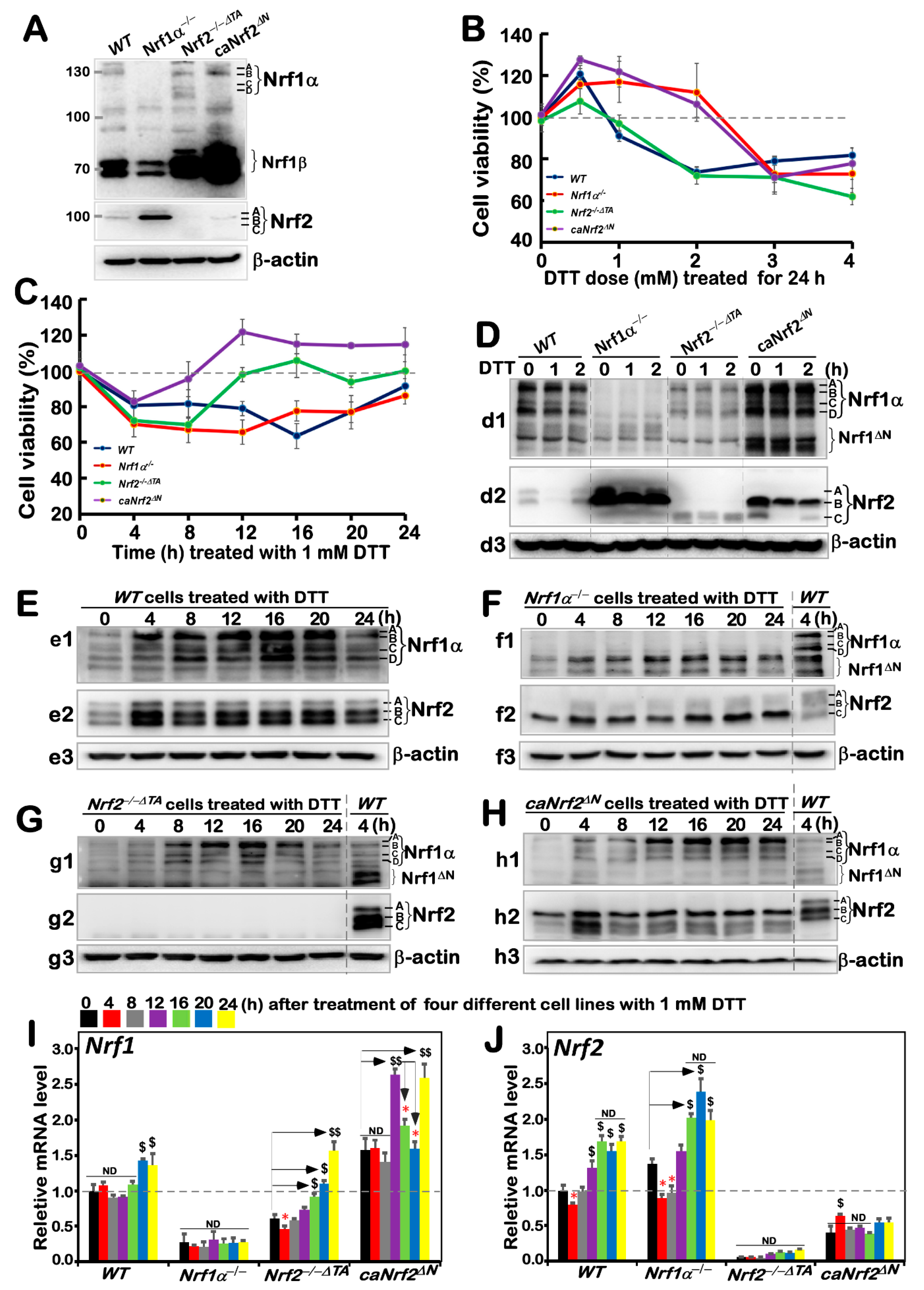
3.1. Distinct Intervening Effects of DTT on Different Genotypic Cell Survival and Nrf1 and Nrf2 Expression
Before this experiment, we verified the characteristic proteins of four cell lines with different genotypes, and confirmed them to be true, as reported previously [
20,
24] (
Figure 1A). Besides, as mentioned by Xiang et al. [
26], there were four major isoforms derived from human Nrf1α: A and B represent its full-length glycoprotein and deglycoprotein, respectively, while C and D denote two distinct lengths of its
N-terminally truncated proteins. All these four Nrf1α-derived isoforms were completely deleted upon specific knockout of Nrf1α in
Nrf1α−/− cells (
Figure 1A), but still present in
WT,
Nrf2−/−ΔTA, and
caNrf2ΔN cell lines. Relatively, the short Nrf1β was obviously decreased in
Nrf1α−/− cells, but significantly augmented in both
Nrf2−/−ΔTA, and
caNrf2ΔN cell lines when compared to its equivalent in
WT cells, indicating that basal abundances of Nrf1 and/or its processing may be monitored by Nrf2, besides itself. It is worth noting that Nrf2 was highly expressed in
Nrf1α−/− cell lines, but completely abolished in
Nrf2−/−ΔTA cells. Overall, it is inferable that loss of Nrf1α or Nrf2 may lead to putative inter-regulatory changes in cognate gene expression profiles among these four cell lines, which were used in subsequent experiments.
