Each Cellular Compartment Has a Characteristic Protein Reactive Cysteine Ratio Determining Its Sensitivity to Oxidation
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Proteomic Analysis
2.2. Cell Culture and Treatments
2.3. Reactive Thiol Groups and Protein Staining
2.4. Apoptosis Induction by TRAIL
2.5. Transcription and Immunofluorescence
2.6. Immunostaining and Antibodies
2.7. Protein Carbonylation Analysis
2.8. siRNA
2.9. Microscopy and Image Analysis
3. Results
3.1. Different Cellular Compartments Have Distinct Protein Cys Content
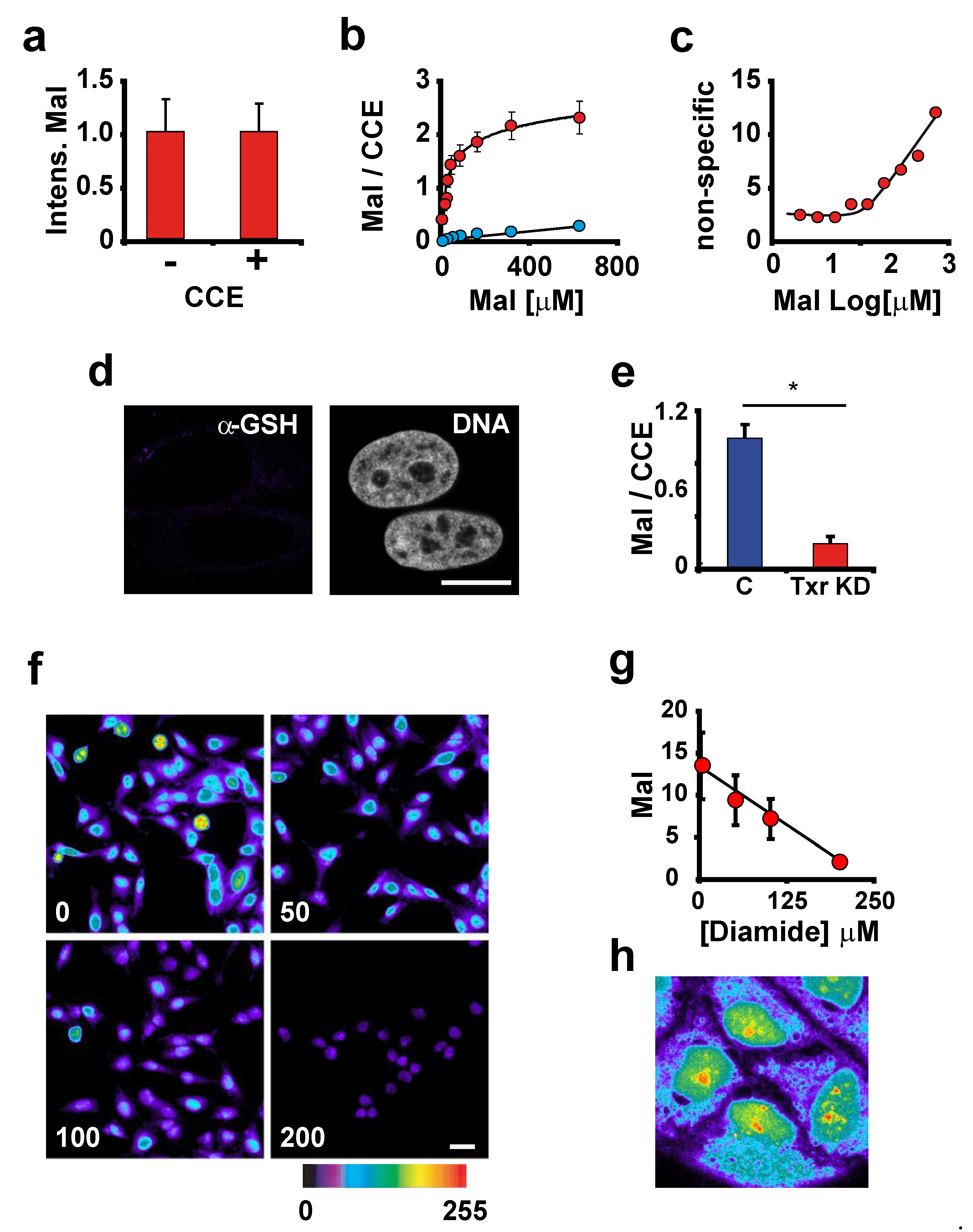
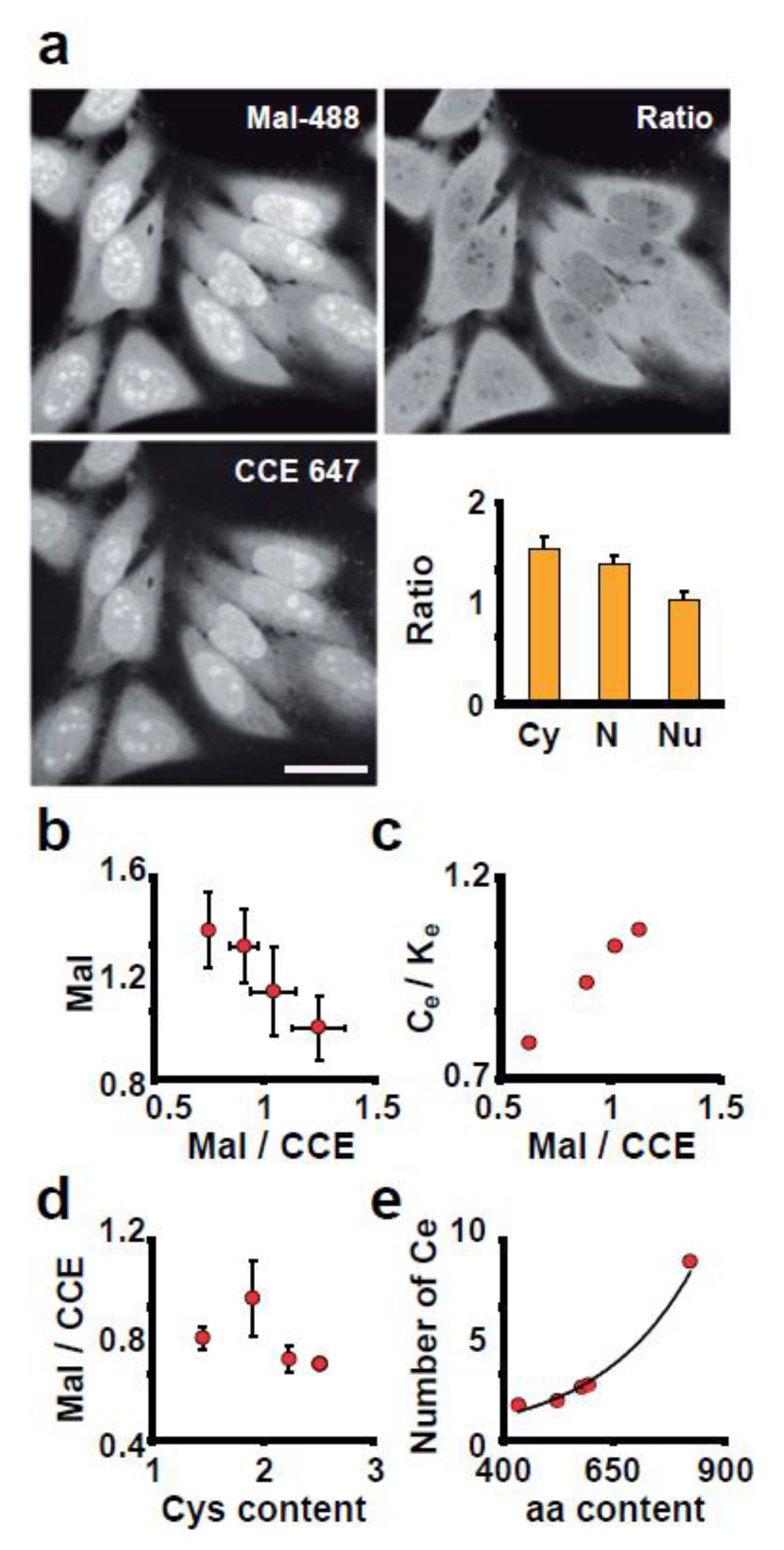
3.2. The Intensity of Maleimide-Alexa Dye Staining Reflects the Amount of Reactive Thiols in Proteins
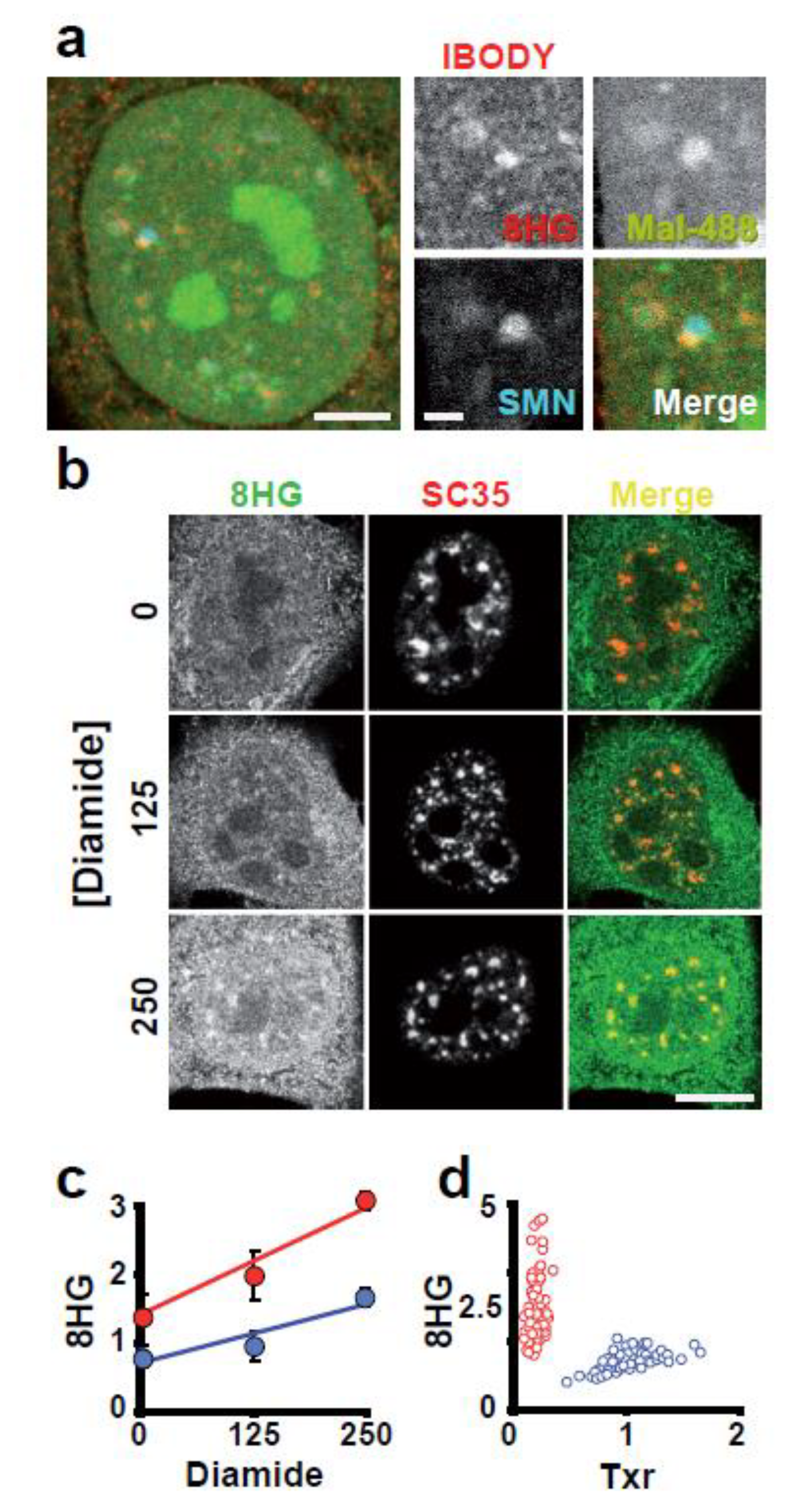
3.3. ROS Sensitivity Is Related to the Distribution of Reactive Thiol Groups in a Compartment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ROS | Reactive Oxygen Species. |
| GSH | Glutathione reduced |
| GSSG | Glutathione oxydized |
| Cys | Cysteine |
| Lys | Lysine |
| Mal-488 | Maleimide-Alexa 488 |
| CCE 647 | Alexa Fluor 647 carboxylic acid succinimidyl ester |
| Trx | Thioredoxin |
| NAC | N-acetyl cysteine |
| TRAIL | TNF-related apoptosis-inducing ligand |
| NEM | N-ethyl maleimide |
| 8HG | 8-hydroxy-Guanosine |
| DRB | 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole |
| DAPI | 4′,6-diamidino-2-phenylindole |
| TRAIL | Tumour necrosis factor-related apoptosis-inducing ligand |
| hnRNP | Heterogeneous nuclear ribonucleoprotein |
| SMN | survival motor neuron protein |
| PML | Promyelocytic leukemia protein |
| PTF | OPT domain |
| OCT1 | Octamer transcription factor 1 |
| UBF | Upstream binding factor |
| BrUTP | Br-Uridine Triphosphate |
| Ce | Exposed Cys |
| Ke | Exposed Lys |
| Nrf2 | Nuclear factor-erythroid 2-related factor 2 |
| PNrf2 | Phospo-Nuclear factor-erythroid 2-related factor 2 |
