Hop Tannins as Multifunctional Tyrosinase Inhibitor: Structure Characterization, Inhibition Activity, and Mechanism †
Abstract
1. Introduction
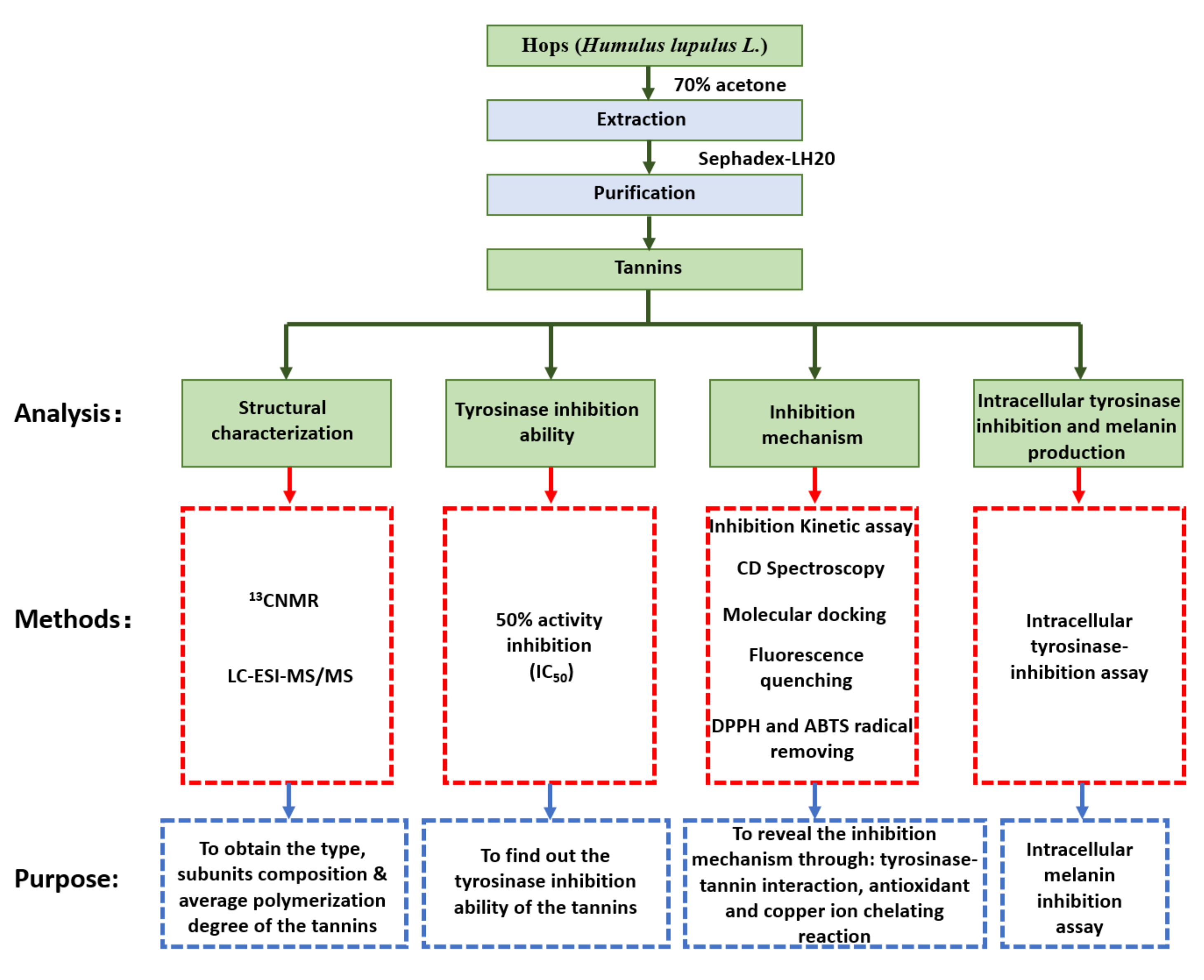
2. Materials and Methods
2.1. Preparation of Hop Tannins
2.2. 13CNMR Analysis of Hops Tannin
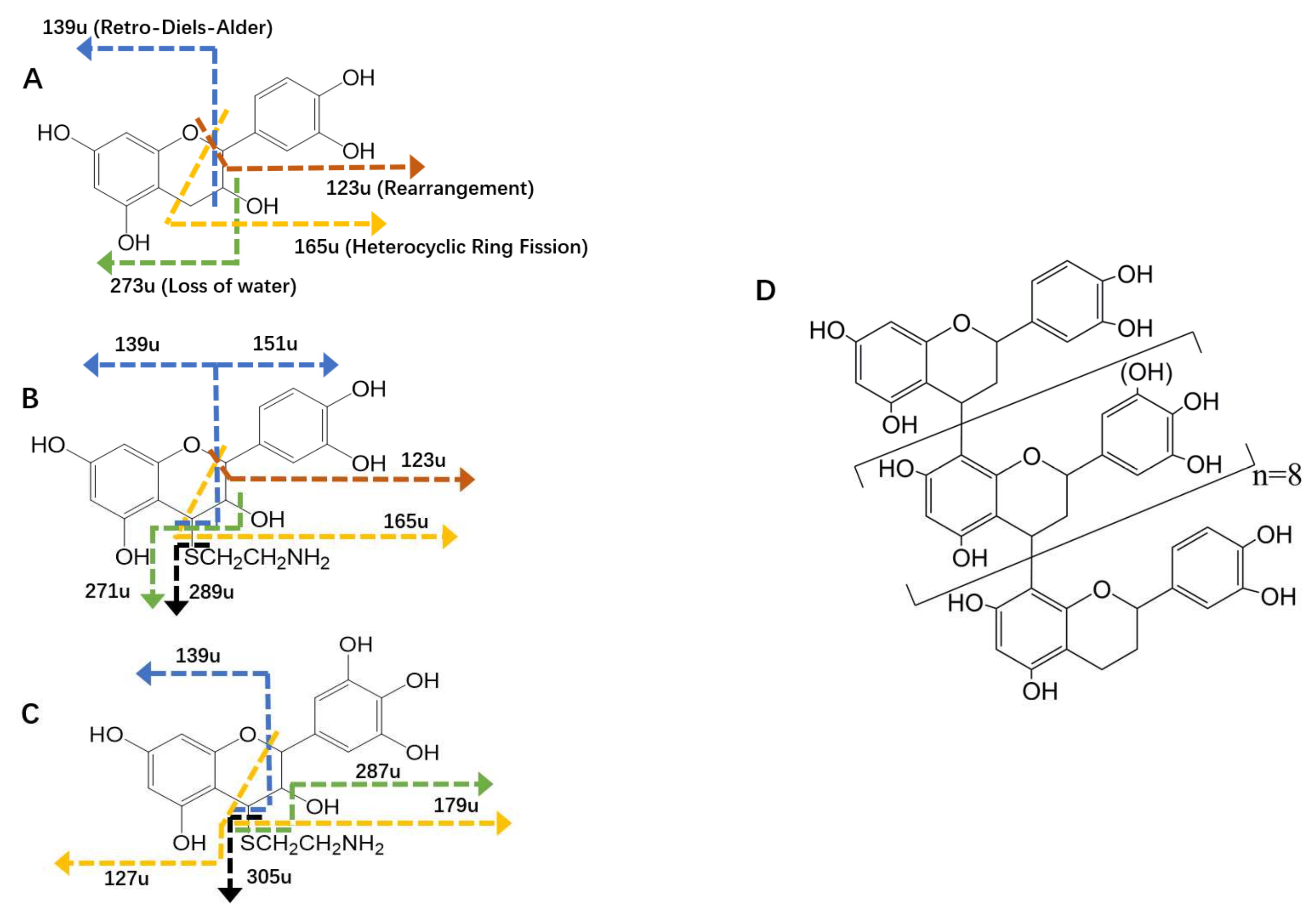
2.3. HPLC–ESI–MS/MS Analysis of Subunits
2.4. Tyrosinase-Inhibition Assay
2.4.1. 50% Activity Inhibition
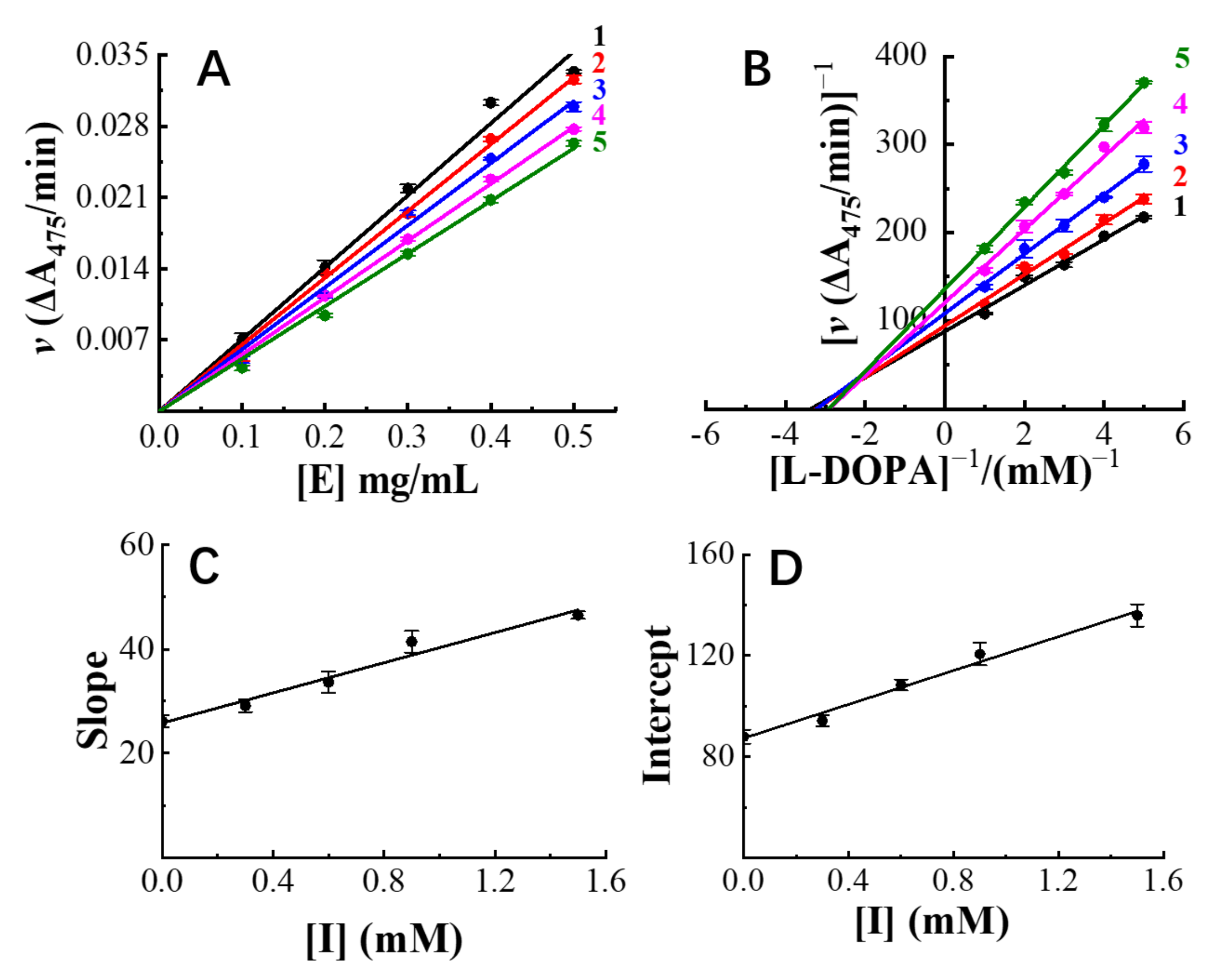
2.4.2. Inhibition Kinetic Assay
2.5. CD Spectroscopy
2.6. In Silico Molecule Docking
2.7. Antioxidant Ability Evaluation
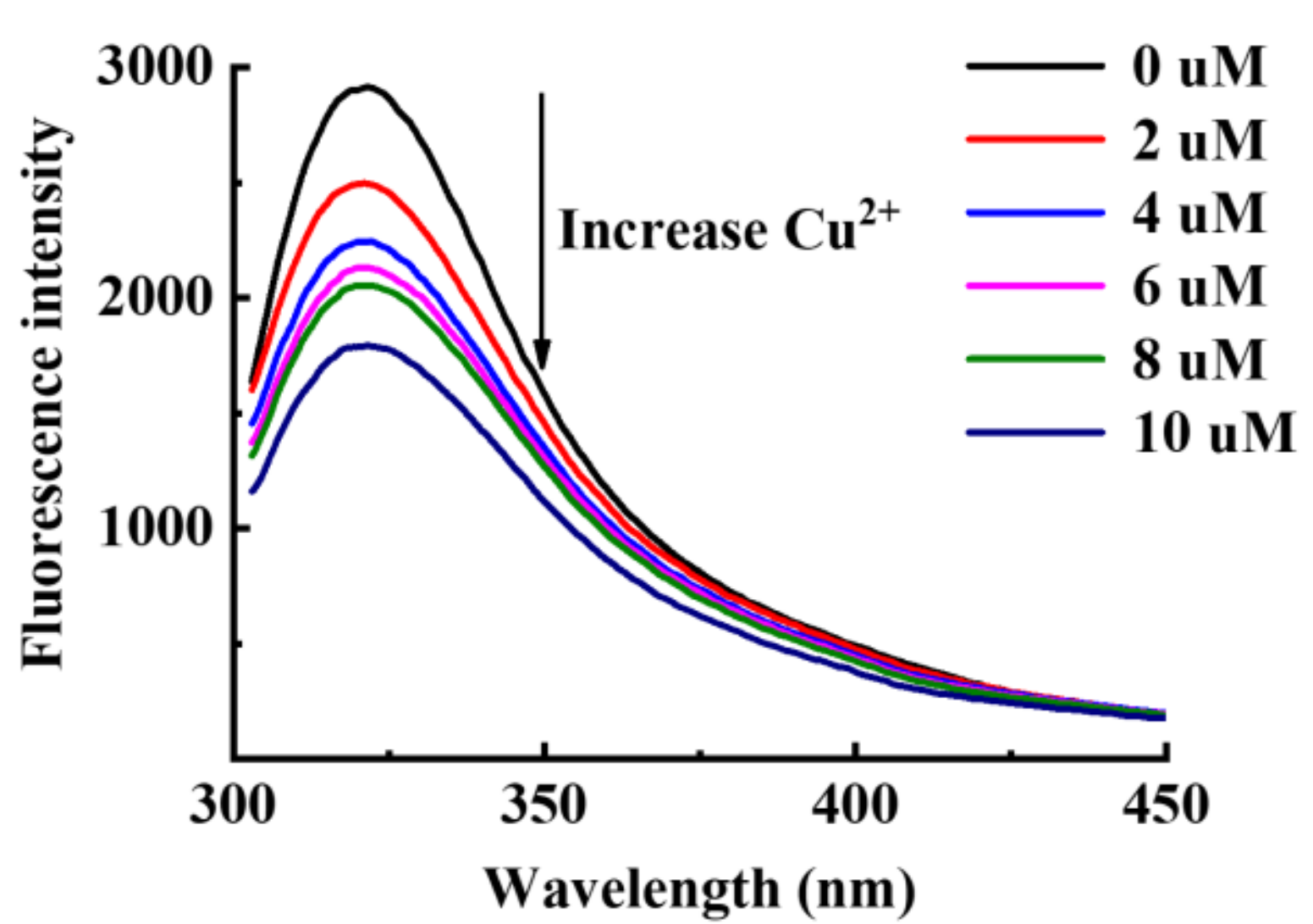
2.8. Fluorescence Quenching Analysis
2.9. Intracellular Tyrosinase Inhibition and Melanin Production
2.9.1. Cell Line and Cell Culture
2.9.2. Intracellular Tyrosinase-Inhibition Assay
2.9.3. Intracellular Melanin Inhibition Assay
3. Results and Discussion
3.1. Structural Characters of Hops Tannin
3.2. Tyrosinase Inhibition Ability of Hop Tannins
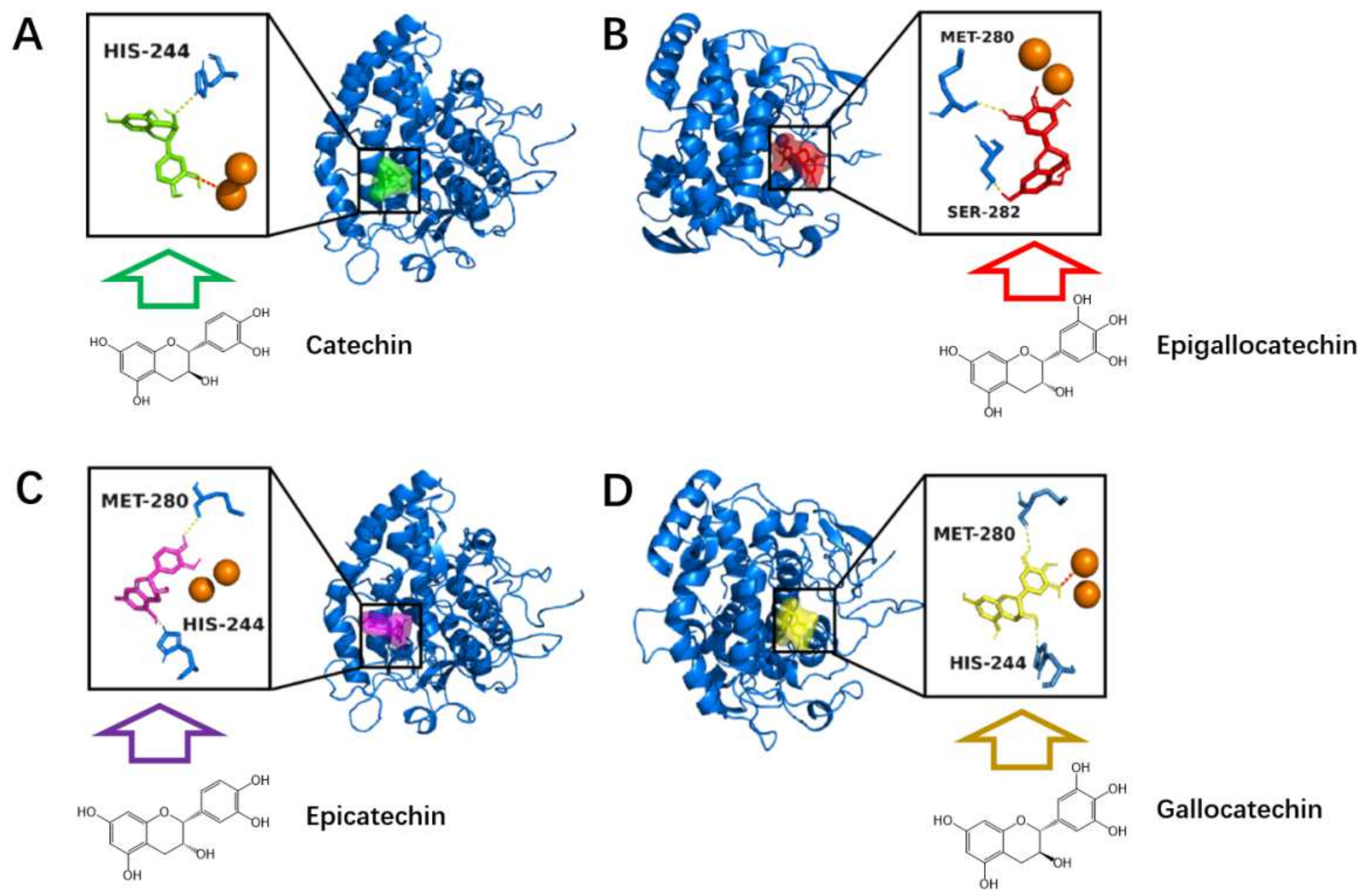
3.3. Inhibition Mechanism Exhibited by Hops Tannin
3.4. Copper-Chelating and Antioxidant Abilities
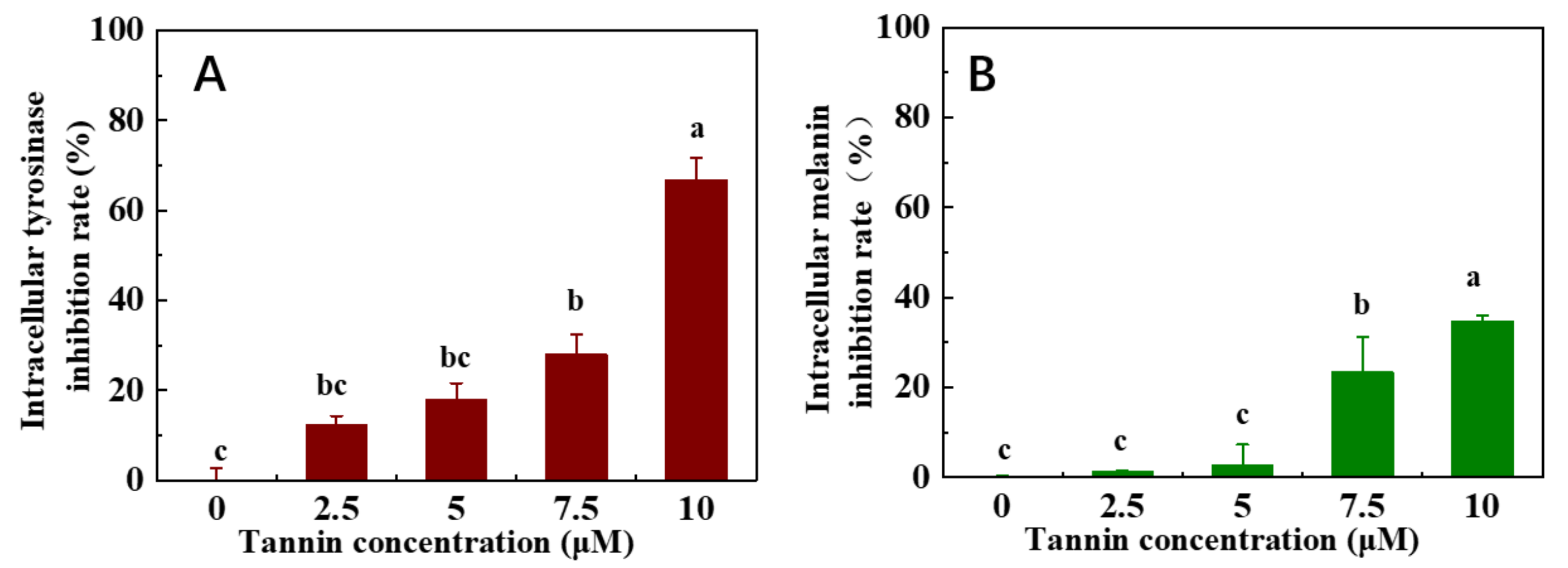
3.5. Cell Viability, Intracellular Tyrosinase Inhibition, and Melanin Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pillaiyar, T.; Namasivayam, V.; Manickam, M.; Jung, S.-H. Inhibitors of Melanogenesis: An Updated Review. J. Med. Chem. 2018, 61, 7395–7418. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.; Wang, S.W.; Li, C.W.; Lin, H.R.; Yang, C.S.; Chu, Y.C.; Lee, T.H.; Chen, J.J. Comparison of Various Solvent Extracts and Major Bioactive Components from Portulaca oleracea for Antioxidant, Anti-Tyrosinase, and Anti-α-Glucosidase Activities. Antioxidants 2022, 11, 398. