Tandem Mass Tagging-Based Quantitative Proteomics Analysis Reveals Damage to the Liver and Brain of Hypophthalmichthys molitrix Exposed to Acute Hypoxia and Reoxygenation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Hypoxia–Reoxygenation Experiment and Sampling
2.3. Brain and Liver Function Tests
2.4. Proteome Extraction and Digestion
2.5. TMT Labeling, Peptide Fractionation, and LC–MS/MS Analysis
2.6. Bioinformatic Analyses
2.7. RNA Extraction and Quantitative Real-Time Reverse-Transcription PCR (qRT–PCR)
2.8. Statistical Analyses
3. Results
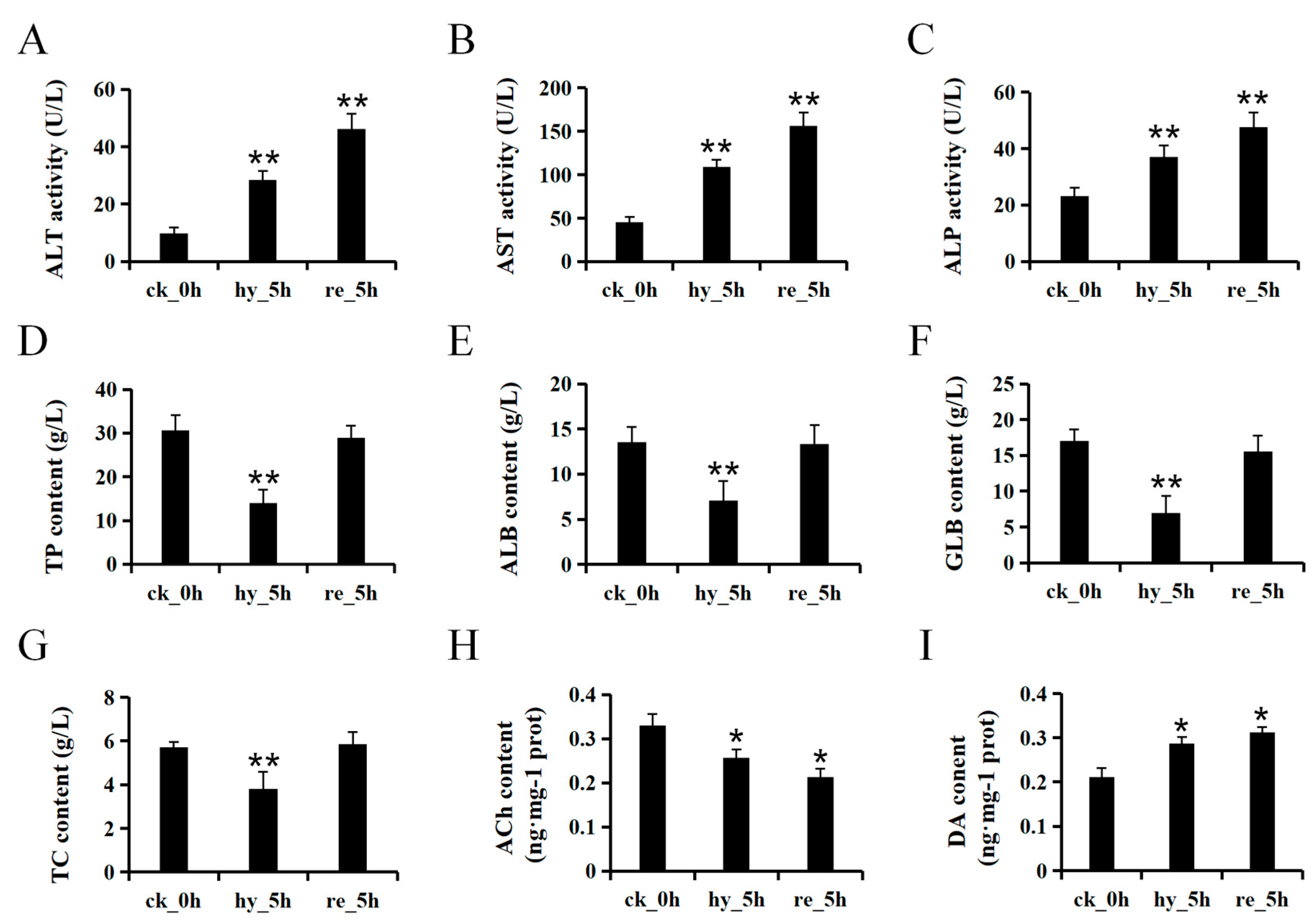
3.1. Acute Hypoxia and Reoxygenation Affects the Physiology of the Liver and Brain in H. molitrix
3.2. Protein Identification, Quantification, and Annotation Analysis in the Liver and Brain of H. molitrix
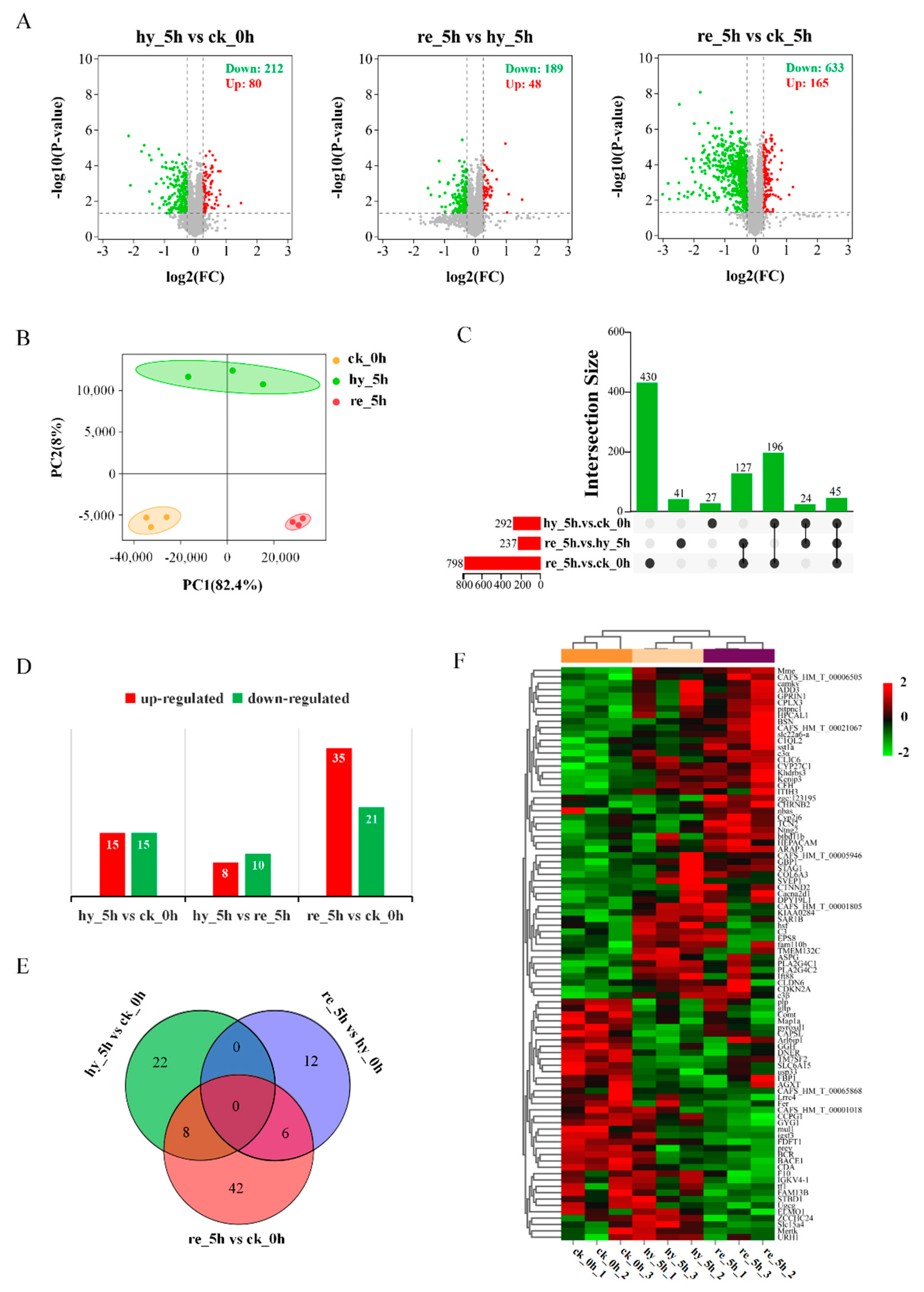
3.3. Proteome Changes in the Liver and Brain in Response to the Acute Hypoxia and Reoxygenation Exposure
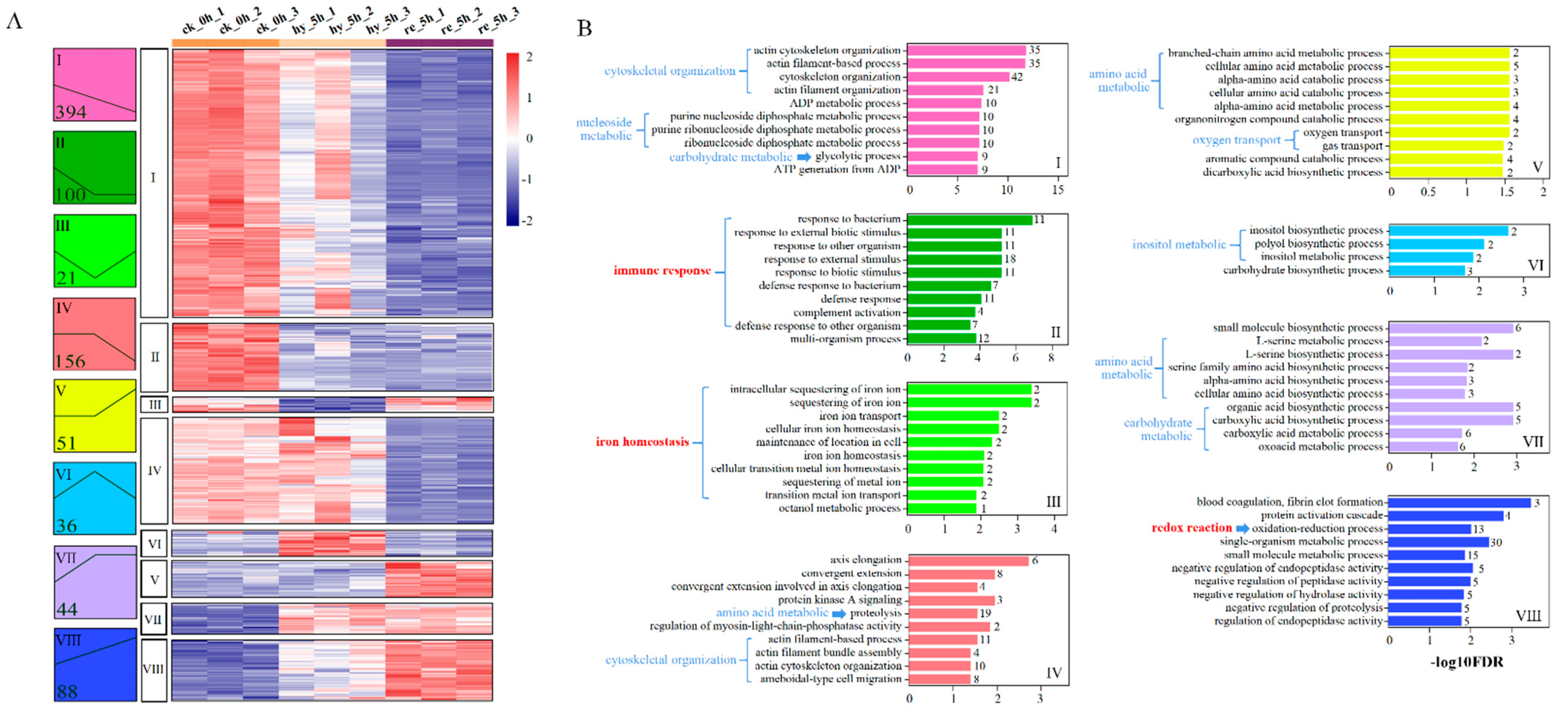
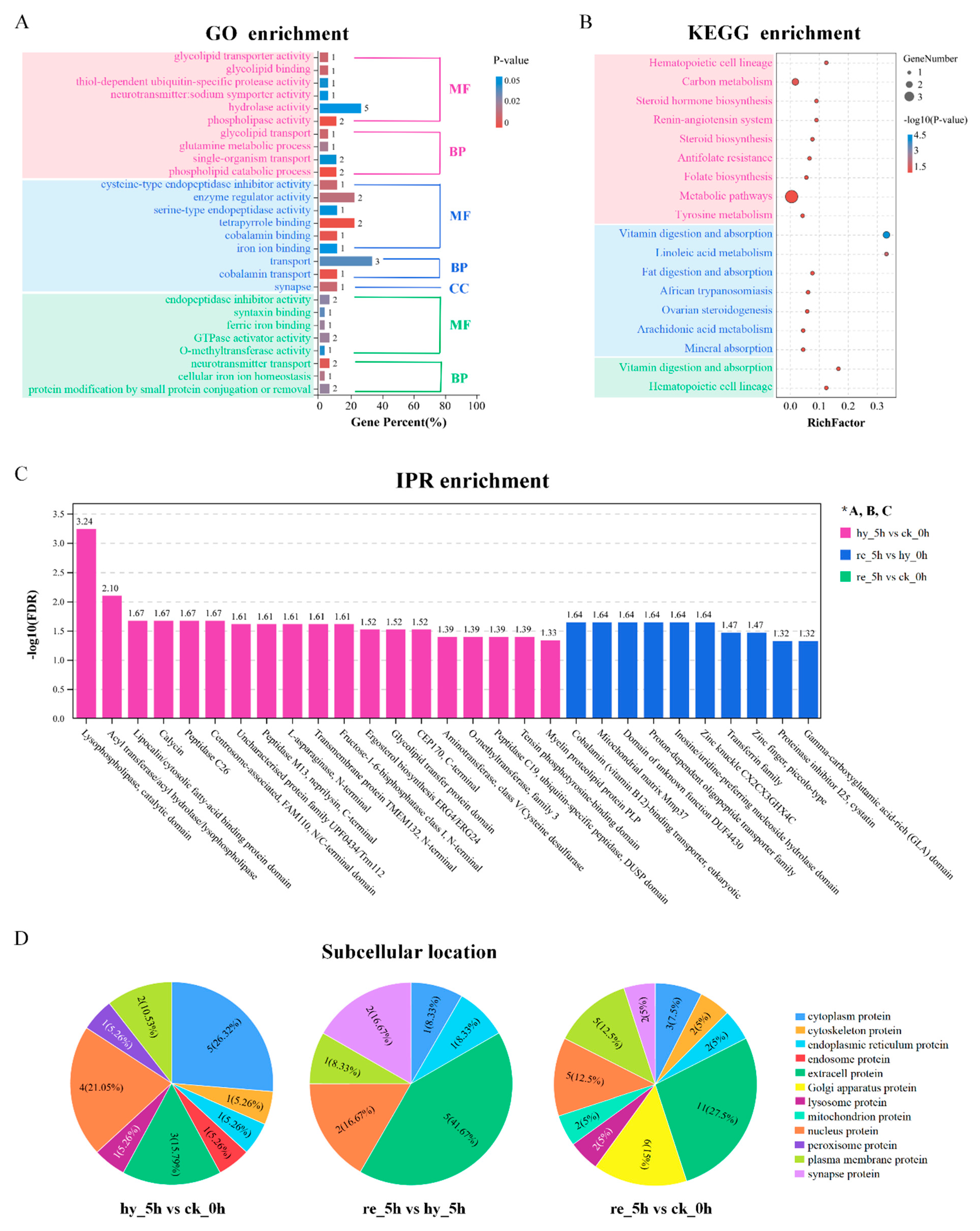
3.4. Bioinformatic Analyses of DAPs in the Liver
3.5. Bioinformatic Analyses of DAPs in the Brain
3.6. Validation of the DAPs Identified from the Proteomics Analysis
4. Discussion
4.1. Hypoxia–Reoxygenation Impairs the Function of Liver and Brain
4.2. Effects of Hypoxia–Reoxygenation on the Immune Response, Iron Homeostasis, and Redox Reactions in the Liver and Brain
4.2.1. Immune Response
4.2.2. Iron Homeostasis
4.2.3. Redox Reactions
4.3. Other Effects of Hypoxia–Reoxygenation on the Liver
4.4. Other Effects of Hypoxia–Reoxygenation on the Brain
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, J.L.; Zhao, L.L.; Wu, H.; Liu, Q.; Liao, L.; Luo, J.; Lian, W.Q.; Cui, C.; Jin, L.; Ma, J.D. Acute hypoxia changes the mode of glucose and lipid utilization in the liver of the largemouth bass (Micropterus salmoides). Sci. Total Environ. 2020, 713, 135157. [Google Scholar] [CrossRef] [PubMed]
- Dan, X.-M.; Yan, G.-J.; Zhang, A.-J.; Cao, Z.-D.; Fu, S.-J. Effects of stable and diel-cycling hypoxia on hypoxia tolerance, postprandial metabolic response, and growth performance in juvenile qingbo (Spinibarbus sinensis). Aquaculture 2014, 428, 21–28. [Google Scholar] [CrossRef]
- D’Avanzo, C.; Kremer, J.N. Diel oxygen dynamics and anoxic events in an eutrophic estuary of Waquoit Bay, Massachusetts. Estuaries 1994, 17, 131–139. [Google Scholar] [CrossRef]
- Diaz, R.J.; Breitburg, D.L. The hypoxic environment. In Fish Physiology; Elsevier: Amsterdam, The Netherlands, 2009; Volume 27, pp. 1–23. [Google Scholar] [CrossRef]
- Yang, H.; Cao, Z.-D.; Fu, S.-J. The effects of diel-cycling hypoxia acclimation on the hypoxia tolerance, swimming capacity and growth performance of southern catfish (Silurus meridionalis). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2013, 165, 131–138. [Google Scholar] [CrossRef]
