutathione S-transferases polymorphism and CRSwNP/sNP [18].
In our previous study, the expression of OxS-related genes in nasal mucosae and NP tissues was analyzed via PCR microarray analysis. We found that 19 genes were significantly upregulated, and 4 were significantly downregulated. Among them, inducible nitric oxide synthase (iNOS) and heme oxygenase 1 (HO-1) were notably upregulated, whereas lactoperoxidase (LPO), myeloperoxidase (MPO), and superoxide dismutase 3 (SOD3) were highly downregulated [12]. In this study, we aimed to determine the oxidant/antioxidant status and gene expression profile in CRSsNP through the systemic transcriptomic analysis approach. Among the 84 OxS genes tested, 24 were overexpressed, while 3 were underexpressed. The results were confirmed by a customized real-time PCR array and RT-PCR analysis, as well as Western blotting. An apparent increase in OxS, including lipid peroxidation and protein tyrosine nitrosylation, was found in CRSsNP. Interestingly, further analysis identified 16 redox differentially expressed genes (DEGs) expressed between CRSsNP and -wNP and revealed that CRSsNP possessed “an adaptive antioxidant defense signature”, whereas CRSwNP displayed “a pro-inflammatory and oxidant pathway”.
2. Materials and Methods
2.1. Materials
The Ab raised against NOS2 (iNOS) (catalog no: 610328) was obtained from BD Biosciences (Becton Drive-Franklin Lakes, NJ, USA). The heme oxygenase-1 (HMOX1; HO-1) (catalog no: ab13243), superoxide dismutase 3 (SOD3) Ab (catalog no: ab21974), and lactoperoxidase (LPO) Ab (catalog no: ab231026) were purchased from Abcam (Cambridge, MA, USA). The Ab for β-actin (catalog no: GTX110564) was purchased from GeneTex, Inc. (Hsinchu, Taiwan).
2.2. Patient Recruitment and Sample Collection
This study was approved by the Ethics Committee of the Shin Kong Wu Ho-Su Memorial Hospital, Taipei, Taiwan (Permission No: 20161210R), and informed consent was obtained from the patients. The nasal mucosa tissues from 24 patients with CRSsNP were collected for this study. CRSsNP was diagnosed based on patient history, local findings from anterior rhinoscopy, nasal endoscopy, and sinus computed tomography. No patient had a history of allergy, asthma, or aspirin sensitivity and none had been treated with either oral or topical antiallergic agents or steroids for at least 2 months. The ethmoidal mucosa and the mucosa around the osteomeatal complex in the CRSsNP group were retrieved during functional endoscopic sinus surgery. In the control group, patients with blockages in their lacrimal drainage systems were free of other nasal diseases, and the control agger nasi sinus cell mucosae were prepared during dacryocystorhinostomy procedures [12]. The patients’ demographic data and the tissue samples used for this study are summarized in Table S1. Briefly, nasal tissue samples were taken from 6 control and 6 CRSsNP patients for the 84-gene real-time PCR microarray analysis. Additionally, 18 independent control and CRSsNP patients’ samples were used for the customized real-time PCR microarrays. The remaining tissue samples from these 24 patients were randomly used for RT-PCR, Western blot, and immunohistochemistry analysis.
2.3. Real-Time PCR Microarrays
We utilized real-time PCR microarray analysis to map the complete gene expression related to OxS. Total RNAs of human and CRSsNP control nasal mucosae were isolated by using an Rneasy Mini kit (Qiagen, Valencia, CA, USA). The cDNA was transcribed using an RT2 Reaction Ready First Strand Synthesis Kit (Qiagen) and was analyzed using the human OxS PCR array (Qiagen). The real-time PCR microarrays used an RT2 SYBR Green Fluor qPCR Mastermix (Qiagen) on a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Data were normalized by using housekeeping genes (β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β2-microglobulin (B2M), ribosomal protein, large, p0 (RPLP0)) and analyzed by comparing the 2−ΔΔCt of the normalized sample. For confirmation/validation of the (non-)altered genes by customized real-time PCR microarrays, we conducted a customized human OxS PCR array from Qiagen. A total of 18 additional and independent human control and CRSsNP nasal tissue samples were used in this confirmation. The specific primers were designed to detect the expression of the non-changed and some significantly altered genes alongside two housekeeping genes, β-actin and RPLP0. The methodology was consistent with the above real-time PCR microarray protocol.
2.4. RT-PCR Analysis of mRNA Expression Levels of DEGs
Changes in the mRNA expression levels of some of the significantly differentially expressed genes observed in the microarrays were examined by using RT-PCR for the individual transcripts. The tissues were processed as homogenates. The primers used are listed in Table 1. Total RNA extraction, 1st-strand cDNA synthesis, and PCR analysis were performed as previously described [19], except the annealing temperature for the PCR was set to 51–61 °C, depending on the sequences of the primers used.

Table 1.
Primers for reverse transcription polymerase chain reaction.
2.5. Tissue Lysate Preparation and Western Blot Analysis
Nasal tissue lysates were prepared as previously described [20]. Total proteins were analyzed on SDS–polyacrylamide gels, electroblotted onto PVDF membranes (EMD Millipore Corporation, Billerica, MA, USA), and then probed using a primary Ab. The immunoblots were developed using Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore). The membranes were stripped with a stripping buffer, washed, and reprobed with the Abs to examine the level of β-actin and then developed.
2.6. Immunohistochemistry
The modifications of 4-hydroxynonenal (4-HNE) and 3-nitrotyrosine in both control and CRSsNP nasal mucosae were assessed using immunohistochemistry (IHC), following a previously described method with slight modifications [21]. In brief, tissue sections were deparaffinized and hydrated in graded ethanol. After washing, the sections were immersed and heated in a water bath for 20 min. The slides were then incubated overnight at 4 °C with primary antibodies specific to 4-HNE or 3-nitrotyrosine (Abcam), following a blocking step with a buffer containing 10% FBS (Thermo Fisher Scientific, Waltham, NY, USA). After washing with TBS, the slides were treated with Super Enhancer and Poly-HRP and developed using one-step 3-amino-9-ethylcarbazole with the Super Sensitive Polymer-HRP IHC Detection System (Biogenex Laboratories, Inc., Fremont, CA, USA) for 5–30 min. Finally, the sections were counterstained with hematoxylin for 20–40 s, washed with tap water, and mounted using 100% glycerol. The quantitation of the staining results was performed using the Invitrogen Celleste 5.0 Image Analysis Software (Thermo Fisher Scientific, Waltham, MA, USA). Three random regions of interest (ROIs) per nasal mucosa were analyzed per patient. Mean optical density (OD) was computed from the positively stained area relative to the total ROI area.