Based on redox characteristics, a reducing compound, DTT, was employed to explore the effect of foreign substances on cell viability of different genotypes by MTT assay, so as to evaluate the formation of formazan precipitates with succinate dehydrogenase in the mitochondria of all living cells only. The results revealed that the viability of all cell lines decreased when they were intervened for 24 h by higher concentrations (3 to 4 mM) of DTT (
Figure 1B), although lower concentrations (0.5–2 mM) of DTT caused modestly enhanced survival of
Nrf1α−/− and
caNrf2ΔN cell lines. By contrast, a significant dose-dependent effect of DTT was manifested in
Nrf2−/−ΔTA, similarly to that obtained from
WT cells. Considering such different survival between cells along with the cytotoxicity of DTT, we selected a more appropriate dose at 1 mM of this compound to continue intervention of all experimental cell lines for distinct periods of time from 0 to 24 h (i.e., 0, 4, 8, 12, 16, 20 or 24 h). As shown in
Figure 1C, the viability of all four cell lines was modestly decreased within 8 h of DTT intervention. Such decreased viability of
Nrf1α−/− and
WT cell lines was maintained for 20 h, after which they gradually recovered from intervention. By contrast, the resilience of
Nrf2−/−ΔTA and
caNrf2ΔN cell lines appeared to be strikingly recovered after 8 h to 12 h intervention with DTT, and then reached or exceeded their basal levels, respectively. However, the overall viability of these cell lines was not very significant, and thus, several time periods of DTT intervention were selected with the reference value for follow-up experiments to explore the regulatory differences between Nrf1 and Nrf2 in mediating the cellular reductive stress responses to intervention of this reducing compound.
The short-term (i.e., 0, 1, 2 h) intervention of DTT did not cause a significant difference in Nrf1 abundances in each of the other three cell lines except
Nrf1α−/− (
Figure 1(d1)), while Nrf2 protein abundances were only modestly decreased in each of the other three cell lines except
Nrf2−/−ΔTA (
Figure 1(d2), although Nrf1 and Nrf2 were rather highly expressed in
Nrf1α−/− and
caNrf2ΔN cell lines), when compared with their respective basal levels (obtained at time point 0). Therefore, the time of intervention was extended from 4 h to 24 h, so as to gain the time-dependent effect of DTT on Nrf1 and Nrf2, as well on their targets.
The resulting data are illustrated in
Figure 1E, revealing significant increases in Nrf1α-derived isoforms, particularly its glycoprotein-A, with the extended time of DTT stimulation from 4 h to 20 h, by comparison with
WTt0 as the vehicle control. DTT-inducible expression of Nrf2 was also increased to a considerably higher level after 4 h of this treatment, and so higher expression was maintained to 24 h before stopping experiments. Such changes in Nrf1 and Nrf2 when
WT cells were exposed to DTT for 4 h (i.e.,
WTt4) served as the ensuing parallel experimental controls. Next, examinations of DTT-treated
Nrf1c cells unraveled that albeit this protein itself was completely lost, highly expressed Nrf2 was further enhanced to much higher levels than that obtained from the
WTt4 (
Figure 1F). In
Nrf2−/−ΔTA cells, although basal expression of Nrf1 seemed weaker than that of
WTt4, its DTT-inducible expression was substantially augmented (
Figure 1G), specifically after this chemical intervention from 8 h to 20 h, whereas Nrf2 was totally abolished. In
caNrf2ΔN cells, the expression levels of Nrf1 and Nrf2 were also significantly incremented by DTT in a time-dependent manner (
Figure 1H). All relevant immunoblots were quantitatively analyzed as shown in
supplemental Figure S1.
Further examinations by RT-qPCR showed that DTT intervention of
WT cells only caused a marginally slower increase in Nrf1 mRNA expression after 20 h stimulation, while Nrf2 mRNA levels were significantly incremented after only 12 h DTT treatment when compared to its basal levels (
Figure 1I,J). In
Nrf1α−/− cells, Nrf2 mRNA expression was also further augmented, although Nrf1 expression was largely abrogated. By contrast, although basal Nrf1 expression in
Nrf2−/−ΔTA cells was prevented, its DTT-stimulated expression was still enhanced after 16 h of this treatment (
Figure 1I). However, much to our surprise, we found that although putative constitutive active protein of Nrf2 was present in
caNrf2ΔN cells, its mRNA expression levels were unaffected by DTT (
Figure 1J), whereas conversely, a striking elevation of Nrf1 mRNA expression occurred after 12 h treatment. Overall, these findings demonstrate that a potential inter-regulatory relationship exists between Nrf1 and Nrf2, albeit under DTT-leading reductive stress conditions.