- Piontkivska, H.; Chung, J.S.; Ivanina, A.V.; Sokolov, E.P.; Techa, S.; Sokolova, I.M. Molecular characterization and mRNA expression of two key enzymes of hypoxia-sensing pathways in eastern oysters Crassostrea virginica (Gmelin): Hypoxia-inducible factor α (HIF-α) and HIF-prolyl hydroxylase (PHD). Comp. Biochem. Physiol. Part D Genom. Proteom. 2011, 6, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Rashid, I.; Nagpure, N.S.; Srivastava, P.; Kumar, R.; Pathak, A.K.; Singh, M.; Kushwaha, B. HRGFish: A database of hypoxia responsive genes in fishes. Sci. Rep. 2017, 7, 42346. [Google Scholar] [CrossRef]
- Bouvry, D.; Planes, C.; Malbert-Colas, L.; Escabasse, V.; Clerici, C. Hypoxia-induced cytoskeleton disruption in alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 2006, 35, 519–527. [Google Scholar] [CrossRef] [Green Version]
- Kang, P.M.; Haunstetter, A.; Aoki, H.; Usheva, A.; Izumo, S. Morphological and molecular characterization of adult cardiomyocyte apoptosis during hypoxia and reoxygenation. Circ. Res. 2000, 87, 118–125. [Google Scholar] [CrossRef]
- Yan, L.; Wang, P.; Zhao, C.; Fan, S.; Lin, H.; Guo, Y.; Ma, Z.; Qiu, L. Toxic responses of liver in Lateolabrax maculatus during hypoxia and re-oxygenation. Aquat. Toxicol. 2021, 236, 105841. [Google Scholar] [CrossRef]
- Nilsson, G.E.; Östlund-Nilsson, S. Does size matter for hypoxia tolerance in fish? Biol. Rev. 2008, 83, 173–189. [Google Scholar] [CrossRef]
- Xiao, W. The hypoxia signaling pathway and hypoxic adaptation in fishes. Sci. China Life Sci. 2015, 58, 148–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.-L.; Mao, M.-G.; Lü, H.-Q.; Wen, S.-H.; Sun, M.-L.; Liu, R.-t.; Jiang, Z.-Q. Digital gene expression analysis of Takifugu rubripes brain after acute hypoxia exposure using next-generation sequencing. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 24, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wu, F.; Xie, S.; Zhang, L. Acute hypoxia and reoxygenation: Effect on oxidative stress and hypoxia signal transduction in the juvenile yellow catfish (Pelteobagrus fulvidraco). Aquaculture 2021, 531, 735903. [Google Scholar] [CrossRef]
- Terui, K.; Enosawa, S.; Haga, S.; Zhang, H.Q.; Kuroda, H.; Kouchi, K.; Matsunaga, T.; Yoshida, H.; Engelhardt, J.F.; Irani, K. Stat3 confers resistance against hypoxia/reoxygenation-induced oxidative injury in hepatocytes through upregulation of Mn-SOD. J. Hepatol. 2004, 41, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, Y.; Xia, Q.; Tu, Z.; Sun, J.; Jing, Q.; Chen, P.; Zhao, X. Propofol Mediated Protection of the Brain from Ischemia/Reperfusion Injury Through the Regulation of Microglial Connexin 43. Front. Cell Dev. Biol. 2021, 9, 1104. [Google Scholar] [CrossRef]
- China Statistics Press. China Fishery Statistical Yearbook 2020; China Statistics Press: Beijing, China, 2020; Volume 2020, pp. 24–34. [Google Scholar]