2.7. Data Analysis
The PCR microarray data underwent a normality test per sample by using the Shapiro–Wilk test, which indicated they followed normal distribution. Protein–protein association networks and functional enrichment analyses of the DEGs were performed using STRING database (version 12.0; https://string-db.org/) [22]. All data are expressed as the mean ± SEM unless otherwise indicated. Differences between groups were compared with an unpaired and parametric Student’s t-test. Differences were considered to be statistically significant at p < 0.05.
3. Results
3.1. Twenty-Seven OxS-Related Genes Are Significantly Altered in CRSsNP Nasal Mucosa Tissues
To explore the changes in OxS-related genes in CRSsNP, a quantitative real-time PCR microarray analysis was performed to analyze 84 genes in the nasal mucosa tissues of control and CRSsNP tissues. Gene expression changes greater than two-fold with a p-value of <0.05 were considered to be significantly differentially expressed in CRSsNP patients. A total of 27 genes were significantly differentially expressed and are designated differentially expressed genes (DEGs), as shown in Table 2. Among the DEGs, 24 were overexpressed (green text/number) and 3 were underexpressed (red text/number). The overexpressed genes were ALB, AKR1C2, BAG2, DUOX1, EPX, GCLC, GCLM, GPX2, GSTP1, HSP90AA1, HMOX1, NQO1, NCF2, NOS2, PRDX1, PRDX3, PRDX5, SLC7A11, SOD1, TXN, TXNRD1, TPO, TTN, and UCP2, and the underexpressed genes were GPX3, LPO, and SOD3.

Table 2.
PCR microarray analysis of 84 OxS genes involved in CRSsNP.
A heat map of the 84 genes provided a whole view of the fold changes in their expression between the control and CRSsNP groups (Figure 1). The two markedly upregulated DEGs (uDEGs) were inducible nitric oxide synthase (iNOS; NOS2) and NAD(P)H dehydrogenase, quinone 1 (NQO1), which were upregulated by more than 17.62 and 7.99 times, respectively. Conversely, the most downregulated DEG (dDEG) was LPO, which was underexpressed by more than 15.51-fold. The uDEGs with greater than 4-fold changes were AKR1C2, GCLM, GPX2, NOS2, and NQO1, whereas the only dDEG with more than 4-fold change was LPO. All of the significant DEGs in CRSsNP that had a greater than two-fold change (black and dashed lines) with a p-value < 0.05 (above the orange line) are shown in a volcano plot (Figure 2).
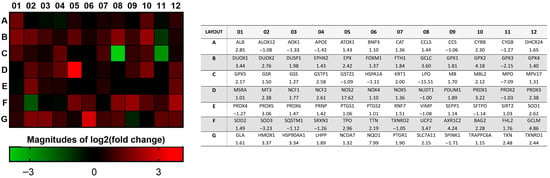
Figure 1.
The fold change and regulation in the OxS gene expression between the control and CRSsNP nasal mucosa tissues. The heat map provides log2 (fold change), whereas the table provides fold regulation data. Note that AKR1C2, GCLM, GPX2, NOS2, NQO1, and LPO were DEGs that changed by more than 4 times.
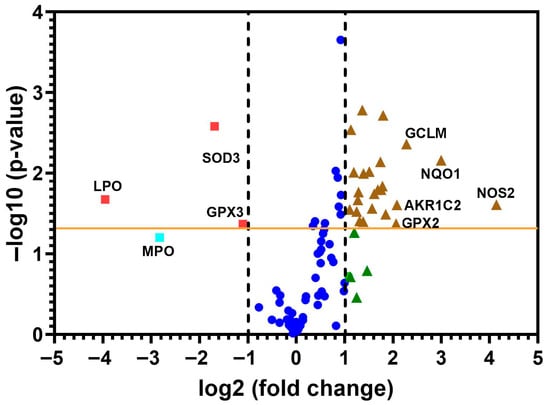
Figure 2.
The volcano plot of the changes in redox gene expressions. The 84 genes were examined from the control and CRSsNP nasal mucosa linked to OxS. The y-axis represents statistical significance, and the x-axis shows fold changes. Two vertical dashed black lines mark the threshold for two-fold changes in gene expression, and the orange line indicates a p-value of 0.05. Genes that showed significant changes are positioned beyond the dashed lines and above the orange line (denoted by brown triangles and red squares). The uDEGs that changed by more than 4 times and all dDEGs were labeled with their respective gene name. Deep blue circles represent genes less than two-fold changes, whereas green triangles and a light blue square are genes more than two-fold changes without statistical significance.
3.2. Confirmation of the DEGs by RT-PCR, Western Blot, and Customized Real-Time PCR Array Analyses
Next, an RT-PCR analysis of the selected individual transcripts of the genes was performed. The data confirmed the significant upregulation of mRNA expression of HSP90AA1 (1.5-fold increase), iNOS (2.1-fold increase), and HMOX1 (also called HO-1) (1.4-fold increase) and the significant downregulation of mRNA expression of SOD3 (reduced to 0.67-fold of control) and GPX3 (reduced to 0.5-fold of control) in the CRSsNP nasal mucosa tissues (Figure 3).
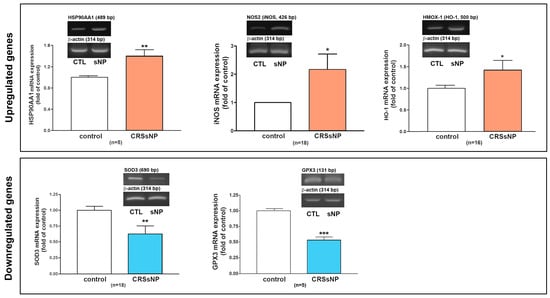
Figure 3.
RT-PCR analysis to verify the mRNA expression of some DEGs. The total RNA was extracted from human control and CRSsNP nasal mucosa tissues and the mRNA expression levels of the indicated genes were determined by RT-PCR analysis. Data are mean ± SEM. CTL: control; sNP: CRSsNP. * p < 0.05, ** p < 0.01, and *** p < 0.001 versus control.