3.2. Differential Expression of Nrf1/2-Mediated Redox Responsive Genes Induced by DTT in Distinct Genotypic Cells
According to the results of transcriptome sequencing analysis of four genotypes of cells by Qiu et al., Nrf1 and Nrf2 have regulatory differences in the expression of GCLC, GCLM, GSR and other oxidative stress related genes [
24]. Additionally, our previous study had shown that tBHQ, as a small molecule antioxidant, stimulates mRNA expression of some
ARE-dependent genes downstream of Nrf1 and Nrf2 (such as
GCLM,
GCLC,
HO1, etc.). Here, we examined whether these ARE-driven genes were affected by intervention of DTT as a reducing agent of sulfhydrylation. As anticipated, RT-qPCR results showed that significant time-dependent increases in DTT-inducible mRNA expression of
GCLM and
GCLC encoding two subunits of GSH synthesis rate-limiting enzyme are of crucial importance in the redox process [
27] in DTT-treated
WT cells (
Figure 2A,B). Knockout of
Nrf1α−/− cells caused a faster significant increase in DTT-inducible expression of
GCLM from 4 h to its maximum at 16 h, which was then maintained to 24 h before stopping experiments (
Figure 2A), while only a modest increase in
GCLC expression was apparently lagged, occurring from 16 h stimulation by DTT (
Figure 2B). By sharp contrast,
Nrf2−/−ΔTA cells only displayed a marginal increase in expression of
GCLM, whereas
GCLC was unaffected by DTT (
Figure 2A,B). Conversely, such DTT-induced expression of
GCLM was completely abolished in
caNrf2ΔN cells, although its basal expression was enhanced, while significantly inducible expression of
GCLC was greatly lagged until 20 h to 24 h of treatment. Collectively, these imply that differential expression of
GCLM and
GCLC is monitored by inter-regulated Nrf1 and Nrf2 in a time-dependent fashion.
Taking
WT (at
t0) as the control, basal protein abundances of GCLC and GCLM were upregulated in
Nrf1α−/− and
caNrf2ΔN cell lines, but significantly down-regulated in
Nrf2−/−ΔTA cells (
Figure 2(c1,c2)). However, no significant changes or even decreases in DTT-inducible GCLC and GCLM proteins for shorter terms of 1 h to 2 h were determined in all examined cell lines (
Figure 2C and
Figure S2). Upon DTT intervention of
WT cells for the longer durations, significant changes in GCLC and GCLM proteins were observed to increment with the increasing time of administration (
Figure 2(d1,d2)). After knockout of
Nrf1α−/−, GCLM changed rather significantly compared to GCLC, with a time-dependent gradual increase (
Figure 2(e1,e2)); knockout of
Nrf2−/−ΔTA caused
GCLM expression to become fainter, while GCLC appeared to be unaltered in comparison to the
WT (at
t4h) control (
Figure 2(f1,f2)). Additionally,
caNrf2ΔN cells had no further stimulated increases in GCLC and GCLM in response to DTT, although their basal levels were higher than that of the
WT (at
t4h) (
Figure 2(g1,g2)).
As mentioned herein, the oxidized glutathione disulfide (GSSG) can be reduced to GSH form by glutathione-disulfide reductase (GSR, a central enzyme of antioxidant defense), in this redox cycle, where glutathione peroxidase 1 (GPX1) can also achieve the purpose of oxidative detoxification in cells by reducing some peroxides. When compared with
WT (at
t0) cells, basal mRNA levels of
GSR and
GPX1 were evidently up-expressed in
Nrf1α−/− and
caNrf2ΔN cell lines, but rather substantially down-expressed in
Nrf2−/−ΔTA cells (
Figure 2H,I). After DTT treatment, transcriptional expression of
GSR in
WT cells was significantly up-regulated in a time-dependent manner from 12 h, whereas inducible expression of
GPX1 was modestly upregulated by this chemical. Intriguingly, a DTT-inducible decrease, rather than increase, of
GSR occurred only with 12 h treatment of
Nrf1α−/− cells (
Figure 2H), as accompanied by obvious down-regulation of
GPX1 (
Figure 2I).
Nrf2−/−ΔTA cells only displayed a lagged increase in mRNA expression of
GSR at 24 h stimulation (
Figure 2H), but biphasic induction of
GPX1 by DTT occurred respectively at 4 h and 20 h, although its basal levels were lowered (
Figure 2I). Conversely,
caNrf2ΔN cells exhibited an obvious time-dependent DTT-inducible decrease in
GSR, along with lagged induction of
GPX1 by this treatment for 20 h to 24 h, which was rather significantly higher than its basal levels (
Figure 2I). Further examinations of GSR and GPX1 proteins revealed no significant changes in their inducible expression upon short-term intervention of all examined cells by DTT, although their basal levels were highly up-expressed in
Nrf1α−/− and
caNrf2ΔN cell lines, but down-regulated in
Nrf2−/−ΔTA cells in comparison to the
WT (at
t0) control (
Figure 2(c3,c4) and
Figure S2). Continued detection of the longer-term DTT intervening effects on
WT cells revealed almost no significantly inducible changes in GSR and GPX1 abundance, except from a marginal induction being lagged at 24 h (
Figure 2(d3,d4)). Similarly to the control
WT (at
t4), almost no induction of GSR by DTT was observed in
Nrf1α−/− cells, but as compared with enhanced expression of GPX1 in a time-dependent manner (
Figure 2(e3,e4)). In
Nrf2−/−ΔTA cells, considerably fainter expression of GSR was not induced by DTT, while GPX1 was also not triggered by this drug (
Figure 2(f3,f4)). However,
caNrf2ΔN cells displayed a modestly stimulated expression of GPX1, but not GSR in response to DTT (
Figure 2(g3,g4)). Together, such distinct expression profiles of GSR and GPX1 at mRNA and protein levels may be attributable to coordinated inter-regulation by Nrf1 and Nrf2.