References
- Toyokuni, S.; Okamoto, K.; Yodoi, J.; Hiai, H. Persistent oxidative stress in cancer. FEBS Lett. 1995, 358, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, J.; Post, J.A. Molecular events associated with reactive oxygen species and cell cycle progression in mammalian cells. Gene 2004, 337, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schafer, F.Q.; Buettner, G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001, 30, 1191–1212. [Google Scholar] [CrossRef] [PubMed]
- LoPachin, R.M.; Gavin, T. Reactions of electrophiles with nucleophilic thiolate sites: Relevance to pathophysiological mechanisms and remediation. Free Radic. Res. 2015, 50, 195–205. [Google Scholar] [CrossRef]
- Go, Y.-M.; Chandler, J.D.; Jones, D.P. The cysteine proteome. Free Radic. Biol. Med. 2015, 84, 227–245. [Google Scholar] [CrossRef]
- Nagahara, N.; Matsumura, T.; Okamoto, R.; Kajihara, Y. Protein cysteine modifications: (1) medical chemistry for proteomics. Curr. Med. Chem. 2009, 16, 4419–4444. [Google Scholar] [CrossRef]
- Hansen, R.E.; Roth, D.; Winther, J.R. Quantifying the global cellular thiol–disulfide status. Proc. Natl. Acad. Sci. USA 2009, 106, 422–427. [Google Scholar] [CrossRef]
- Brandes, N.; Reichmann, D.; Tienson, H.; Leichert, L.I.; Jakob, U. Using Quantitative Redox Proteomics to Dissect the Yeast Redoxome. J. Biol. Chem. 2011, 286, 41893–41903. [Google Scholar] [CrossRef]
- Go, Y.-M.; Jones, D.P. The Redox Proteome. J. Biol. Chem. 2013, 288, 26512–26520. [Google Scholar] [CrossRef]
- Jones, D.P.; Go, Y.-M. Mapping the cysteine proteome: Analysis of redox-sensing thiols. Curr. Opin. Chem. Biol. 2011, 15, 103–112. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Carroll, K.S. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef]