To further investigate whether the protein DEG products are correspondingly changed in the nasal mucosa tissues of CRSsNP, the control and CRSsNP nasal mucosae were analyzed via Western blot (WB) analysis. Figure 4 shows two representative gel images for NOS2 (130 kDa in a non-reduced form), HMOX1 (32 kDa), SOD3 (32 kDa), LPO (80 kDa), and β-actin (45 kDa) expression in the nasal mucosae of controls and CRSsNP patients via WB analysis (left panels). The quantitative densitometric analysis of all collected samples (n = 8–10) indicated significantly increased levels of both NOS2 and HMOX-1 in the CRSsNP nasal samples, with approximately 1.8- and 1.3-fold increases, respectively. A reduced expression of SOD3 and LPO protein was found in CRSsNP, with about 0.5- and 0.6-fold of control (right panels). This indicates that the DEGs’ corresponding proteins were simultaneously up- and downregulated.
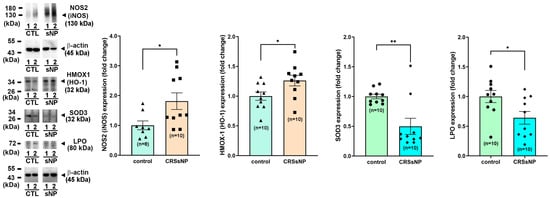
Figure 4.
The protein expression levels of the randomly selected DEGs. The protein levels of the altered genes in the control and CRSsNP nasal mucosae were analyzed by Western blotting followed by densitometric analysis. The representative and quantitative results are shown (n = 8–10 for control and n = 10 for CRSsNP). Data are mean ± SEM. * p < 0.05 and ** p < 0.01 versus control.
Next, the two non-significantly changed genes (DUSP1 and PRDX4) and four DEGs (LPO, SOD3, NOS2, and HMOX1) were further selected to confirm and validate the DEGs in 18 additional patients’ nasal tissue samples using the customized real-time PCR array analysis (Table 3), confirming that the non-significantly changed genes remained unchanged, and the DEGs remained significantly changed.

Table 3.
Confirmation of the expression levels of some randomly selected genes by customized PCR microarray.
3.3. The DEGs Lead to the Presence of Oxidative/Nitrosative Damage in CRSsNP Nasal Mucosa
We have identified 24 and 3 genes to be up- and downregulated, respectively, in CRSsNP. An in-depth literature search was performed to analyze the possible effects of ROS (Table 4a) and RNS (Table 4b) on antioxidant defense and prooxidant activity in tissues in relation to these DEGs. Briefly, as shown in Table 4a, the overexpressed PRDX1/3/5, GPX2, SOD1, TXN, and TXNRD increase ROS detoxification [23], and the GCLC, GCLM, and SLC7A11 enhance glutathione (GSH) synthesis [24]. In contrast, the downregulated GPX3 and SOD3 decrease extracellular ROS clearance and can increase extracellular OxS risk [25,26], and the upregulated DUOX1, NOS2, and NCF2 can lead to ROS/RNS generation, including H2O2, NO, and superoxide [27]. The increased EPX causes oxidative burst activity, and decreased LPO expression lowers H2O2 conversion (H2O2↑) and antimicrobial defense [28]. Table 4b shows that NOS2 can be a direct NO donor and enhances RNS production [29]. GCLC, GCLM, SLC7A11, TXN, and TXNRD1 serve as an S-nitrosoglutathione (GSNO) pool [24,30], and TXN and TXNRD1 mediate protein trans-nitrosylation and denitrosylation [31]. Moreover, DUOX1, NCF2, EPX, and CYBB mediate ROS-NO crosstalk [32]. These could contribute to RNS production, nitrosyl group formation, and protein nitrosylation.

Table 4.
(a) The putative OxS status based on analysis of the DEGs identified in CRSsNP; (b) The putative nitrosative stress (NsS) status based on analysis of the DEGs identified in CRSsNP.
The analysis in Table 4 only reflects the possible imbalance of antioxidant defense and ROS/RNS-mediated OxS damage due to DEGs, but their association with the overall OxS status in CRSsNP nasal tissue samples still needs to be explored. It has been reported that the various degradation products of lipid peroxidation include α- and β-unsaturated hydroxyalkenal, as well as 4-hydroxy-2,3-trans-nonenal (4-HNE) [33], whereas 3-nitrotyrosine results from the nitration of both protein-bound and free tyrosine residues by reactive peroxynitrite molecules [34]. Therefore, 4-HNE and 3-nitrotyrosine modifications were assayed via IHC using the respective specific antibodies. As shown in Figure 5, while only a little 4-HNE staining was found in the control nasal mucosae, a lot of positive staining for 4-HNE (deep red color) was observed in the CRSsNP nasal mucosa tissues. Positive staining was found in the epithelium, subepithelial stromal cells, glands, and some infiltrated leukocytes. Meanwhile, the 3-nitrotyrosine modifications were also enhanced in the CRSsNP nasal mucosae, which were located in the infiltrated leukocytes underneath the epithelium infiltrated leukocytes but were expressed in the epithelium, glands, and some infiltrated leukocytes. Taken together, our results reveal a significant increase in oxidative and nitrosative stress in CRSsNP, including lipid peroxidation and protein tyrosine nitrosylation.
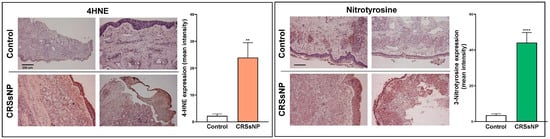
Figure 5.
The increase in OxS and NsS in CRSsNP nasal mucosae. The control and CRSsNP (SNP) nasal mucosae were analyzed by IHC to determine the status of OxS and NsS by using anti-4-HNE Ab, anti-3-nitrotyrosine Ab, and their corresponding nonimmune IgG (NIgG). The images were captured under a phase-contrast microscope. A (deep) red color indicates positive staining of 4-HNE or 3-nitrotyrosine. Scale bar: 200 μm. The quantitation of the staining results was performed by measuring the mean intensity of three positive staining areas of each tissue sample using the Invitrogen Celleste 5.0 Image Analysis Software (n = 9). ** p < 0.01 and **** p < 0.0001 versus control.