The expression of downstream genes
NQO1 and
HO-1 closely related to
Nrf1 and
Nrf2 was also increased with time-dependent induction of DTT in
WT cells (
Figure 2J,K). When compared with the
WTt0 control, basal mRNA expression levels of
NQO1 and
HO-1 were upregulated in
Nrf1α−/− cells, but down-regulated in
Nrf2−/−ΔTA cells, along with almost no changes in
caNrf2ΔN cells. The inducible expression of
HO-1 was further augmented by DTT stimulation of
Nrf1α−/− cells for 12 h to 24 h (
Figure 2K), whereas
NQO1 was largely unaffected by this chemical, except for a marginal increase lagged at 20 h (
Figure 2J). Intriguingly,
NQO1 and
HO-1 were roughly unaltered or down-regulated by DTT, except for a slightly stimulated expression lagged at 24 h of this treatment in
Nrf2−/−ΔTA and
caNrf2ΔN cell lines. Further investigation of NQO1 and HO-1 revealed a largely similar trend of changes in their protein expression to that of mRNAs. In comparison to
WT (at
t0), basal NQO1 and HO1 expression abundances were enhanced in
Nrf1α−/− cells, but suppressed in either
Nrf2−/−ΔTA or
caNrf2ΔN cells (
Figure 2(c5,c6)). Short-term DTT intervention of examined cells for 1 h to 2 h also revealed no significant changes or even a modestly decreased trend in NQO1 and HO1 proteins. However, long-term intervention of
WT cells with DTT from 4 h to 24 h led to gradually incremented abundances of NQO1 and HO-1 in a time-dependent fashion (
Figure 2(d4,d6)). Further comparison with the
WT (at
t4) control indicated that DTT intervention of
Nrf1α−/− cells for 4 h to 24 h also induced a gradual increased expression trend for NQO1, while HO-1 reached a relative inducible expression peak at 12 h and then gradually weakened (
Figure 2(e5,e6)). In
Nrf2−/−ΔTA cells, NQO1 and HO-1 expression levels were not only weaker than those in
WT (at
t4), but also not induced by DTT (
Figure 2(f5,f6)). Similarly,
caNrf2ΔN cells also manifested no significant changes in NQO1 and HO-1 in response to DTT, with the exception of only slight HO-1 induction at 24 h (
Figure 2(g5,g6)).
Herein, we also examined the expression of
TALDO (encoding a key enzyme in the non-oxidative pentose phosphate pathway to yield NADPH so as to maintain a reduced state of glutathione, thus protecting cells from oxygen free radicals [
28]),
TKT (encoding thiamine dependent enzyme to guide excess phosphate sugar to glycolysis in the pentose phosphate pathway), and
MT1E and
MT2 (two members the metal sulfur family that act as antioxidants in the steady-state control of metals in cells and prevent the production of hydroxyl radicals [
29]). As shown, biphasic changes in
MT1E mRNA expression were observed in
WT cells, which was first decreased, then gradually recovered and even increased as the time of DTT intervention was extended to 24 h of its maximum response. Loss of
Nrf1α−/− led to a complete abolishment of basal and DTT-inducible mRNA expression of
MT1E (
Figure 2L). Intriguingly, similar results were obtained from
caNrf2ΔN cells. Conversely, loss of
Nrf2−/−ΔTA led to a substantial increase in basal
MT1E expression, but its DTT-inducible expression changes were triphasic, which was first inhibited from 4 h to 12 h, then recovered at 16 h and induced to the maximum at 20 h, but finally returned to its basal level (
Figure 2L). For
MT2, only a lagged DTT-stimulated increase occurred at 24 h treatment of
WT cells (
Figure 2M). When compared with the
WTt0 control, basal MT2 expression was significantly up-regulated in
Nrf1α−/− and
caNrf2ΔN cell lines. A biphasic change in its DTT-inducible expression was exhibited in
Nrf1α−/− cells. Similarly lagged DTT-triggering expression of MT2 was also observed in
caNrf2ΔN cells. Conversely,
Nrf2−/−ΔTA cells gave a marginal increase in basal MT2 levels; its DTT-stimulated expression was completely abolished (
Figure 2M). Further examination revealed that basal mRNA expression levels of
TALDO and
TKT were up-regulated in
Nrf1α−/− and
caNrf2ΔN, rather than
Nrf2−/−ΔTA, cell lines, when compared with the
WTt0 control (
Figure 2N,O). Similar changes in
TALDO protein levels were obtained (
Figure 2C). Interestingly, significant increases in
TALDO and
TKT expression levels were stimulated by DTT in
WT cells. Knockout of
Nrf1α−/− or
Nrf2−/−ΔTA still gave rise to lagged induction of
TALDO and
TKT within 16 h to 24 h after DTT intervention (
Figure 2N,O), while
caNrf2ΔN displayed a biphasic change in
TALDO and TKT expression levels that was first decreased, and then recovered and increased during DTT stimulation with an inducible maximum occurring at 24 h (
Figure 2N,O). However, little or no effect of DTT was observed on TALDO protein expression in all examined cells (
Figure 2C–G).