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Neupane, T.; Baral, R.; Jee, J.-G. Hydroxamic Acid as a Potent Metal-Binding Group for Inhibiting Tyrosinase. Antioxidants 2022, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.-S.; Wu, J.-Y.; Chang, C.-Y. A New Natural Antioxidant Biomaterial from Cinnamomum osmophloeum Kanehira Leaves Represses Melanogenesis and Protects against DNA Damage. Antioxidants 2019, 8, 474. [Google Scholar] [CrossRef]
- Zolghadri, S.; Bahrami, A.; Khan, M.T.H.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 279–309. [Google Scholar] [CrossRef]
- Satooka, H.; Kubo, I. Resveratrol as a kcat type inhibitor for tyrosinase: Potentiated melanogenesis inhibitor. Bioorg Med. Chem. 2012, 20, 1090–1099. [Google Scholar] [CrossRef]
- Mutahir, S.; Khan, M.A.; Khan, I.U.; Yar, M.; Ashraf, M.; Tariq, S.; Ye, R.-L.; Zhou, B.-J. Organocatalyzed and mechanochemical solvent-free synthesis of novel and functionalized bis-biphenyl substituted thiazolidinones as potent tyrosinase inhibitors: SAR and molecular modeling studies. Eur. J. Med. Chem. 2017, 134, 406–414. [Google Scholar] [CrossRef]
- Favre, E.; Daina, A.; Carrupt, P.A.; Nurisso, A. Modeling the met form of human tyrosinase: A refined and hydrated pocket for antagonist design. Chem. Biol. Drug Des. 2014, 84, 206–215. [Google Scholar] [CrossRef]
- Criton, M.; Le Mellay-Hamon, V. Analogues of N-hydroxy-N′-phenylthiourea and N-hydroxy-N′-phenylurea as inhibitors of tyrosinase and melanin formation. Bioorgan. Med. Chem. Lett. 2008, 18, 3607–3610. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Biswas, R.; Sharma, A.; Banerjee, S.; Biswas, S.; Katiyar, C.K. Validation of medicinal herbs for anti-tyrosinase potential. J. Herb. Med. 2018, 14, 1–16. [Google Scholar] [CrossRef]