- Xiong, Y.; Uys, J.D.; Tew, K.D.; Townsend, D.M.; Arora, D.; Jain, P.; Singh, N.; Kaur, H.; Bhatla, S.C.; Giustarini, D.; et al. S-Glutathionylation: From Molecular Mechanisms to Health Outcomes. Antioxidants Redox Signal. 2011, 15, 233–270. [Google Scholar] [CrossRef]
- Jones, D.P.; Go, Y.-M. Redox compartmentalization and cellular stress. Diabetes Obes. Metab. 2010, 12, 116–125. [Google Scholar] [CrossRef]
- The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011, 39, D214–D219. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Petersen, B.; Petersen, T.N.; Andersen, P.; Nielsen, M.; Lundegaard, C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 2009, 9, 51. [Google Scholar] [CrossRef]
- das Neves, R.P.; Jones, N.S.; Andreu, L.; Gupta, R.; Enver, T.; Iborra, F.J. Connecting Variability in Global Transcription Rate to Mitochondrial Variability. PLoS Biol. 2010, 8, e1000560. [Google Scholar] [CrossRef]
- Iborra, F.J.; Buckle, V. Wide confocal cytometry: A new approach to study proteomic and structural changes in the cell nucleus during the cell cycle. Histochemistry 2008, 129, 45–53. [Google Scholar] [CrossRef]
- Horowitz, M.P.; Milanese, C.; Di Maio, R.; Hu, X.; Montero, L.M.; Sanders, L.H.; Tapias, V.; Sepe, S.; van Cappellen, W.A.; Burton, E.A.; et al. Single-Cell Redox Imaging Demonstrates a Distinctive Response of Dopaminergic Neurons to Oxidative Insults. Antioxidants Redox Signal. 2011, 15, 855–871. [Google Scholar] [CrossRef]
- Jakoby, W.B.; Griffith, O.W. Sulfur and sulfur amino acids. In Methods in Enzymology; Springer: Berlin/Heidelberg, Germany, 1987; Volume 143, pp. 1–582. [Google Scholar]
- Arnér, E.S.J.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. BioChem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef]