3.3. Differential Requirements of Nrf1 and Nrf2 for the ER Stress-Responsive Genes Stimulated by DTT
It was reported that Nrf2 is significantly up-regulated by aggregated β-amyloid-mediated ER stress and activated by PERK as a canonical ER stress sensor [
30,
31]. Our previous work had shown differential and integral roles of Nrf1 and Nrf2 in mediating the unfolded protein response (i.e., UPR
ER) to the classic ER stressor tunicamycin [
20]. Herein, we examined whether Nrf1 and Nrf2 are required for monitoring putative ER stress response to DTT, which interferes with disulfide bond formation during protein folding towards maturation. Thus, it was reasoned that one of the first targets of oxidative protein folding attacked by DTT should be protein disulfide isomerase (PDI, which can also act as a reductase to cut those protein disulfide bonds attached to the cell surface, aside from an ER chaperone to inhibit misfolded protein aggregation [
32,
33]). As expected, DTT-stimulated mRNA expression levels of
PDI were up-regulated in all four examined cell lines, even upon loss of
Nrf1α−/− or
Nrf2−/−ΔTA, but its basal enhancement occurred in
Nrf1α−/− and
caNrf2ΔN, rather than
Nrf2−/−ΔTA, cell lines (
Figure 3A). Such a rebound effect on transcriptional expression of
PDI is likely controlled by a feedback loop coordinated with Nrf1 and Nrf2. However, its protein expression abundances were significantly incremented by DTT in
Nrf1α−/− cells, but only modestly enhanced in the other three cell lines (
Figure 3J–M). This implies the possibly enhanced stability of PDI may be attributable to
Nrf1α−/−-impaired proteasomal degradation, particularly under reductive stress conditions.
Next, to our surprise, we found that mRNA expression levels of GRP78 (a pivotal partner with three ER sensors, PERK, IRE1 and ATF6) were rapidly increased to its maximum induction by 4 h intervention of DTT in all four examined cell lines, and then gradually decreased to its basal levels or to rather lower extents, as stimulation time was extended to 24 h (
Figure 3B). Similar biphasic changes in GRP78 protein abundance were also determined in these four cell lines (
Figure 3(j2–m2)). However, it is notable that the extents of DTT-inducible mRNA and protein expression in
Nrf1α−/− cells were rather lower than those obtained from
WT,
Nrf2−/−ΔTA and
caNrf2ΔN, cell lines, but almost no differences in induction of GRP78 were observed between
Nrf2−/−ΔTA and
caNrf2ΔN cell lines. This indicates that Nrf1, rather than Nrf2, is required for bidirectional regulation of GRP78 at distinct levels in response to DTT.
By further examination of three ER stress responsive genes, it was revealed that a modest bimodal induction of
PERK by DTT occurred early at 4 h and later after 16 h of this chemical treatment, respectively, in
WT cells, and such bimodality was further augmented in
Nrf2−/−ΔTA cells (
Figure 3C), but substantially blunted in
Nrf1α−/− cells. Of note, the early peak was abolished by knockout of
Nrf1α, while the latter peak was abrogated and even suppressed by
caNrf2ΔN. This implies that Nrf1 and Nrf2 bi-directionally positively and negatively regulate expression of
PERK, particularly its lagged induction by DTT, respectively. In
WT cells, phosphorylated protein of p-PERK was significantly stimulated by DTT at 4 h and then decreased to rather lower extents (
Figure 3(j4)). A similar, but modest, induction pattern of p-PERK by DTT was manifested in
caNrf2ΔN cells (
Figure 3(m4)), but this induction seemed to be abolished by knockout of
Nrf1α−/− (
Figure 3(k4)). Conversely, in
Nrf2−/−ΔTA cells, DTT-inducible expression of p-PERK was gradually incremented from 12 h to 24 h of its maximum (
Figure 3(l4)). In addition, no significant changes in total PERK were observed in all four examined cell lines, except for partial attenuation by knockout of
Nrf1α−/− within an indicated period of time (
Figure 3J–M and
Figure S3). Collectively, induction of the PERK signaling by DTT is also positively and negatively monitored by Nrf1 and Nrf2, respectively.