- Li, C.; Wang, J.; Chen, J.; Schneider, K.; Veettil, R.K.; Elmer, K.R.; Zhao, J. Native bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix populations in the Pearl River are threatened by Yangtze River introductions as revealed by mitochondrial DNA. J. Fish Biol. 2020, 96, 651–662. [Google Scholar] [CrossRef]
- Feng, C.; Li, X.; Sha, H.; Luo, X.; Zou, G.; Liang, H. Comparative transcriptome analysis provides novel insights into the molecular mechanism of the silver carp (Hypophthalmichthys molitrix) brain in response to hypoxia stress. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 41, 100951. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, J.; Wen, X.; Zhao, C.; Zhang, H.; Li, X.; Yin, S. Comparative iTRAQ-Based Quantitative Proteomic Analysis of Pelteobagrus vachelli Liver under Acute Hypoxia: Implications in Metabolic Responses. Proteomics 2017, 17, 1700140. [Google Scholar] [CrossRef]
- Li, X.; Li, F.; Zou, G.; Feng, C.; Sha, H.; Liu, S.; Liang, H. Physiological responses and molecular strategies in heart of silver carp (Hypophthalmichthys molitrix) under hypoxia and reoxygenation. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100908. [Google Scholar] [CrossRef]
- Bárcena, B.; Salamanca, A.; Pintado, C.; Mazuecos, L.; Villar, M.; Moltó, E.; Bonzón-Kulichenko, E.; Vázquez, J.; Andrés, A.; Gallardo, N. Aging Induces Hepatic Oxidative Stress and Nuclear Proteomic Remodeling in Liver from Wistar Rats. Antioxidants 2021, 10, 1535. [Google Scholar] [CrossRef]
- Henry, M.L.; Velez-Irizarry, D.; Pagan, J.D.; Sordillo, L.; Gandy, J.; Valberg, S.J. The Impact of N-Acetyl Cysteine and Coenzyme Q10 Supplementation on Skeletal Muscle Antioxidants and Proteome in Fit Thoroughbred Horses. Antioxidants 2021, 10, 1739. [Google Scholar] [CrossRef] [PubMed]
- McBride, Z.; Chen, D.; Reick, C.; Xie, J.; Szymanski, D.B. Global analysis of membrane-associated protein oligomerization using protein correlation profiling. Mol. Cell. Proteom. 2017, 16, 1972–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Alonso, R.; Bustos, F.; Budzyk, M.; Kumar, P.; Helbig, A.O.; Hukelmann, J.; Lamond, A.I.; Lanner, F.; Zhou, H.; Petsalaki, E. Phosphoproteomics identifies a bimodal EPHA2 receptor switch that promotes embryonic stem cell differentiation. Nat. Commun. 2020, 11, 1357. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, M.; Ling, C.; Sha, H.; Zou, G.; Liang, H. Molecular Characterization and Response of Prolyl Hydroxylase Domain (PHD) Genes to Hypoxia Stress in Hypophthalmichthys molitrix. Animals 2022, 12, 131. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, W.; Qiao, H.; Jiang, S.; Xiong, Y.; Jin, S.; Gong, Y.; Fu, H. Integrated Metabolomics and Transcriptomic Analysis of Hepatopancreas in Different Living Status Macrobrachium nipponense in Response to Hypoxia. Antioxidants 2022, 11, 36. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yee, P.P.; Wei, Y.; Kim, S.-Y.; Lu, T.; Chih, S.Y.; Lawson, C.; Tang, M.; Liu, Z.; Anderson, B.; Thamburaj, K. Neutrophil-induced ferroptosis promotes tumor necrosis in glioblastoma progression. Nat. Commun. 2020, 11, 5424. [Google Scholar] [CrossRef]