3.4. The Analysis by STRING Reveals a Novel Adaptive Antioxidant Defense Imbalance Signature in CRSsNP but a Pro-Inflammatory/Oxidant Pathway in CRSwNP
The gene expression profile in CRSsNP was compared with that found in CRSwNP [12]. A shown in Figure 6A, 24 uDEGs and 3 dDEGs were found in CRSsNP, whereas 19 and 4 were identified in CRSwNP [12]. In Figure 6B, the Venn diagram shows that there were 17 DEGs completely identical/overlapping in CRSsNP and -wNP. However, 16 DEGs were quite different between CRSsNP and -wNP, in which 10 DEGs were unique in CRSsNP and only 6 DEGs were particular to CRSwNP (panel B). To further pinpoint the DEG association with OxS in CRSsNP and -wNP, the total 27 and 23 DEGs of CRSsNP and -wNP were subjected to analysis by the STRING protein interaction network and gene ontology (GO) enrichment analysis. In Figure S1, a highly dense antioxidant hub was found in CRSsNP. Moreover, the presence of GCLC, GCLM, GSTP1, TXNRD1, and GPX2 in CRSsNP highly suggested strong GSH- and thiol-based antioxidant detoxification potential. On the contrary, in wNP, the CYBB (NOX2), MPO, and PTGS2 were tightly connected to iron metabolism (FTH1, HMOX1), favoring an inflammatory oxidative burst (panel A). The GO enrichment analysis indicated that CRSsNP and -wNP shared two identities in the top three biological processes and four identities in the top five molecular functions. It was noted that the “response to toxic substance” and “thioredoxin peroxidase activity” were unique in CRSsNP in GO biological process and molecular function (panel B, box and highlighted region).
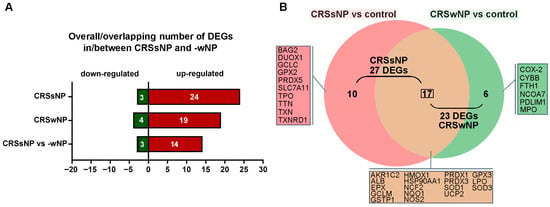
Figure 6.
Analysis of OxS-related DEGs in CRSsNP and -wNP. (A) Overall/overlapping DEGs (Up- and downregulated genes) in/between nasal tissue samples of CRSsNP versus control and CRSwNP versus control. (B) The Venn diagram shows the number and genes of (un)intersective DEGs between CRSsNP versus control and CRSwNP versus control.
Surprisingly, if the overlapping 17 DEGs were excluded and only the unique 10 and 6 DEGs in CRSsNP and -wNP were, respectively, subjected to analysis by the STRING, further differences were revealed. In Figure 7, in CRSsNP, the cluster genes, like TXNRD1, PRDX5, TXN, GPX2, GCLC, SLC7A11, formed a highly interconnected and tight core network, indicating strong functional interdependence, especially within antioxidant defense pathways including the thioredoxin system (TXN, TXNRD1), peroxiredoxin system (PRDX5), and GSH system (GCLC, GPX2, SLC7A11) (highlighted by a circle). The peripheral yet connected DUOX1 and TPO reflect a separate role in extracellular ROS production and possibly in thyroid hormone biosynthesis, which likely bridged intracellular ROS detoxification and extracellular ROS generation roles. The isolated nodes (BAG2, TTN) suggest different or secondary roles in CRSsNP and may be non-redox-related. Therefore, the network suggests “a dominant, adaptive antioxidant defense signature” with additional roles in extracellular redox signaling. Conversely, in CRSwNP, the core triangle cluster of PTGS2, MPO, and CYBB (NOX2) with FTH1 at the top indicates centrality in ROS/RNS generation (CYBB via NADPH oxidase and PTGS2) and neutrophil- and prooxidant-mediated microbial killing (MPO) with iron metabolism (FTH1) bridging the gap, suggesting “a prooxidant-inflammatory axis” (highlighted by a circle). Both PDLIM1 and NCOA7 were unconnected and might play non-core roles, perhaps linked to transcriptional regulation or structural modulation under CRSwNP.
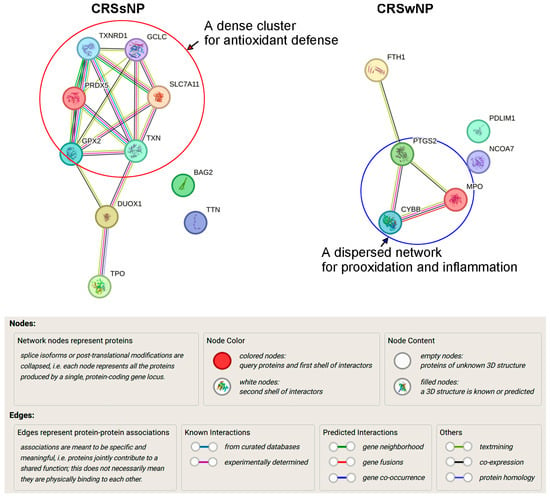
Figure 7.
Analysis of protein–protein interaction. The 10 and 6 distinct DEGs of CRSsNP and -wNP obtained from the Venn diagram were subjected to analysis by the STRING Database (Version 12.0) for determining the protein–protein interactions. The explanation of the nodes, lines, and edges in the interaction network is provided. The circles highlighting the proteins located in the core cluster/network of the interactions suggest an antidefense pathway in CRSsNP but a prooxidant and inflammatory pathway in CRSwNP.
4. Discussion
Low levels of ROS/RNS production are required to maintain physiological functions such as signal transduction and host defense, but a higher level of ROS/RNS due to an imbalance between oxidant and antioxidant defense systems can produce OxS, damaging cellular macromolecules [35,36]. In this study, we demonstrated that out of the 84 genes examined, 24 and 3 OxS-related genes were significantly up- and downregulated in CRSsNP, respectively (Table 2). The results were confirmed and validated by the customized PCR array, RT-PCR, and Western blotting (Table 3 and Figure 3 and Figure 4). Further analysis suggested participation of these DEGs in prooxidant (ROS/RNS production) and antioxidant defense (Table 4), and OxS was higher in CRSsNP nasal mucosae, indicating that the DEGs substantially led to an increase in OxS and NsS in CRSsNP (Figure 5). Notably, in comparison with the findings of our previous study [12], we not only identified the unique genes for CRSsNP and -wNP but also demonstrated that CRSsNP possesses “a dominant, adaptive antioxidant defense signature”, while CRSwNP prefers “a pro-inflammatory and oxidant pathway” (Figure 6, Figure 7 and Figure S1).