The downstream eIF2α-ATF4-CHOP of PERK signaling was also examined herein. The results showed only marginal induction of
eIF2α and
CHOP at mRNA expression levels after 16 h or early at 4 h of DTT intervention, respectively, along with no induction or even decreases in
ATF4 in
WT cells (
Figure 3D–F). By contrast, basal and/or DTT-inducible
eIF2α expression levels were repressed by
Nrf1α−/−, but enhanced by
Nrf2−/−ΔTA, even though unaffected by
caNrf2ΔN (
Figure 3D). In addition, the phosphorylated eIF2α expression was modestly induced by DTT in
Nrf2−/−ΔTA cells, but no marked changes were observed in the other three cell lines (
Figure 3J–M and
Figure S3). These indicate that Nrf1 and Nrf2 regulate eIF2α in a way similar to their monitoring of PERK expression. However, DTT intervention led to significant down-regulation of
ATF4 in
Nrf1α−/− and
caNrf2ΔN cell lines when compared with
WT cells, although upregulation of its basal mRNA expression levels in
Nrf1α−/− and
caNrf2ΔN cells occurred with their suppression in
Nrf2−/−ΔTA cells (
Figure 3E). However, no striking changes in ATF4 protein levels were observed in all examined cells (
Figure 3J–M). Hence, it is inferable that Nrf2 may be required for DTT-stimulated trans-repression of
ATF4. Moreover, only early induction of
CHOP by DTT occurred, to a lower degree, at 4 h treatment of
WT cells and was further amplified in
Nrf2−/−ΔTA and
caNrf2ΔN, but not in
Nrf1α−/− cell lines, although its basal expression was also upregulated in
Nrf1α−/− cells (
Figure 3F). Additionally, no significant changes in CHOP protein levels were determined in all four cell lines (
Figure 3J–M). This implies that Nrf1 is likely required for early induction of
CHOP by DTT. Overall, these findings indicate differential and integral roles of Nrf1 and Nrf2 in monitoring the PERK signaling to its downstream eIF2α-ATF4-CHOP pathway.
A bimodal of the IRE1-XBP1 signaling at their mRNA levels was also induced by DTT in
WT cells. Such induction of
IRE1 was significantly enhanced in
Nrf2−/−ΔTA cells, though its basal levels were down-regulated (
Figure 3G). The early peak of
IRE1 induction by DTT was abrogated or suppressed in
Nrf1α−/− or
caNrf2ΔN cells, but its later peak was unaffected or augmented in the two cell lines, respectively. The marked early peak of
XBP1 induced by DTT was observed in
WT cells, decreased by
Nrf2−/−ΔTA and abolished by
Nrf1α−/− or
caNrf2ΔN (
Figure 3H), whereas the secondary peak was also abolished by
caNrf2ΔN, but unaffected by
Nrf1α−/− or Nrf2−/−ΔTA. Furthermore, time-dependent increments of
ATF6 mRNA expression occurred from 12 h to 24 h in DTT-treated
WT,
Nrf1α−/− or
Nrf2−/−ΔTA, but not
caNrf2ΔN cells (
Figure 3I). In addition, no obvious changes in IRE1, p-IRE1 and XBP1 proteins were observed in all four examined cell lines, but ATF6 protein levels were, to different extents, enhanced by DTT in
WT,
Nrf1α−/−,
caNrf2ΔN, rather than
Nrf2−/−ΔTA, cell lines (
Figure 3J–M). Taken together, these findings demonstrate that Nrf1 and Nrf2 differentially regulate the ER-stress responsive genes to DTT.
3.4. Distinct Intracellular Redox Changes among Different Genotypic Cell Lines in Response to DTT
As a collective term, ROS include superoxide anion, hydrogen peroxide, and all other oxygenated active substances, but are still hard to be accurately detected, because they have strong oxidation activity to be exerted for a short retention time while they are easy to be removed by antioxidants. Therefore, visualization of ROS by fluorescence microscopy and its quantification by flow cytometry are usually used to detect their changing status evaluated by the green fluorescence raised from the DCFH dye reaction with intracellular ROS, to assess the difference in antioxidant capacity [
34]. The results showed that basal ROS levels in
Nrf1α−/− cells were obviously higher than that of
WTt0, while a slight increase in the yield of ROS was observed after 4 h stimulation by DTT, before being decreased to lower extents than its basal levels (
Figure 4A–C). Such a slight DTT-stimulated rise in ROS was also observed in
WT cells before being decreased and then recovered to its basal levels. By contrast, basal ROS levels were also apparently increased in
Nrf2−/−ΔTA cells, but this status seemed to be unaffected by DTT intervention (
Figure 4A–C). Intriguingly, no differences in basal and DTT-stimulated ROS levels in
caNrf2ΔN cells were determined by comparison to
WT controls. Besides, similar changes in the intensity of fluorescence arising from intracellular DCFH-DA dye observed by microscopy were also fully consistent with the results as described above (
Figure 4D). Altogether, these findings demonstrate that although both Nrf1 and Nrf2 are responsible for endogenous antioxidant cytoprotection, Nrf1 rather than Nrf2 is required for DTT-stimulated antioxidant defense response.