- Kosower, N.S.; Kosower, E.M. Diamide: An oxidant probe for thiols. Methods Enzymol. 1995, 251, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Weerapana, E.; Wang, C.; Simon, G.M.; Richter, F.; Khare, S.; Dillon, M.B.D.; Bachovchin, D.A.; Mowen, K.; Baker, D.; Cravatt, B.F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W.; Park, S.C.; Kim, J.-H.; Kim, I.-K.; Han, K.S.; Kim, K.Y.; Lee, W.B.; Jung, Y.-K.; Kim, S.S. The involvement of oxidative stress in tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in HeLa cells. Cancer Lett. 2002, 182, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Kong, A.-N. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 2009, 48, 91–104. [Google Scholar] [CrossRef]
- Mayer, C.; Grummt, I. Cellular Stress and Nucleolar Function. Cell Cycle 2005, 4, 1036–1038. [Google Scholar] [CrossRef]
- Munnamalai, V.; Suter, D.M. Reactive oxygen species regulate F-actin dynamics in neuronal growth cones and neurite outgrowth. J. Neurochem. 2009, 108, 644–661. [Google Scholar] [CrossRef]
- Grek, C.L.; Zhang, J.; Manevich, Y.; Townsend, D.M.; Tew, K.D. Causes and Consequences of Cysteine S-Glutathionylation. J. Biol. Chem. 2013, 288, 26497–26504. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Rossi, R.; Colombo, G.; Giustarini, D.; Milzani, A. Protein S-glutathionylation: A regulatory device from bacteria to humans. Trends Biochem. Sci. 2009, 34, 85–96. [Google Scholar] [CrossRef]
- Bellomo, G.; Vairetti, M.; Stivala, L.; Mirabelli, F.; Richelmi, P.; Orrenius, S. Demonstration of nuclear compartmentalization of glutathione in hepatocytes. Proc. Natl. Acad. Sci. USA 1992, 89, 4412–4416. [Google Scholar] [CrossRef]
- Voehringer, D.W.; McConkey, D.J.; McDonnell, T.J.; Brisbay, S.; Meyn, R.E. Bcl-2 expression causes redistribution of glutathione to the nucleus. Proc. Natl. Acad. Sci. USA 1998, 95, 2956–2960. [Google Scholar] [CrossRef]
- Meyer, A.J.; Dick, T.P. Fluorescent Protein-Based Redox Probes. Antioxidants Redox Signal. 2010, 13, 621–650. [Google Scholar] [CrossRef]