Regarding the distinct signature/pathway in CRSsNP and -wNP, it was found that a total of 16 unique DEGs were distinct between them (Figure 6). In sNP, these DEGs, such as GPX2, PRDX5, TXN, and TXNRD1, are all central to reducing ROS production [23,26,37,38], and GCLC and SLC7A11 are crucial in GSH biosynthesis and cystine uptake, which enhances cellular redox buffering capacity [24,39], suggesting strong intracellular antioxidant reinforcement, especially via GSH and thioredoxin systems. On the other hand, CYBB (as a NOX2 subunit), PTGS2 (COX-2), FTH1, and PRDX3 formed a linear connection in wNP, highly suggesting prooxidant pathways and inflammation as COX-2 produces prostaglandins (inflammatory ROS) [40], CYBB generates superoxide in phagocytes [41], FTH1 (Ferritin Heavy Chain) regulates iron storage and prevents Fenton reaction-mediated oxidative damage [42,43], and NCOA7 has oxidant detoxification roles in lysosomes [44,45]. Therefore, it can be concluded that CRSsNP has a signature of adaptive antioxidant responses, whereas CRSwNP has highlighted inflammatory ROS production (prooxidant) pathways and perhaps less reliance on antioxidant reinforcement. Nevertheless, one may question such a signature, as the presence of OxS and NsS in CRSsNP seems contradictory. This can be explained by the inconsistently translated protein amounts of these DEGs and a compensatory (secondary) antioxidant response to the ongoing OxS, which is known as “antioxidant response element (ARE)-driven compensation”, especially by Nrf2 [46]. Moreover, the signature may involve a tissue protective mechanism to OxS and NsS.
Among the overlapping DEGs between CRSsNP and -wNP, the upregulated NOS2, NQO1, AKR1C2, GPX2, and GCLM and the downregulated LPO were notable and showed more than 4-fold changes. NOS2 is known as an inducible NO synthase (iNOS) and is well known to produce NO [47]. There are many studies indicating that NOS2 is more highly expressed in NPs than in normal cases [48,49,50,51,52]. However, few studies have reported overexpression of NOS2 in CRSsNP. Recently, it was shown that CRSwNP patients exhibited decreased nasal NO despite elevated NOS2 mRNA expression, implying that lowered nasal NO production in CRSwNP may not be related to NOS expression [51]. This may raise a question about whether the increase in NOS2 expression correlates with the nasal NO production in CRSsNP. This is highly speculative, since NOS2 is a major donor for NO and RNS and protein nitrosylation has been found to be higher in CRSsNP (Figure 5). NQO1, belonging to the NAD(P)H dehydrogenase (quinone) family, is a FAD-binding protein family that prevents the one-electron reduction in quinones that results in the production of radical species [53,54]. It has been shown that NQO1 knockdown enhances ROS production and diminishes cell proliferation, but its overexpression increases proliferation in glioblastoma cells [55]. Moreover, GPX2 belongs to the glutathione peroxidase family [26]. These gene products are responsible for removing ROS and other harmful substances and facilitating anti-OxS. As to GCLM and AKR1C2, they are the first-rate limiting enzymes of GSH synthesis and are involved in eliminating ROS and regulating tumor invasion, migration, and other malignant phenotypes [56]. Among these upregulated DEGs, HMOX-1 and GPX2 were located at the core of network interaction, whereas AKR1C2, GCLM, NQO1, and NOS2 were at the periphery (Figure S1). The reasons why LPO was downregulated in CRSsNP and -wNP remain to be explored. LPO is synthesized and secreted from epithelial cells [28,57]. It is unlikely that the downregulation resulted from shedding/damage of the epithelium. Our recent unpublished data found that a bacterial cell wall component downregulates basal constitutive LPO production in nasal epithelial cells, suggesting that its expression can be tightly regulated in nasal epithelial cells in a CRS microbial infection microenvironment.
Our analysis of the literature indicated the putative pro- and antioxidant activities of 27 dysregulated DEGs (Table 4), and OxS and NsS levels were demonstrated to be higher in the CRSsNP nasal mucosae (Figure 5). The reaction between O2•− and nitric oxide (NO•) may produce ONOO−, and their decomposition then causes some highly oxidizing intermediates, including NO2•, OH•, CO3•−, and NO3− [58,59]. The nitric oxide and peroxynitrite molecules contribute to the nitrosylation of protein tyrosine [34]. The upregulation of DUOX1, NCF2, and NOS2 (iNOS) may contribute to ROS, O2•−, and NO production. For example, NCF2 is part of the NADPH oxidase complex (NOX2) and can produce superoxide, which reacts with NO to form peroxynitrite [4]. BAG2 has been shown to protect neurons against 1-methyl-4-phenylpyridinium-induced OxS in an in vitro cell model of Parkinson’s disease [60]. In addition, numerous initiators of lipid peroxidation, such as 4-HNE in biological systems, are often hydroxyl radical (OH•), ozone (O3), nitrogen oxide (NO), nitrogen dioxide (NO2), and sulfur dioxide (SO2) [61], which may result from the downregulation of GPX3, LPO, and SOD3. The increase in ROS production may also lead to the upregulation of several antioxidants to clear ROS, such as TXN and TXNRD1 (reduction in disulfides (S-S) within oxidized cellular proteins), PRDX1, 3, and -5 (detoxifying H2O2), HMOX-1 (degradation of cellular heme against OxS), GCLM, AKR1C2, SLC7A11, NQO1, and GSTP1 for GSH synthesis and function.