Further experiments revealed that an intrinsic significant augmentation in the proportion of GSSG to GSH was de facto in
Nrf1α−/− cells, whereas
Nrf2−/−ΔTA cells only had a modestly enhanced ratio of GSSG to GSH, when compared with the
WTt0 control (
Figure 4E). Upon stimulation of DTT for 4 h, an inducible elevated rate of GSSG to GSH was examined only in
WT and
Nrf2−/−ΔTA, but not
Nrf1α−/− or caNrf2ΔN cells, which seemed to be the highly endogenous level obtained from
Nrf1α−/− cells. When stimulation with DTT extended to 24 h, such DTT-stimulated elevation descended closely to basal levels (
Figure 4E). Notably, DTT intervention of
Nrf1α−/− cells for 4 h to 24 h resulted in substantially decreased rates of GSSG to GSH, but no changes in the GSSG to GSH ratio were observed in
caNrf2ΔN cells. From these, it is inferable to be attributable to aberrant accumulation of hyperactive Nrf2 in
Nrf1α−/− cells insomuch as to reinforce its antioxidant and detoxifying cytoprotection against DTT, whereas
caNrf2ΔN cells with a genetic deletion of the
N-terminal Keap1-binding domain lost their powerful response to the redox-sensing by Keap1.
Further experimental evidence was also obtained from flow cytometry analysis of cell apoptosis, revealing the lowest apoptosis level among untreated
WT cells, while a relatively higher number of
Nrf1α−/− cells underwent apoptosis (
Figure 4F,G). Almost similar apoptosis of
Nrf2−/−ΔTA and
caNrf2ΔN cells was close to that of
WT cells, but much lower than that of
Nrf1α−/− cells (
Figure 4F). However, DTT stimulation caused significant decreases in apoptosis of
Nrf1α−/− cells, but slightly increased apoptosis of
Nrf2−/−ΔTA or
caNrf2ΔN cells at 24 h after this chemical treatment, along with no changes in
caNrf2ΔN cells (
Figure 4G). This demonstrates that accumulated Nrf2 in
Nrf1α−/− cells still exerted its intrinsic cytoprotective effect against DTT-induced apoptosis, but this effect was lost in both
Nrf2−/−ΔTA and
caNrf2ΔN cell lines (the former
Nrf2−/−ΔTA lacks its transactivation domain to regulate its target genes, while the latter
caNrf2ΔN lacks its responsive interaction with the redox-sensing Keap1).
3.5. The Redox Status of Cys342 and Cys640 in Nrf1 Is Required for Their Protein Stability and Trans-Activity
Our previous studies showed that Nrf1 undergoes a variety of post-translational modifications, such as glycosylation, deglycosylation, ubiquitination and phosphorylation, as well as selectively proteolytic processing [
13]. Here, we investigated which cysteine (Cys) residues within Nrf1 can directly sense the reductive stressor DTT (that can reduce protein disulfide bond (-
S-
S-) to sulfhydryl (-
SH) and also oxidize itself into six-membered ring, to assist in maintaining protein function [
35]). A schematic shows mutagenesis mapping of four Cys residues of Nrf1 into serines (i.e., C342S, C521S, C533S and C640S) within its NST, Neh6L and bZIP domains, respectively (
Figure 5A). Next, the pulse-chase experiments revealed that a considerable portion of the full-length glycoproteins of two mutants, Nrf1
C342S and Nrf1
C640S, were rapidly converted into their deglycoproteins and then proteolytically processed to disappear in a faster manner than those equivalents arising from wild-type Nrf1 (
Figure 5B). Protein stability was evaluated by Western blotting to measure the half-lives of Nrf1
C342S, Nrf1
C640S and wild-type Nrf1, which were calculated for their glycoprotein turnover to be 0.36 h (21.6 min), 0.35 h (21 min) and 1.27 h (76.2 min) (
Figure 5C), and for their deglycoprotein turnover to be 0.45 h (27 min), 0.66 h (39.6 min) and 1.58 h (94.8 min) (
Figure 5D), respectively, after treatment with CHX (to inhibit the nascent polypeptide synthesis). By contrast, the stability of another two mutants Nrf1
C521S and Nrf1
C533S were only marginally affected, when compared with that of wild-type Nrf1 (
Figure 5B–D).
Further examination of a bi-Cys mutant Nrf1
C342/640S revealed that the half-lives of its glycoprotein and deglycoprotein turnover were calculated to be 0.39 h (23.4 min) and 1.10 h (66.0 min), respectively, after CHX treatment (
Figure 5E,F). Upon addition of proteasomal inhibitor MG132 plus CHX, the half-life of this mutant glycoprotein was only modestly extended to 1.13 h (67.8 min), just because it had to be deglycosylated during the pulse-chase experiments. Rather, the half-life of this mutant deglycoprotein was significantly extended to over 4 h before stopping this experiment (
Figure 5E,F). This implies that this deglycoprotein turnover is quality-controlled by proteasome-mediated degradation pathway. Interestingly, transactivation activity of
ARE-driven luciferase reporter gene regulated by Nrf1 was significantly inhibited by its mutants Nrf1
C342S, Nrf1
C640S or Nrf1
C342/640S (
Figure 5G). Therefore, it is inferable that the redox state of both Cys342 and Cys640 residues in Nrf1 could be required for its protein stability and transcriptional activity.