- Back, P.; De Vos, W.H.; Depuydt, G.G.; Matthijssens, F.; Vanfleteren, J.R.; Braeckman, B.P. Exploring real-time in vivo redox biology of developing and aging Caenorhabditis elegans. Free Radic. Biol. Med. 2012, 52, 850–859. [Google Scholar] [CrossRef]
- Marino, S.M.; Gladyshev, V.N. Cysteine Function Governs Its Conservation and Degeneration and Restricts Its Utilization on Protein Surfaces. J. Mol. Biol. 2010, 404, 902–916. [Google Scholar] [CrossRef]
- Kemp, M.; Go, Y.-M.; Jones, D.P. Nonequilibrium thermodynamics of thiol/disulfide redox systems: A perspective on redox systems biology. Free Radic. Biol. Med. 2008, 44, 921–937. [Google Scholar] [CrossRef]
- Requena, J.R.; Chao, C.C.; Levine, R.L.; Stadtman, E.R. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 69–74. [Google Scholar] [CrossRef]
- Honda, K.; Smith, M.A.; Zhu, X.; Baus, D.; Merrick, W.C.; Tartakoff, A.M.; Hattier, T.; Harris, P.L.; Siedlak, S.L.; Fujioka, H.; et al. Ribosomal RNA in Alzheimer Disease Is Oxidized by Bound Redox-active Iron. J. Biol. Chem. 2005, 280, 20978–20986. [Google Scholar] [CrossRef]
- Forneris, F.; Binda, C.; Battaglioli, E.; Mattevi, A. LSD1: Oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem. Sci. 2008, 33, 181–189. [Google Scholar] [CrossRef]
- Karytinos, A.; Forneris, F.; Profumo, A.; Ciossani, G.; Battaglioli, E.; Binda, C.; Mattevi, A. A Novel Mammalian Flavin-dependent Histone Demethylase. J. Biol. Chem. 2009, 284, 17775–17782. [Google Scholar] [CrossRef]
- Boisvert, F.-M.; Lamond, A.I. p53-Dependent subcellular proteome localization following DNA damage. Proteomics 2010, 10, 4087–4097. [Google Scholar] [CrossRef]
- Boisvert, F.-M.; Van Koningsbruggen, S.; Navascués, J.; Lamond, A. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.-M.; Lamond, A.I. The Nucleolus under Stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Rainwater, R.; Parks, D.; Anderson, M.E.; Tegtmeyer, P.; Mann, K. Role of Cysteine Residues in Regulation of p53 Function. Mol. Cell. Biol. 1995, 15, 3892–3903. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Diaz, C.; Frugier, T.; Melki, J. Spinal muscular atrophy. Semin. Pediatr. Neurol. 2002, 9, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Ottinger, E.; Cho, S.; Dreyfuss, G. Inactivation of the SMN Complex by Oxidative Stress. Mol. Cell 2008, 31, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, H.; Fujikane, A.; Ito, R.; Matsumoto, M.; Nakayama, K.I.; Sekiguchi, M. Human proteins that specifically bind to 8-oxoguanine-containing RNA and their responses to oxidative stress. Biochem. Biophys. Res. Commun. 2010, 403, 220–224. [Google Scholar] [CrossRef]
- Nakielny, S.; Dreyfuss, G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 1996, 134, 1365–1373. [Google Scholar] [CrossRef]
- Saitoh, N.; Spahr, C.S.; Patterson, S.D.; Bubulya, P.; Neuwald, A.F.; Spector, D.L. Proteomic Analysis of Interchromatin Granule Clusters. Mol. Biol. Cell 2004, 15, 3876–3890. [Google Scholar] [CrossRef]
- Li, Z.; Wu, J.; DeLeo, C.J. RNA damage and surveillance under oxidative stress. IUBMB Life 2006, 58, 581–588. [Google Scholar] [CrossRef]
- Lee, K.-M.; Hsu, I.-W.; Tarn, W.-Y. TRAP150 activates pre-mRNA splicing and promotes nuclear mRNA degradation. Nucleic Acids Res. 2010, 38, 3340–3350. [Google Scholar] [CrossRef]
- Markovic, J.; Borrás, C.; Ortega, A.; Sastre, J.; Viña, J.; Pallardó, F.V. Glutathione Is Recruited into the Nucleus in Early Phases of Cell Proliferation. J. Biol. Chem. 2007, 282, 20416–20424. [Google Scholar] [CrossRef]
- Markovic, J.; Mora, N.J.; Broseta, A.M.; Gimeno, A.; De-La-Concepción, N.; Viña, J.; Pallardó, F.V. The Depletion of Nuclear Glutathione Impairs Cell Proliferation in 3t3 Fibroblasts. PLoS ONE 2009, 4, e6413. [Google Scholar] [CrossRef]