- Kim, Y.J.; Uyama, H. Tyrosinase inhibitors from natural and synthetic sources: Structure, inhibition mechanism and perspective for the future. Cell. Mol. Life Sci. 2005, 62, 1707–1723. [Google Scholar] [CrossRef]
- Jung, J.H.; Jin, S.G. Microneedle for transdermal drug delivery: Current trends and fabrication. J. Pharm. Investig. 2021, 51, 503–517. [Google Scholar] [CrossRef]
- Halder, J.; Gupta, S.; Kumari, R.; Das Gupta, G.; Rai, V.K. Microneedle Array: Applications, Recent Advances, and Clinical Pertinence in Transdermal Drug Delivery. J. Pharm. Innov. 2020, 16, 558–565. [Google Scholar] [CrossRef]
- Lamberti, L.; Grillo, G.; Gallina, L.; Carnaroglio, D.; Chemat, F.; Cravotto, G. Microwave-Assisted Hydrodistillation of Hop (Humulus lupulus L.) Terpenes: A Pilot-Scale Study. Foods 2021, 10, 2726. [Google Scholar] [CrossRef]
- Sanz, V.; Torres, M.D.; Vilariño, J.M.L.; Domínguez, H. What is new on the hop extraction? Trends Food Sci. Technol. 2019, 93, 12–22. [Google Scholar] [CrossRef]
- Karabín, M.; Hanko, V.; Nešpor, J.; Jelínek, L.; Dostálek, P. Hop tannin extract: A promising tool for acceleration of lautering. J. Inst. Brew. 2018, 124, 374–380. [Google Scholar] [CrossRef]
- Hough, J.S.; Briggs, D.E.; Stevens, R.; Young, T.W. The Chemistry of Hop Constituents. In Malting and Brewing Science: Volume II Hopped Wort and Beer; Hough, J.S., Briggs, D.E., Stevens, R., Young, T.W., Eds.; Springer: Boston, MA, USA, 1982; pp. 422–455. [Google Scholar]
- Liu, J.; Chen, Y.; Zhang, X.; Zheng, J.; Wang, J.; Hu, W.; Teng, B. Tyrosinase inhibition ability provided by hop tannins: A mechanistic investigation. In Proceedings of the The 2nd International Electronic Conference on Foods, Future Foods and Food Technologies for a Sustainable World, Online, 15–30 October 2021. [Google Scholar]