- Long, Y.; Yan, J.; Song, G.; Li, X.; Li, X.; Li, Q.; Cui, Z. Transcriptional events co-regulated by hypoxia and cold stresses in Zebrafish larvae. BMC Genom. 2015, 16, 385. [Google Scholar] [CrossRef] [Green Version]
- Shin, D.; Kim, E.H.; Lee, J.; Roh, J.-L. Nrf2 inhibition reverses resistance to GPX4 inhibitor-induced ferroptosis in head and neck cancer. Free Radic. Biol. Med. 2018, 129, 454–462. [Google Scholar] [CrossRef]
- Sheng, Y.; Hua, Z.Y.; Yang, Z.; Wei, X.L.; Sheng, Y.J.; Jia, H.L.; Jun, Q. Effects of acute hypoxic stress on biochemical parameters, immune regulation and metabolic capacity of the blood in genetically improved farmed tilapia (GIFT, Oreochromis niloticus). J. Appl. Ichthyol. 2019, 35, 978–986. [Google Scholar] [CrossRef]
- Williams, K.J.; Cassidy, A.A.; Verhille, C.E.; Lamarre, S.G.; MacCormack, T.J. Diel cycling hypoxia enhances hypoxia tolerance in rainbow trout (Oncorhynchus mykiss): Evidence of physiological and metabolic plasticity. J. Exp. Biol. 2019, 222, jeb206045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.W.; Houlihan, D.F.; Nilsson, G.E.; Brechin, J. Tissue-specific changes in protein synthesis rates in vivo during anoxia in crucian carp. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 1996, 271, R897–R904. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, A.A.; Driedzic, W.R.; Campos, D.; Heinrichs-Caldas, W. Protein synthesis is lowered by 4EBP1 and eIF2-alpha signaling while protein degradation may be maintained in fasting, hypoxic Amazonian cichlids Astronotus ocellatus. J. Exp. Biol. 2018, 221, jeb167601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, J.G. Physiological, behavioral and biochemical adaptations of intertidal fishes to hypoxia. J. Exp. Biol. 2011, 214, 191–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, H.; Liu, G.; Lu, T.; Sun, L. Developmental neurotoxicity of Microcystis aeruginosa in the early life stages of zebrafish. Ecotoxicol. Environ. Saf. 2018, 151, 35–41. [Google Scholar] [CrossRef]
- Gibson, G.E.; Duffy, T.E. Impaired synthesis of acetylcholine by mild hypoxic hypoxia or nitrous oxide. J. Neurochem. 1981, 36, 28–33. [Google Scholar] [CrossRef]
- Wu, Q.; Yan, W.; Liu, C.; Li, L.; Yu, L.; Zhao, S.; Li, G. Microcystin-LR exposure induces developmental neurotoxicity in zebrafish embryo. Environ. Pollut. 2016, 213, 793–800. [Google Scholar] [CrossRef]
- Broderick, P.A.; Gibson, G.E. Dopamine and serotonin in rat striatum duringin vivo hypoxic-hypoxia. Metab. Brain Dis. 1989, 4, 143–153. [Google Scholar] [CrossRef]
- Meyer-Lindenberg, A.; Kohn, P.D.; Kolachana, B.; Kippenhan, S.; McInerney-Leo, A.; Nussbaum, R.; Weinberger, D.R.; Berman, K.F. Midbrain dopamine and prefrontal function in humans: Interaction and modulation by COMT genotype. Nat. Neurosci. 2005, 8, 594–596. [Google Scholar] [CrossRef]