Next, the effect of DTT-induced redox stress on Nrf1 stability was further determined. The results showed that DTT enhanced accumulation of all the endogenous Nrf1-drived proteins, and they were further accumulated by MG132 plus DTT (
Figure 5(h1)). However, the half-lives of Nrf1 glycoprotein (i.e., Nrf1-G), deglycoprotein (i.e., Nrf1-D) and its processed protein (i.e., Nrf1-P) were measured to be 0.28 h (16.8 min), 1.30 h (78 min), and 1.32 h (79.2 min), respectively, after DTT co-treatment of cells with CHX (
Figure 5(h2–h4)). Upon addition of MG132 to DTT/CHX-treated cells, the half-life of Nrf1-G was slightly extended to 0.60 h (36 min), whereas the half-lives of Nrf1-D and Nrf1-P were strikingly prolonged to over 4 h after stopping experiments. Further determination of Nrf1 protein turnover was carried out by Western blotting of ectopically expressed isoforms of this CNC-bZIP factor and its bi-Cys mutant Nrf1
C342/640S, which were resolved by gradient LDS-NuPAGE gels containing 4–12% polyacrylamides (
Figure S4). The results revealed that abundance of Nrf1 was enhanced by DTT (
Figure S4A). In the presence of DTT, the half-lives of Nrf1-G, -D and -P were calculated to be 0.18 h (10.8 min), 1.06 h (63.6 min) and 0.74 h (44.4 min), respectively, after CHX treatment (
Figure S4(a2–a4)). By contrast, Nrf1
C342/640S appeared to be unaffected by this chemical (
Figure S4(b1)), with distinct half-lives of its isoforms that were slightly changed to be 0.24 h (14.4 min), 0.64 h (38.4 min) and 0.57 h (34.2 min), respectively after co-treatment of DTT with CHX (
Figure S4(b2–b4)). Notably, the glycoprotein half-life of Nrf1 or Nrf1
C342/640S was only modestly extended by addition of MG132, to 0.30 h (18 min) or 0.50 h (30 min), respectively, but their deglycoproteins and proteolytic proteins were all significantly prolonged to over 4 h. Taken together, these data indicate that the redox state of Cys342 and Cys640 residues in Nrf1 is not only required for its protein stability, but that both may also serve as a redox sensor for the DTT stressor.
3.6. Biphasic Effects of DTT on Transcriptional Expression of Nrf1-Target Proteasomal Genes
It was previously reported that Nrf1, rather than Nrf2, plays an essential role in controlling transcriptional expression of all proteasomal subunit genes [
36]. Such expression of the proteasomal genes regulated by Nrf1 is required for the ER-associated degradation (ERAD), as accompanied by induction of three classical response pathways driven by the ER stress-sensing genes (i.e.,
PERK,
IRE1 and
ATF6). Therefore, we explored whether DTT-stimulated Nrf1 and Nrf2 are also required for the expression of key proteasomal (
PSM) genes along with ER signaling networks. The RT-qPCR results showed significant decreases in basal mRNA expression levels of all three examined genes
PSMB5,
PSMB6 and
PSMB7 in
Nrf1α−/− cells (
Figure 6A), while basal
PSMB5 and
PSMB7 expression levels were also partially decreased in
Nrf2−/−ΔTA or
caNrf2ΔN cells, when compared with
WTt0 controls. This implies that except for Nrf1, Nrf2 is also partially involved in regulating transcription of some PSM genes (e.g.,
PSMB5 and
PSMB7) through its
N-terminal Keap1-binding domain, in addition to its transactivation domain.
Interestingly, DTT stimulation of
Nrf1α−/− cells (with accumulation of hyperactive Nrf2) caused evident increases in
PSMB5,
PSMB6 and
PSMB7 at mRNA and protein expression levels close to basal
WTt0 controls (
Figure 6A or
Figure 6B,(b5–b7)). This indicates such up-regulation of proteasomal genes by DTT may occur through Nrf1-independent and/or Nrf2-dependent pathways. In
WT cells, DTT caused partial decreases in
PSMB5 and
PSMB7, and also biphasic changes (i.e., decreased early, then recovered and even elevated) in
PSMB6 (
Figure 6A), but their protein abundances were almost unaltered (
Figure 6(b1–b3)). Similarly, DTT-inducible biphasic expression of all three examined genes was also observed in
Nrf2−/−ΔTA or
caNrf2ΔN cell lines, with no obvious changes in their protein levels. Together, these findings suggest transcriptional expression of proteasomal genes is also tightly regulated by other not-yet-identified factors (e.g., Bach1), along with Nrf1 and Nrf2, particularly under DTT-stimulated stress conditions, although the basal proteasomal expression is predominantly governed by Nrf1, as well partially by Nrf2.