- Liu, J.; Liu, Y.; He, X.; Teng, B.; McRae, J.M. Valonea Tannin: Tyrosinase Inhibition Activity, Structural Elucidation and Insights into the Inhibition Mechanism. Molecules 2021, 26, 2747. [Google Scholar] [CrossRef]
- Teng, B.; Hayasaka, Y.; Smith, P.A.; Bindon, K.A. Effect of Grape Seed and Skin Tannin Molecular Mass and Composition on the Rate of Reaction with Anthocyanin and Subsequent Formation of Polymeric Pigments in the Presence of Acetaldehyde. J. Agric. Food Chem. 2019, 67, 8938–8949. [Google Scholar] [CrossRef]
- Teng, B.; Jian, X.; Gao, Y.; Chen, W. Comparison of polyflavonoids in bayberry tanning effluent and commercial bayberry tannin: Prerequisite information for vegetable tanning effluent recycling. J. Clean. Prod. 2016, 112, 972–979. [Google Scholar] [CrossRef]
- Teng, B.; Hayasaka, Y.; Smith, P.A.; Bindon, K.A. Precipitation of Tannin-Anthocyanin Derivatives in Wine is Influenced by Acetaldehyde Concentration and Tannin Molecular Mass with Implications for the Development of Nonbleachable Pigments. J. Agric. Food Chem. 2021, 69, 4804–4815. [Google Scholar] [CrossRef]
- Kennedy, J.A.; Taylor, A.W. Analysis of proanthocyanidins by high-performance gel permeation chromatography. J. Chromatogr. A 2003, 995, 99–107. [Google Scholar] [CrossRef]
- Chai, W.M.; Wei, M.K.; Wang, R.; Deng, R.G.; Zou, Z.R.; Peng, Y.Y. Avocado Proanthocyanidins as a Source of Tyrosinase Inhibitors: Structure Characterization, Inhibitory Activity, and Mechanism. J. Agric. Food Chem. 2015, 63, 7381–7387. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.