It may be a limitation that this study used a small number of six control and CRSsNP samples for transcriptomic profiling (PCR microarray analysis), but the effect sample sizes were calculated to be 10, 21, and 37 based on a formal power analysis (see the calculations in Supplementary data). In this regard, six of the (non)DEGs were further confirmed and validated by the customized PCR microarray with an “independent” 18 patients (cohort). We showed the same (un)changed trend in these genes as our initial profiling and with smaller p-values in an appropriate effect sample size (>n = 14 by the formal power analysis with σ = 0.75). Due to limited tissue specimens, it should be mentioned that the sample size in initial profiling did not reach the calculated effect size. The small size may limit power [only detects very large but may miss moderate effects (false negatives)] and reduce robustness. In addition, the small set of significant hits may be overestimated. The authors would also like to address the limitation of using different sinus subsites for control and CRSsNP samples (which have different epithelial cell compositions and basal gene expression), while most studies adopt nasal mucosae from nasal septum deviation surgery and agger nasi mucosa removed during dacryocystorhinotomy as a control.
For a quick overview of our findings, a side-by-side table is provided to summarize the overlapping and unique DEGs, their biological functions, and possible clinical implications (Table S2). Our STRING analysis suggested CRSwNP with a prooxidant–inflammatory axis, which is more closely associated with type 2 inflammatory pathways and explained the favorable response of these patients to biologic therapies and surgery [62,63]. However, an important clinical nuance is that a subset of CRSsNP patients may also exhibit type 2-high endotypes despite the absence of visible NPs [64]. This biological heterogeneity may help explain the remarkable response to biologic therapies observed in some CRSsNP patients. Therefore, the findings of this study highlight the need for endotype-driven treatment approaches rather than relying solely on the phenotypic classification of CRS.
5. Conclusions
In this study, we used a systems biology approach to profile OxS-related genes and identified 27 DEGs and the status of oxidative stress in CRSsNP nasal mucosa tissues. The DEGs were verified and confirmed by a customized PCR microarray, RT-PCR, and Western blotting. More importantly, we compared the DEGs in CRSsNP with previously reported CRSwNP DEGs and found up to 16 genes distinct between them. We showed here that CRSsNP possesses “an adaptive antioxidant defense signature”, while CRSwNP tends to exhibit “a pro-inflammatory/oxidant pathway”, highlighting the unique redox gene sets and pathway differences between CRSsNP and -wNP.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antiox14111292/s1. Table S1: The patients’ information and tissue samples used in the analysis. Table S2: The unique/overlapping DEGs of CRSsNP and -wNP and their biological functions and possible clinical implications. Figure S1: The protein interaction network and gene ontology (GO) enrichment analysis of the overall 27 and 23 DEGs of CRSsNP and -wNP by the STRING database.
Author Contributions
Conceptualization, Y.-J.T. and W.-B.W.; methodology, Y.-J.T., J.-M.S. and W.-B.W.; software, J.-M.S.; validation, Y.-J.T. and W.-B.W.; formal analysis, Y.-J.T., J.-M.S. and W.-B.W.; investigation, Y.-J.T., J.-M.S. and W.-B.W.; resources, Y.-J.T.; data curation, Y.-J.T., M.-C.M. and W.-B.W.; writing—original draft preparation, Y.-J.T., M.-C.M. and W.-B.W.; writing—review and editing, M.-C.M. and W.-B.W.; supervision, W.-B.W.; project administration, W.-B.W.; funding acquisition, Y.-J.T. and W.-B.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Technology in Taiwan, grant number MOST 105-2320-B-030-005 and National Science and Technology Council in Taiwan, grant number NSTC 112-2320-B-030-004-MY3, and Shin Kong Wu Ho-Su Memorial Hospital, grant number 112-SKH-FJU-06.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Shin Kong Wu Ho-Su Memorial Hospital, Taipei, Taiwan; ethical approval number: 20161210R. The approval date was 9 February 2017.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent was obtained from the patient(s) to publish this paper.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
References
- Duracková, Z. Some current insights into oxidative stress. Physiol. Res. 2010, 59, 459–469. [Google Scholar] [CrossRef]
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative stress: Harms and benefits for human health. Oxidative Med. Cell. Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef]
- Sato, H.; Shibata, M.; Shimizu, T.; Shibata, S.; Toriumi, H.; Ebine, T.; Kuroi, T.; Iwashita, T.; Funakubo, M.; Kayama, Y.; et al. Differential cellular localization of antioxidant enzymes in the trigeminal ganglion. Neuroscience 2013, 248, 345–358. [Google Scholar] [CrossRef]
- Pacher, P.; Beckman, J.S.; Liaudet, L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007, 87, 315–424. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Tan, B.; Yin, Y.; Blachier, F.; Tossou, M.C.; Rahu, N. Oxidative stress and inflammation: What polyphenols can do for us? Oxidative Med. Cell. Longev. 2016, 2016, 7432797. [Google Scholar] [CrossRef]
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta 2013, 1830, 3217–3266. [Google Scholar] [CrossRef]
- Van Bruaene, N.; Derycke, L.; Perez-Novo, C.A.; Gevaert, P.; Holtappels, G.; De Ruyck, N.; Cuvelier, C.; Van Cauwenberge, P.; Bachert, C. Tgf-beta signaling and collagen deposition in chronic rhinosinusitis. J. Allergy Clin. Immunol. 2009, 124, 253–259, 259.e251–252. [Google Scholar] [CrossRef] [PubMed]
