Integrated Metabolomics and Transcriptomic Analysis of Hepatopancreas in Different Living Status Macrobrachium nipponense in Response to Hypoxia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Gather
2.2. Metabolites Extraction
2.3. GC-TOF-MS Analysis
2.4. Data Preprocessing and Annotation
2.5. Data Validation
2.5.1. RNA-Seq by qRT-PCR
2.5.2. Identification of Key Metabolites in M. nipponense
2.6. Statistical Analysis
3. Results
3.1. Principal Component Analysis (PCA) Analysis of Metabolism Profile
3.2. Significant Metabolites in Response to Hypoxia
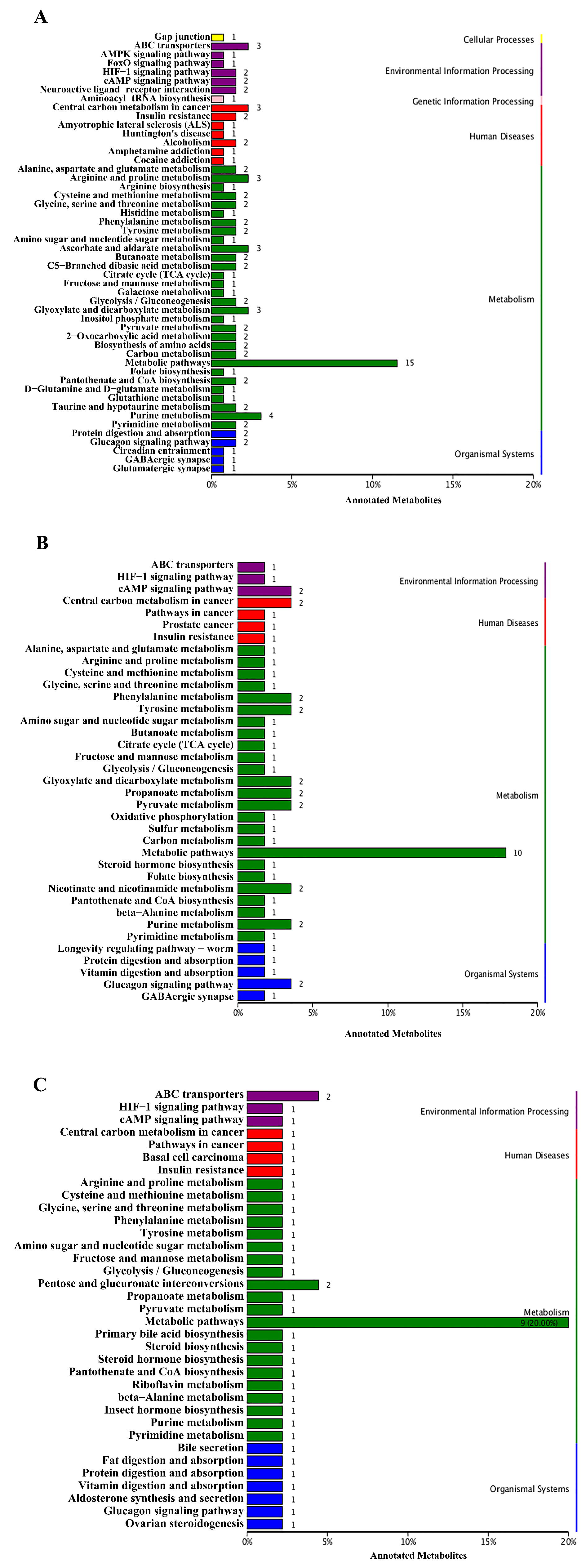
3.3. KEGG Pathway Analysis
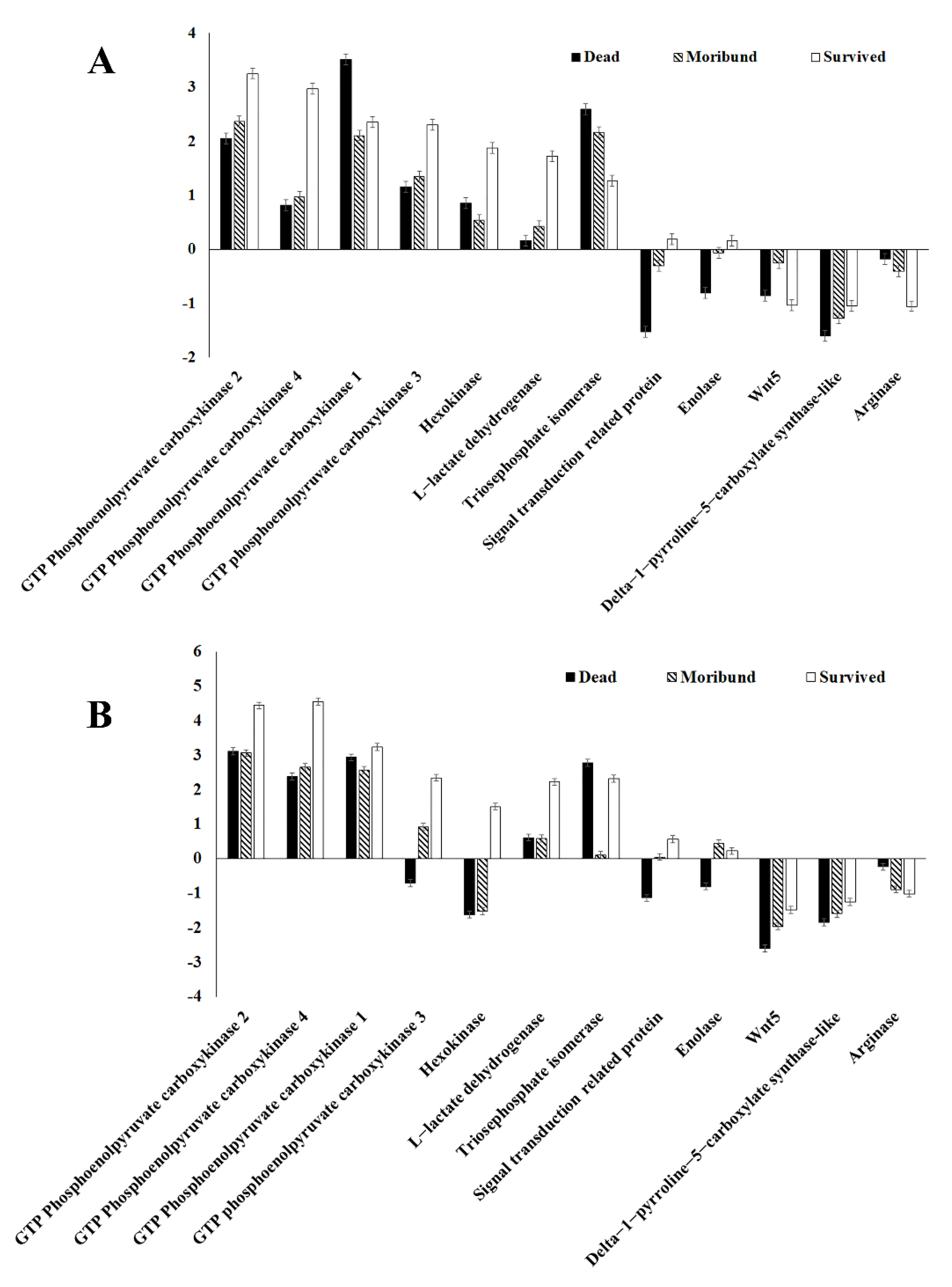
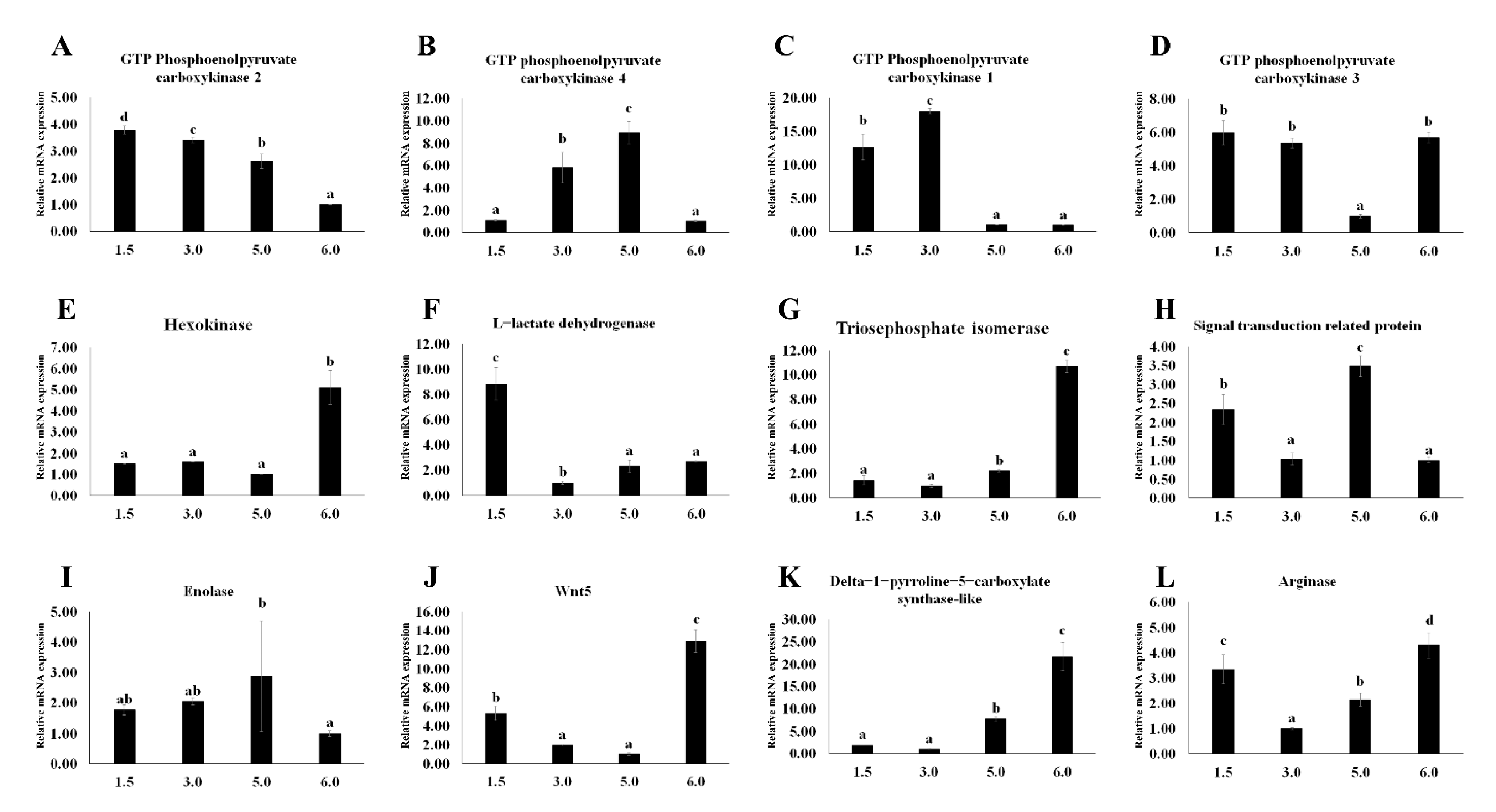
3.4. Gene Expression Validation
3.5. Subsequent Experimental Verification under Hypoxia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garlo, E.V.; Milstein, C.B.; Jahn, A.E. Impact of hypoxic conditions in the vicinity of little egg inlet, New Jersey in summer 1976. Estuar. Coast. Mar. Sci. 1979, 8, 421–432. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, C.; Zhang, D.; Jiang, H.; Peng, S.; Liu, Y.; Zhao, K.; Wang, C.; Chen, L. Analysis of the erythropoietin of a tibetan plateau schizothoracine fish (Gymnocypris dobula) reveals enhanced cytoprotection function in hypoxic environments. BMC Evol. Biol. 2016, 16, 11. [Google Scholar] [CrossRef] [Green Version]
- Law, S.H.; Wu, R.S.; Ng, P.K.; Richard, M.; Kong, R.Y. Cloning and expression analysis of two distinct HIF-alpha isoforms–gcHIF-1alpha and gcHIF-4alpha–from the hypoxia-tolerant grass carp, Ctenopharyngodon idellus. BMC Mol. Biol. 2006, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Rojas, D.A.; Perez-Munizaga, D.A.; Centanin, L.; Antonelli, M.; Wappner, P.; Allende, M.L.; Reyes, A.E. Cloning of hif-1a and hif-2a and mRNA expression pattern during development in zebrafish. Gene Expr. Patterns 2007, 7, 339–345. [Google Scholar] [CrossRef]
- Saydur Rahman, M.; Thomas, P. Molecular cloning, characterization and expression of two hypoxia-inducible factor alpha subunits, HIF-1αa and HIF-2α, in a hypoxia-tolerant marine teleost, Atlantic croaker (Micropogonias undulatus). Gene 2007, 396, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Chen, L.P.; Zhang, J.; Chen, C.; Wei, X.L.; Gul, Y.; Wang, W.M.; Wang, H.L. Molecular characterization and expression analysis of three