-M.; Huang, Q.; Lin, M.-Z.; Ou-Yang, C.; Huang, W.-Y.; Wang, Y.-X.; Xu, K.-L.; Feng, H.-L. Condensed Tannins from Longan Bark as Inhibitor of Tyrosinase: Structure, Activity, and Mechanism. J. Agric. Food Chem. 2018, 66, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Heitz, M.P.; Rupp, J.W. Determining mushroom tyrosinase inhibition by imidazolium ionic liquids: A spectroscopic and molecular docking study. Int. J. Biol. Macromol. 2018, 107, 1971–1981. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Nakashima, K.K.; Bastos, E.L. Rationale on the High Radical Scavenging Capacity of Betalains. Antioxidants 2019, 8, 222. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free. Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Si, C.L.; Wu, L.; Shen, T.; Huang, X.F.; Du, Z.G.; Ren, X.D.; Luo, X.G.; Hu, W.C. Recovery of Low-molecular Weight Galloyltannins from Agricultural Residue of Juglans sigillata Dode Seed Husks and their Tyrosinase Inhibitory Effect. Bioresources 2014, 9, 2226–2236. [Google Scholar] [CrossRef]
- Chai, W.M.; Wang, R.; Wei, M.K.; Zou, Z.R.; Deng, R.G.; Liu, W.S.; Peng, Y.Y. Proanthocyanidins Extracted from Rhododendron pulchrum Leaves as Source of Tyrosinase Inhibitors: Structure, Activity, and Mechanism. PLoS ONE 2015, 10, e0145483. [Google Scholar] [CrossRef]
- Czochanska, Z.; Foo, L.Y.; Newman, R.H.; Porter, L.J. Polymeric proanthocyanidins. Stereochemistry, structural units, and molecular weight. J. Chem. Soc. Perkin Trans. 1 1980, 34, 2278–2286. [Google Scholar] [CrossRef]
- Davis, A.L.; Cai, Y.; Davies, A.P.; Lewis, J.R. 1H and 13C NMR Assignments of Some Green Tea Polyphenols. Magn. Reson. Chem. 1996, 34, 887–890. [Google Scholar] [CrossRef]