- Dufour, S.; Sebert, M.E.; Weltzien, F.A.; Rousseau, K.; Pasqualini, C. Neuroendocrine control by dopamine of teleost reproduction. J. Fish Biol. 2010, 76, 129–160. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Tawwab, M.; Monier, M.N.; Hoseinifar, S.H.; Faggio, C. Fish response to hypoxia stress: Growth, physiological, and immunological biomarkers. Fish Physiol. Biochem. 2019, 45, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Holland, M.C.H.; Lambris, J.D. The complement system in teleosts. Fish Shellfish Immunol. 2002, 12, 399–420. [Google Scholar] [CrossRef] [Green Version]
- Dunkelberger, J.R.; Song, W.-C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riihilä, P.; Nissinen, L.; Farshchian, M.; Kallajoki, M.; Kivisaari, A.; Meri, S.; Grénman, R.; Peltonen, S.; Peltonen, J.; Pihlajaniemi, T. Complement component C3 and complement factor B promote growth of cutaneous squamous cell carcinoma. Am. J. Pathol. 2017, 187, 1186–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnhold, J. The dual role of myeloperoxidase in immune response. Int. J. Mol. Sci. 2020, 21, 8057. [Google Scholar] [CrossRef]
- Chadzinska, M.; Baginski, P.; Kolaczkowska, E.; Savelkoul, H.F.; Lidy Verburg-van Kemenade, B. Expression profiles of matrix metalloproteinase 9 in teleost fish provide evidence for its active role in initiation and resolution of inflammation. Immunology 2008, 125, 601–610. [Google Scholar] [CrossRef]
- Yang, C.; Chen, L.; Su, J.; Feng, X.; Rao, Y. Two novel homologs of high mobility group box 3 gene in grass carp (Ctenopharyngodon idella): Potential roles in innate immune responses. Fish Shellfish Immunol. 2013, 35, 1501–1510. [Google Scholar] [CrossRef]
- Nogueras-Ortiz, C.J.; Mahairaki, V.; Delgado-Peraza, F.; Das, D.; Avgerinos, K.; Eren, E.; Hentschel, M.; Goetzl, E.J.; Mattson, M.P.; Kapogiannis, D. Astrocyte-and neuron-derived extracellular vesicles from Alzheimer’s disease patients effect complement-mediated neurotoxicity. Cells 2020, 9, 1618. [Google Scholar] [CrossRef]
- Sarma, J.V.; Ward, P.A. The complement system. Cell Tissue Res. 2011, 343, 227–235. [Google Scholar] [CrossRef]
- Ponka, P.; Beaumont, C.; Richardson, D.R. Function and regulation of transferrin and ferritin. Semin. Hematol. 1998, 35, 35–54. [Google Scholar] [PubMed]
- Devenport, S.N.; Shah, Y.M. Functions and implications of autophagy in colon cancer. Cells 2019, 8, 1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chepelev, N.L.; Willmore, W.G. Regulation of iron pathways in response to hypoxia. Free Radic. Biol. Med. 2011, 50, 645–666. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhong, X.-P.; Qiao, Z.-X.; Gui, J.-F. Inductive transcription and protective role of fish heme oxygenase-1 under hypoxic stress. J. Exp. Biol. 2008, 211, 2700–2706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morishita, H.; Mizushima, N. Diverse cellular roles of autophagy. Annu. Rev. Cell Dev. Biol. 2019, 35, 453–475. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by lipid peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Luo, M.; Zhang, K.; Zhang, J.; Gao, T.; Connell, D.O.; Yao, F.; Mu, C.; Cai, B.; Shang, Y. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020, 11, 433. [Google Scholar] [CrossRef] [Green Version]