- Takabayashi, T.; Kato, A.; Peters, A.T.; Hulse, K.E.; Suh, L.A.; Carter, R.; Norton, J.; Grammer, L.C.; Cho, S.H.; Tan, B.K.; et al. Excessive fibrin deposition in nasal polyps caused by fibrinolytic impairment through reduction of tissue plasminogen activator expression. Am. J. Respir. Crit. Care Med. 2013, 187, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Van Bruaene, N.; Bachert, C. Tissue remodeling in chronic rhinosinusitis. Curr. Opin. Allergy Clin. Immunol. 2011, 11, 8–11. [Google Scholar] [CrossRef]
- Tai, J.; Shin, J.M.; Park, J.; Han, M.; Kim, T.H. Oxidative stress and antioxidants in chronic rhinosinusitis with nasal polyps. Antioxidants 2023, 12, 195. [Google Scholar] [CrossRef]
- Mihalj, H.; Butković, J.; Tokić, S.; Štefanić, M.; Kizivat, T.; Bujak, M.; Baus Lončar, M.; Mihalj, M. Expression of oxidative stress and inflammation-related genes in nasal mucosa and nasal polyps from patients with chronic rhinosinusitis. Int. J. Mol. Sci. 2022, 23, 5521. [Google Scholar] [CrossRef]
- Tsai, Y.J.; Hsu, Y.T.; Ma, M.C.; Wu, C.K.; Luo, S.D.; Wu, W.B. Transcriptomic analysis of genes associated with oxidative stress in chronic rhinosinusitis patients with nasal polyps: Identifying novel genes involved in nasal polyposis. Antioxidants 2022, 11, 1899. [Google Scholar] [CrossRef] [PubMed]
- Topal, O.; Kulaksızoglu, S.; Erbek, S.S. Oxidative stress and nasal polyposis: Does it affect the severity of the disease? Am. J. Rhinol. Allergy 2014, 28, e1–e4. [Google Scholar] [CrossRef]
- Lin, H.; Ba, G.; Tang, R.; Li, M.; Li, Z.; Li, D.; Ye, H.; Zhang, W. Increased expression of txnip facilitates oxidative stress in nasal epithelial cells of patients with chronic rhinosinusitis with nasal polyps. Am. J. Rhinol. Allergy 2021, 35, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Taş, B.M.; Tuna, A.; Başaran Kankılıç, G.; Koçak, F.M.; Şencan, Z.; Cömert, E.; Bayar Muluk, N. Role of microplastics in chronic rhinosinusitis without nasal polyps. Laryngoscope 2023, 134, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.Y.; Nayak, J.V.; Bravo, D.T.; Le, W.; Nguyen, A.; Edward, J.A.; Hwang, P.H.; Illek, B.; Fischer, H. Expression of dual oxidases and secreted cytokines in chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2013, 3, 376–383. [Google Scholar] [CrossRef]
- Fordham, M.T.; Mulligan, J.K.; Casey, S.E.; Mulligan, R.M.; Wang, E.W.; Sansoni, E.R.; Schlosser, R.J. Reactive oxygen species in chronic rhinosinusitis and secondhand smoke exposure. Otolaryngol. Head Neck Surg. 2013, 149, 633–638. [Google Scholar] [CrossRef]
- Fruth, K.; Best, N.; Amro, M.; Ingel, K.; Gosepath, J.; Mann, W.J.; Brieger, J. No evidence for a correlation of glutathione s-tranferase polymorphisms and chronic rhinosinusitis. Rhinology 2011, 49, 180–184. [Google Scholar] [CrossRef]
- Lai, T.-H.; Shieh, J.-M.; Tsou, C.-J.; Wu, W.-B. Gold nanoparticles induce heme oxygenase-1 expression through nrf2 activation and bach1 export in human vascular endothelial cells. Int. J. Nanomed. 2015, 10, 5925–5939. [Google Scholar] [CrossRef]
- Tsai, Y.-J.; Chi, J.C.-Y.; Hao, C.-Y.; Wu, W.-B. Peptidoglycan induces bradykinin receptor 1 expression through toll-like receptor 2 and nf-κb signaling pathway in human nasal mucosa-derived fibroblasts of chronic rhinosinusitis patients. J. Cell. Physiol. 2018, 233, 7226–7238. [Google Scholar] [CrossRef]
- Tsai, Y.J.; Hao, C.Y.; Chen, C.L.; Wu, P.H.; Wu, W.B. Expression of long pentraxin 3 in human nasal mucosa fibroblasts, tissues, and secretions of chronic rhinosinusitis without nasal polyps. J. Mol. Med. 2020, 98, 673–689. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The string database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Rhee, S.G.; Chae, H.Z.; Kim, K. Peroxiredoxins: A historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005, 38, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Asp. Med. 2009, 30, 42–59. [Google Scholar] [CrossRef]
- Marklund, S.L. Extracellular superoxide dismutase and other superoxide dismutase isoenzymes in tissues from nine mammalian species. Biochem. J. 1984, 222, 649–655. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta 2013, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, J.D. Nox enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004, 4, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Arnhold, J. Heme peroxidases at unperturbed and inflamed mucous surfaces. Antioxidants 2021, 10, 1805. [Google Scholar] [CrossRef]
- Bogdan, C. Nitric oxide and the immune response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef]
- Benhar, M.; Forrester, M.T.; Stamler, J.S. Protein denitrosylation: Enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 721–732. [Google Scholar] [CrossRef]
- Nagarajan, N.; Sadoshima, J. Regulation of protein nitrosylation by thioredoxin 1. In Biochemistry of Oxidative Stress: Physiopathology and Clinical Aspects; Gelpi, R.J., Boveris, A., Poderoso, J.J., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 163–175. [Google Scholar]
- Bedard, K.; Krause, K.H. The nox family of ros-generating nadph oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Csala, M.; Kardon, T.; Legeza, B.; Lizák, B.; Mandl, J.; Margittai, É.; Puskás, F.; Száraz, P.; Szelényi, P.; Bánhegyi, G. On the role of 4-hydroxynonenal in health and disease. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 826–838. [Google Scholar] [CrossRef]
- Bandookwala, M.; Sengupta, P. 3-nitrotyrosine: A versatile oxidative stress biomarker for major neurodegenerative diseases. Int. J. Neurosci. 2020, 130, 1047–1062. [Google Scholar] [CrossRef]
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef]
- Gornicka, A.; Morris-Stiff, G.; Thapaliya, S.; Papouchado, B.G.; Berk, M.; Feldstein, A.E. Transcriptional profile of genes involved in oxidative stress and antioxidant defense in a dietary murine model of steatohepatitis. Antioxid. Redox Signal 2011, 15, 437–445. [Google Scholar] [CrossRef]