- Gu, L.; Kelm, M.A.; Hammerstone, J.F.; Beecher, G.; Holden, J.; Haytowitz, D.; Prior, R.L. Screening of Foods Containing Proanthocyanidins and Their Structural Characterization Using LC-MS/MS and Thiolytic Degradation. J. Agric. Food Chem. 2003, 51, 7513–7521. [Google Scholar] [CrossRef]
- Mouls, L.; Fulcrand, H. UPLC-ESI-MS study of the oxidation markers released from tannin depolymerization: Toward a better characterization of the tannin evolution over food and beverage processing. J. Mass Spectrom. 2012, 47, 1450–1457. [Google Scholar] [CrossRef]
- Cren-Olivé, C.; Déprez, S.; Lebrun, S.; Coddeville, B.; Rolando, C. Characterization of methylation site of monomethylflavan-3-ols by liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2000, 14, 2312–2319. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V. Skin whitening agents: Medicinal chemistry perspective of tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 32, 403–425. [Google Scholar] [CrossRef]
- Sakuma, K.; Ogawa, M.; Sugibayashi, K.; Yamada, K.; Yamamoto, K. Relationship between tyrosinase inhibitory action and oxidation-reduction potential of cosmetic whitening ingredients and phenol derivatives. Arch. Pharm. Res. 1999, 22, 335–339. [Google Scholar] [CrossRef]
- Whiteley, C.G. Enzyme kinetics: Partial and complete uncompetitive inhibition. Biochem. Educ. 2000, 28, 144–147. [Google Scholar] [CrossRef]
- Parvez, S.; Kang, M.; Chung, H.S.; Bae, H. Naturally occurring tyrosinase inhibitors: Mechanism and applications in skin health, cosmetics and agriculture industries. Phytother. Res. 2007, 21, 805–816. [Google Scholar] [CrossRef]