- Yagoda, N.; von Rechenberg, M.; Zaganjor, E.; Bauer, A.J.; Yang, W.S.; Fridman, D.J.; Wolpaw, A.J.; Smukste, I.; Peltier, J.M.; Boniface, J.J. RAS–RAF–MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 2007, 447, 865–869. [Google Scholar] [CrossRef] [Green Version]
- Doll, S.; Proneth, B.; Tyurina, Y.Y.; Panzilius, E.; Kobayashi, S.; Ingold, I.; Irmler, M.; Beckers, J.; Aichler, M.; Walch, A. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. [Google Scholar] [CrossRef]
- Blomhoff, R. Dietary antioxidants and cardiovascular disease. Curr. Opin. Lipidol. 2005, 16, 47–54. [Google Scholar] [CrossRef]
- Zhang, G.; Mao, J.; Liang, F.; Chen, J.; Zhao, C.; Yin, S.; Wang, L.; Tang, Z.; Chen, S. Modulated expression and enzymatic activities of Darkbarbel catfish, Pelteobagrus vachelli for oxidative stress induced by acute hypoxia and reoxygenation. Chemosphere 2016, 151, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gozzelino, R.; Jeney, V.; Soares, M.P. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 323–354. [Google Scholar] [CrossRef] [Green Version]
- McKinnon, R.A.; Sorich, M.J.; Ward, M.B. Cytochrome P450 part 1: Multiplicity and function. J. Pharm. Pract. Res. 2008, 38, 55–57. [Google Scholar] [CrossRef]
- Costa, C.; Catania, S.; De Pasquale, R.; Stancanelli, R.; Scribano, G.; Melchini, A. Exposure of human skin to benzo [a] pyrene: Role of CYP1A1 and aryl hydrocarbon receptor in oxidative stress generation. Toxicology 2010, 271, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yan, B.; Gao, F.; Li, Q.; Meng, X.; Chen, P.; Zhou, L.; Deng, W.; Li, C.; Xu, W. PSCs Reveal PUFA-provoked mitochondrial stress as a central node potentiating RPE degeneration in Bietti’s crystalline dystrophy. Mol. Ther. 2020, 28, 2642–2661. [Google Scholar] [CrossRef]
- Tanaka, Y.; Aleksunes, L.M.; Yeager, R.L.; Gyamfi, M.A.; Esterly, N.; Guo, G.L.; Klaassen, C.D. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J. Pharmacol. Exp. Ther. 2008, 325, 655–664. [Google Scholar] [CrossRef]
- Lornage, X.; Schartner, V.; Balbueno, I.; Biancalana, V.; Willis, T.; Echaniz-Laguna, A.; Scheidecker, S.; Quinlivan, R.; Fardeau, M.; Malfatti, E. Clinical, histological, and genetic characterization of PYROXD1-related myopathy. Acta Neuropathol. Commun. 2019, 7, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennati, A.M.; Schiavoni, G.; Franken, S.; Piobbico, D.; Della Fazia, M.A.; Caruso, D.; De Fabiani, E.; Benedetti, L.; Cusella De Angelis, M.G.; Gieselmann, V. Disruption of the gene encoding 3β-hydroxysterol Δ14-reductase (Tm7sf2) in mice does not impair cholesterol biosynthesis. FEBS J. 2008, 275, 5034–5047. [Google Scholar] [CrossRef]