- Feldherr, C.; Akin, D. The location of the transport gate in the nuclear pore complex. J. Cell Sci. 1997, 110, 3065–3070. [Google Scholar] [CrossRef]
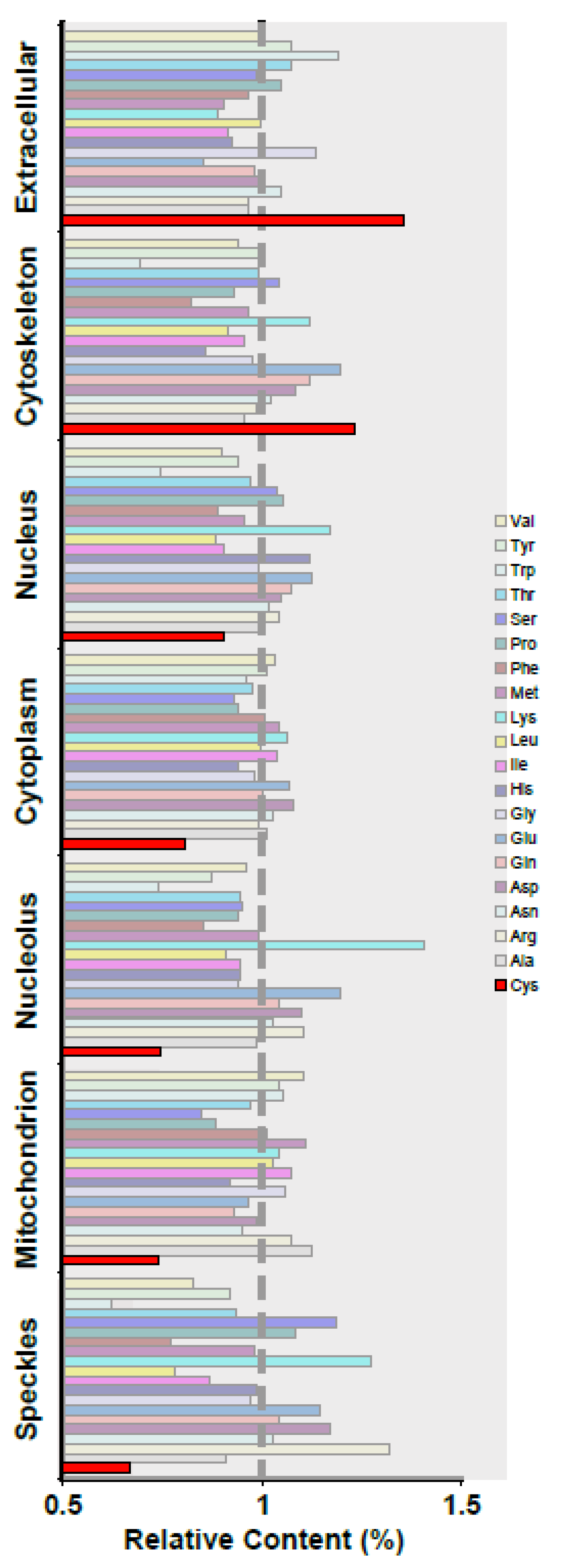



| Cellular Component | GO Accession | Size | aa Average per Protein |
|---|---|---|---|
| SC35 Nuclear Speckles | GO:0016607 | 129 | 720.74 |
| Nucleolus | GO:0005730 | 520 | 581.21 |
| Nucleus | GO:0005634 | 5523 | 624.19 |
| Mitochondria | GO:0005739 | 1353 | 429.08 |
| Cytoskeleton | GO:0005856 | 1593 | 830.95 |
| Cytoplasm | GO:0005737 | 8375 | 595.86 |
| Extracellular | GO:0005576 | 2227 | 521.05 |
| Proteome | Non available | 20,237 | 557.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neves, R.P.d.; Chagoyen, M.; Martinez-Lorente, A.; Iñiguez, C.; Calatrava, A.; Calabuig, J.; Iborra, F.J. Each Cellular Compartment Has a Characteristic Protein Reactive Cysteine Ratio Determining Its Sensitivity to Oxidation. Antioxidants 2023, 12, 1274. https://doi.org/10.3390/antiox12061274
Neves RPd, Chagoyen M, Martinez-Lorente A, Iñiguez C, Calatrava A, Calabuig J, Iborra FJ. Each Cellular Compartment Has a Characteristic Protein Reactive Cysteine Ratio Determining Its Sensitivity to Oxidation. Antioxidants. 2023; 12(6):1274. https://doi.org/10.3390/antiox12061274
Chicago/Turabian StyleNeves, Ricardo Pires das, Mónica Chagoyen, Antonio Martinez-Lorente, Carlos Iñiguez, Ana Calatrava, Juana Calabuig, and Francisco J. Iborra. 2023. "Each Cellular Compartment Has a Characteristic Protein Reactive Cysteine Ratio Determining Its Sensitivity to Oxidation" Antioxidants 12, no. 6: 1274. https://doi.org/10.3390/antiox12061274
APA StyleNeves, R. P. d., Chagoyen, M., Martinez-Lorente, A., Iñiguez, C., Calatrava, A., Calabuig, J., & Iborra, F. J. (2023). Each Cellular Compartment Has a Characteristic Protein Reactive Cysteine Ratio Determining Its Sensitivity to Oxidation. Antioxidants, 12(6), 1274. https://doi.org/10.3390/antiox12061274