- Soares, D.M.M.; Gonçalves, L.C.P.; Machado, C.O.; Esteves, L.C.; Stevani, C.V.; Oliveira, C.C.; Dörr, F.A.; Pinto, E.; Adachi, F.M.M.; Hotta, C.T.; et al. L-DOPA dioxygenase of the fly agaric toadstool: Revision of the dodA gene sequence and mechanism of enzymatic pigment production. bioRxiv 2020. [Google Scholar] [CrossRef]
- Haslam, E. Natural Polyphenols (Vegetable Tannins) as Drugs: Possible Modes of Action. J. Nat. Prod. 1996, 59, 205–215. [Google Scholar] [CrossRef]
- Chang, T.-S. An Updated Review of Tyrosinase Inhibitors. Int. J. Mol. Sci. 2009, 10, 2440–2475. [Google Scholar] [CrossRef]
- Frazier, R.A.; Deaville, E.R.; Green, R.J.; Stringano, E.; Willoughby, I.; Plant, J.; Mueller-Harvey, I. Interactions of tea tannins and condensed tannins with proteins. J. Pharm. Biomed. Anal. 2010, 51, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ping, Y.; Ejima, H.; Alt, K.; Meissner, M.; Richardson, J.J.; Yan, Y.; Peter, K.; von Elverfeldt, D.; Hagemeyer, C.E.; et al. Engineering Multifunctional Capsules through the Assembly of Metal–Phenolic Networks. Angew. Chem. Int. Ed. 2014, 53, 5546–5551. [Google Scholar] [CrossRef] [PubMed]
- Ros, J.R.; Rodríguez-López, J.N.; García-Cánovas, F. Effect of L-ascorbic acid on the monophenolase activity of tyrosinase. Biochem. J. 1993, 295, 309–312. [Google Scholar] [CrossRef] [PubMed]
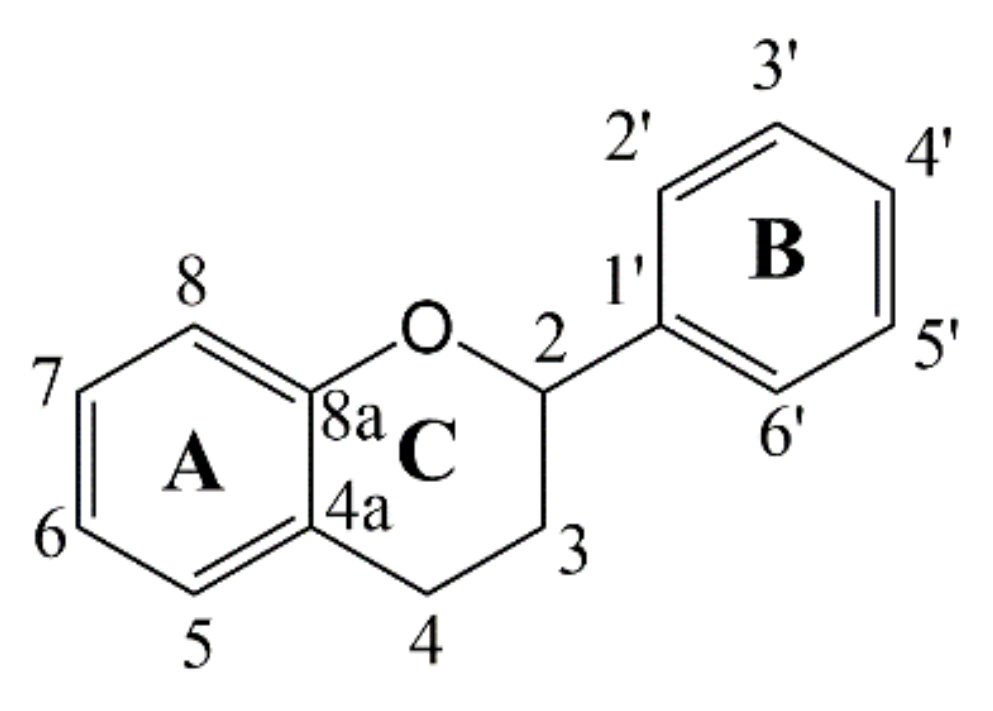
| A-Ring (Phenolic Carbons) | |||||
|---|---|---|---|---|---|
| C-5, C-7 | C-6 | C-8 | C-4a | C-8a | |
| 156.7 | 102.3 | 97.9 | 107.6 | 155.1 | |
| B-ring (phenolic carbons) | |||||
| C-1′ | C-2′ | C-3′ | C-4′ | C-5′ | C-6′ |
| 132.6 | 115.3 | 145.4 | 145.9 | 116.2 | 119.4 |
| C-ring (pyrenoid carbons) | |||||
| C-2 (trans) | C-2 (cis) | C-3 (extension) | C-3 (terminal) | C-4 | |
| 77.2 | 73.1 | 70.7 | 66.7 | 56.0 | |
| Precursor Ions | Retention Time (min) | Fragment Ions | Proposed Structure |