- Ding, J.; Liu, C.; Luo, S.; Zhang, Y.; Gao, X.; Wu, X.; Shen, W.; Zhu, J. Transcriptome and physiology analysis identify key metabolic changes in the liver of the large yellow croaker (Larimichthys crocea) in response to acute hypoxia. Ecotoxicol. Environ. Saf. 2020, 189, 109957. [Google Scholar] [CrossRef]
- Barnett, P. Somatostatin and somatostatin receptor physiology. Endocrine 2003, 20, 255–264. [Google Scholar] [CrossRef]
- Lin, X.; Mattjus, P.; Pike, H.M.; Windebank, A.J.; Brown, R.E. Cloning and expression of glycolipid transfer protein from bovine and porcine brain. J. Biol. Chem. 2000, 275, 5104–5110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nashabat, M.; Maegawa, G.; Nissen, P.H.; Nexo, E.; Al-Shamrani, H.; Al-Owain, M.; Alfadhel, M. Long-term outcome of 4 patients with transcobalamin deficiency caused by 2 novel TCN2 mutations. J. Pediatric Hematol./Oncol. 2017, 39, e430–e436. [Google Scholar] [CrossRef]
- Smith, D.E.; Clémençon, B.; Hediger, M.A. Proton-coupled oligopeptide transporter family SLC15: Physiological, pharmacological and pathological implications. Mol. Asp. Med. 2013, 34, 323–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simm, F.; Griesbeck, A.; Choukair, D.; Weiß, B.; Paramasivam, N.; Klammt, J.; Schlesner, M.; Wiemann, S.; Martinez, C.; Hoffmann, G.F. Identification of SLC20A1 and SLC15A4 among other genes as potential risk factors for combined pituitary hormone deficiency. Genet. Med. 2018, 20, 728–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hägglund, M.G.; Roshanbin, S.; Löfqvist, E.; Hellsten, S.V.; Nilsson, V.C.; Todkar, A.; Zhu, Y.; Stephansson, O.; Drgonova, J.; Uhl, G.R. B0AT2 (SLC6A15) is localized to neurons and astrocytes, and is involved in mediating the effect of leucine in the brain. PLoS ONE 2013, 8, e58651. [Google Scholar] [CrossRef] [Green Version]
- Kohli, M.A.; Lucae, S.; Saemann, P.G.; Schmidt, M.V.; Demirkan, A.; Hek, K.; Czamara, D.; Alexander, M.; Salyakina, D.; Ripke, S. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron 2011, 70, 252–265. [Google Scholar] [CrossRef] [Green Version]
- Reim, K.; Regus-Leidig, H.; Ammermüller, J.; El-Kordi, A.; Radyushkin, K.; Ehrenreich, H.; Brandstätter, J.H.; Brose, N. Aberrant function and structure of retinal ribbon synapses in the absence of complexin 3 and complexin 4. J. Cell Sci. 2009, 122, 1352–1361. [Google Scholar] [CrossRef] [Green Version]


| ID | Protein Name | Annotation | |
|---|---|---|---|
| immune response (19) | CAFS_HM_T_00069923 | C3 | Complement C3 (Fragment) |
| CAFS_HM_T_00069924 | C3 | Complement C3 (Fragment) | |
| CAFS_HM_T_00051668 | CFB | Complement factor B | |
| CAFS_HM_T_00051663 | CFB | Complement factor B | |
| CAFS_HM_T_00004034 | MPO | Myeloperoxidase | |
| CAFS_HM_T_00004821 | mmp13 | Collagenase 3 (Fragment) | |
| CAFS_HM_T_00038844 | MMP9 | Matrix metalloproteinase-9 | |