- Yoshihara, E.; Masaki, S.; Matsuo, Y.; Chen, Z.; Tian, H.; Yodoi, J. Thioredoxin/Txnip: Redoxisome, as a redox switch for the pathogenesis of diseases. Front. Immunol. 2014, 4, 2013. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, C.; Franckenstein, D.; Schmidt, M.; Gehrmann, M.; Hermes, M.; Geppert, B.; Schormann, W.; Maccoux, L.J.; Schug, M.; Schumann, A.; et al. Role of thioredoxin reductase 1 and thioredoxin interacting protein in prognosis of breast cancer. Breast Cancer Res. 2010, 12, R44. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.L.; Deme, J.C.; Kolokouris, D.; Kuteyi, G.; Biggin, P.C.; Lea, S.M.; Newstead, S. Molecular basis for redox control by the human cystine/glutamate antiporter system xc−. Nat. Commun. 2021, 12, 7147. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, Y.; Liu, M.; Qin, X.; Yu, X.; Zhao, H.; Li, X.; Li, W. Cox-2 is required to mediate crosstalk of ros-dependent activation of MAPK/NF-κb signaling with pro-inflammatory response and defense-related no enhancement during challenge of macrophage-like cell line with giardia duodenalis. PLoS Neglected Trop. Dis. 2022, 16, e0010402. [Google Scholar] [CrossRef] [PubMed]
- Hervé, C.; Tonon, T.; Collén, J.; Corre, E.; Boyen, C. Nadph oxidases in eukaryotes: Red algae provide new hints! Curr. Genet. 2006, 49, 190–204. [Google Scholar] [CrossRef]
- Guo, J.; Xu, N.; Yao, Y.; Lin, J.; Li, R.; Li, J.-W. Efficient expression of recombinant human heavy chain ferritin (fth1) with modified peptides. Protein Expr. Purif. 2017, 131, 101–108. [Google Scholar] [CrossRef]
- Enami, S.; Sakamoto, Y.; Colussi, A.J. Fenton chemistry at aqueous interfaces. Proc. Natl. Acad. Sci. USA 2014, 111, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Castroflorio, E.; den Hoed, J.; Svistunova, D.; Finelli, M.J.; Cebrian-Serrano, A.; Corrochano, S.; Bassett, A.R.; Davies, B.; Oliver, P.L. The ncoa7 locus regulates v-atpase formation and function, neurodevelopment and behaviour. Cell Mol. Life Sci. 2021, 78, 3503–3524. [Google Scholar] [CrossRef]
- Wang, L.; Wei, Z.; Wu, Y.; Hu, X.; Zhou, L.; Zhao, M.; Sun, A.; Shao, G.; Yang, W.; Lin, Q. Nuclear receptor coactivator 7 (ncoa7) protects cancer cells from oxidative damage through its erbd domain. Cell Signal 2024, 124, 111382. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the keap1-nrf2-are pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Anavi, S.; Tirosh, O. Inos as a metabolic enzyme under stress conditions. Free Radic. Biol. Med. 2020, 146, 16–35. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, C.; Apa, D.D.; Pata, Y.S.; Görür, K.; Akbaş, Y. Expression of inducible nitric oxide synthase in antrochoanal polyps. Int. J. Pediatr. Otorhinolaryngol. 2003, 67, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kakuta, S. Expression and localization of the inducible isoform of nitric oxide synthase in nasal polyps. Nippon. Jibiinkoka Gakkai Kaiho 2002, 105, 873–881. [Google Scholar] [CrossRef]
- Tang, B.; Tu, J.; Zhang, M.; Zhang, Z.; Yu, J.; Shen, L.; Luo, Q.; Ye, J. Diagnostic value and underlying mechanism of nasal nitric oxide in eosinophilic chronic rhinosinusitis with nasal polyps. Mol. Immunol. 2023, 159, 1–14. [Google Scholar] [CrossRef]
- Wu, V.; Cusimano, M.; Marsden, P.; Lee, J.M. Levels of nasal nitric oxide and nitric oxide synthase expression in chronic rhinosinusitis with nasal polyposis. Int. Forum Allergy Rhinol. 2024, 14, 127–129. [Google Scholar] [CrossRef]
- Yoshimura, T.; Moon, T.C.; St Laurent, C.D.; Puttagunta, L.; Chung, K.; Wright, E.; Yoshikawa, M.; Moriyama, H.; Befus, A.D. Expression of nitric oxide synthases in leukocytes in nasal polyps. Ann. Allergy Asthma Immunol. Off. Publ. Am. Coll. Allergy Asthma Immunol. 2012, 108, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Begleiter, A.; Fourie, J. Induction of nqo1 in cancer cells. Methods Enzymol. 2004, 382, 320–351. [Google Scholar] [PubMed]
- Lee, W.S.; Ham, W.; Kim, J. Roles of nad(p)h:Quinone oxidoreductase 1 in diverse diseases. Life 2021, 11, 1301. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Lei, K.; Xiang, D.; Ye, K. Nqo1 is regulated by pten in glioblastoma, mediating cell proliferation and oxidative stress. Oxidative Med. Cell. Longev. 2018, 2018, 9146528. [Google Scholar] [CrossRef]
- Wang, Z.; Feng, Y.; Song, J.; Sun, D.; Zhang, Y.Y. Function, drug resistance and prognostic effect of akr1c2 in human cancer. Neoplasma 2023, 70, 319–332. [Google Scholar] [CrossRef]
- Zamocky, M.; Jakopitsch, C.; Furtmüller, P.G.; Dunand, C.; Obinger, C. The peroxidase-cyclooxygenase superfamily: Reconstructed evolution of critical enzymes of the innate immune system. Proteins 2008, 72, 589–605. [Google Scholar] [CrossRef]
- Fukai, T.; Ushio-Fukai, M. Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal 2011, 15, 1583–1606. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ros damage and regulating ros signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Qin, L.; Guo, J.; Zheng, Q.; Zhang, H. Bag2 structure, function and involvement in disease. Cell. Mol. Biol. Lett. 2016, 21, 18. [Google Scholar] [CrossRef]
- Bilska-Wilkosz, A.; Iciek, M.; Górny, M. Chemistry and biochemistry aspects of the 4-hydroxy-2,3-trans-nonenal. Biomolecules 2022, 12, 145. [Google Scholar] [CrossRef]
- Giri, S.; Schneider, A.L.; Tan, B.K. Chronic rhinosinusitis: Future treatments and unmet needs. J. Allergy Clin. Immunol. 2022, 150, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Hamada, K.; Oishi, K.; Chikumoto, A.; Murakawa, K.; Ohteru, Y.; Matsuda, K.; Uehara, S.; Suetake, R.; Ohata, S.; Murata, Y.; et al. Impact of sinus surgery on type 2 airway and systemic inflammation in asthma. J. Asthma 2021, 58, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Delemarre, T.; Holtappels, G.; De Ruyck, N.; Zhang, N.; Nauwynck, H.; Bachert, C.; Gevaert, E. Type 2 inflammation in chronic rhinosinusitis without nasal polyps: Another relevant endotype. J. Allergy Clin. Immunol. 2020, 146, 337–343.e336. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).