|---|---|---|---|
| 291 | 29.0 | 273, 165, 139, 123 | (epi) catechin |
| 51.3 | |||
| 366 | 20.5 | 289, 271, 165, 151, 139, 123 | (epi) catechin-cysteamine |
| 35.5 | |||
| 382 | 9.9 | 305, 287, 179, 139, 127 | (epi) gallocatechin-cysteamine |
| 22.8 |
| Concentration of Hop Tannins | α-Helix | β-Sheet | β-Turn | Random Coil |
|---|---|---|---|---|
| 0 | 61.85 | 7.87 | 16.71 | 13.57 |
| 5 μM | 65.20 | 6.89 | 16.61 | 11.30 |
| 10 μM | 67.23 | 6.91 | 16.78 | 9.18 |
| DPPH· (IC50 μM) | ABTS·+ (IC50 μM) | |
|---|---|---|
| Hops tannin | 1.17 ± 0.08 b | 1.52 ± 0.02 b |
| Ascorbic acid | 22.04 ± 0.28 a | 17.83 ± 0.30 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Chen, Y.; Zhang, X.; Zheng, J.; Hu, W.; Teng, B. Hop Tannins as Multifunctional Tyrosinase Inhibitor: Structure Characterization, Inhibition Activity, and Mechanism. Antioxidants 2022, 11, 772. https://doi.org/10.3390/antiox11040772
Liu J, Chen Y, Zhang X, Zheng J, Hu W, Teng B. Hop Tannins as Multifunctional Tyrosinase Inhibitor: Structure Characterization, Inhibition Activity, and Mechanism. Antioxidants. 2022; 11(4):772. https://doi.org/10.3390/antiox11040772
Chicago/Turabian StyleLiu, Jiaman, Yanbiao Chen, Xinxin Zhang, Jie Zheng, Weiying Hu, and Bo Teng. 2022. "Hop Tannins as Multifunctional Tyrosinase Inhibitor: Structure Characterization, Inhibition Activity, and Mechanism" Antioxidants 11, no. 4: 772. https://doi.org/10.3390/antiox11040772
APA StyleLiu, J., Chen, Y., Zhang, X., Zheng, J., Hu, W., & Teng, B. (2022). Hop Tannins as Multifunctional Tyrosinase Inhibitor: Structure Characterization, Inhibition Activity, and Mechanism. Antioxidants, 11(4), 772. https://doi.org/10.3390/antiox11040772