| CAFS_HM_T_00009179 | rnasel3 | Ribonuclease-like 3 | |
| CAFS_HM_T_00024834 | HMGB3 | High-mobility group protein B3 | |
| CAFS_HM_T_00051562 | RHOG | Rho-related GTP-binding protein RhoG | |
| CAFS_HM_T_00059585 | LYZL1 | Lysozyme-like protein 1 | |
| CAFS_HM_T_00068155 | EFNB1 | Ephrin-B1 | |
| CAFS_HM_T_00070745 | CASP1 | Caspase-1 | |
| CAFS_HM_T_00085126 | CASP1 | Caspase-1 | |
| CAFS_HM_T_00075145 | GLUL | Glutamine synthetase | |
| CAFS_HM_T_00078382 | dapl1-b | Death-associated protein-like 1-B | |
| CAFS_HM_T_00086469 | alcama | CD166 antigen homolog A | |
| CAFS_HM_T_00063121 | -- | Histone H1 | |
| CAFS_HM_T_00071322 | -- | Nattectin | |
| iron homeostasis (2) | CAFS_HM_T_00047854 | FerH | Ferritin, heavy subunit |
| CAFS_HM_T_00015421 | FerM | Ferritin, middle subunit | |
| redox reaction (13) | CAFS_HM_T_00005616 | Gpx4 | Phospholipid hydroperoxide glutathione peroxidase, mitochondrial |
| CAFS_HM_T_00018176 | HO-1 | Heme oxygenase-1 | |
| CAFS_HM_T_00009561 | CYP1A1 | Cytochrome P450 1A1 | |
| CAFS_HM_T_00072318 | CYP3A27 | Cytochrome P450 3A27 | |
| CAFS_HM_T_00037413 | CYP4V2 | Cytochrome P450 4V2 | |
| CAFS_HM_T_00069566 | CYP4V3 | Cytochrome P450 4V3 | |
| CAFS_HM_T_00076880 | CYP8B1 | Sterol 12α-hydroxylase | |
| CAFS_HM_T_00031504 | CYP39A1 | Oxysterol 7α-hydroxylase | |
| CAFS_HM_T_00048267 | ACOX3 | Peroxisomal acyl-coenzyme A oxidase 3 | |
| CAFS_HM_T_00021903 | DEGS1 | Sphingolipid delta(4)-desaturase DES1 | |
| CAFS_HM_T_00044883 | HSD11B2 | Corticosteroid 11-beta-dehydrogenase isozyme 2 | |
| CAFS_HM_T_00037168 | Tdo2a | Tryptophan 2,3-dioxygenase A | |
| CAFS_HM_T_00086195 | Tmem195 | Alkylglycerol monooxygenase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Feng, C.; Sha, H.; Zhou, T.; Zou, G.; Liang, H. Tandem Mass Tagging-Based Quantitative Proteomics Analysis Reveals Damage to the Liver and Brain of Hypophthalmichthys molitrix Exposed to Acute Hypoxia and Reoxygenation. Antioxidants 2022, 11, 589. https://doi.org/10.3390/antiox11030589
Li X, Feng C, Sha H, Zhou T, Zou G, Liang H. Tandem Mass Tagging-Based Quantitative Proteomics Analysis Reveals Damage to the Liver and Brain of Hypophthalmichthys molitrix Exposed to Acute Hypoxia and Reoxygenation. Antioxidants. 2022; 11(3):589. https://doi.org/10.3390/antiox11030589
Chicago/Turabian StyleLi, Xiaohui, Cui Feng, Hang Sha, Tong Zhou, Guiwei Zou, and Hongwei Liang. 2022. "Tandem Mass Tagging-Based Quantitative Proteomics Analysis Reveals Damage to the Liver and Brain of Hypophthalmichthys molitrix Exposed to Acute Hypoxia and Reoxygenation" Antioxidants 11, no. 3: 589. https://doi.org/10.3390/antiox11030589
APA StyleLi, X., Feng, C., Sha, H., Zhou, T., Zou, G., & Liang, H. (2022). Tandem Mass Tagging-Based Quantitative Proteomics Analysis Reveals Damage to the Liver and Brain of Hypophthalmichthys molitrix Exposed to Acute Hypoxia and Reoxygenation. Antioxidants, 11(3), 589. https://doi.org/10.3390/antiox11030589