hypoxia-inducible factor alpha subunits, HIF-1a/2a/3a of the hypoxia-sensitive freshwater species, Chinese sucker. Gene 2012, 498, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yang, M.; Fu, H.; Sun, S.; Qiao, H.; Zhang, W.; Gong, Y.; Jiang, S.; Xiong, Y.; Jin, S. Molecular cloning, expression, and in situ hybridization analysis of MnGPx-3 and MnGPx-4 from oriental river prawn, Macrobrachium nipponense, in response to hypoxia and reoxygenation. PLoS ONE 2020, 15, e0229171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Yang, M.; Fu, H.; Sun, S.; Qiao, H.; Zhang, W.; Gong, Y.; Jiang, S.; Xiong, Y.; Jin, S. Molecular cloning and expression of MnGST-1 and MnGST-2 from oriental river prawn, Macrobrachium nipponense, in response to hypoxia and reoxygenation. Int. J. Mol. Sci. 2018, 19, 3102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.F.; Tsai, S.Y.; Ma, M.C.; Wu, M.S. Hypoxic preconditioning enhances renal superoxide dismutase levels in rats. J. Physiol. 2003, 552, 561–569. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saetan, W.; Ye, M.; Lin, X.; Zhang, Y.; Huang, Y.; Du, T.; Li, G.; Tian, C. Comparative transcriptome analysis of heart tissue in response to hypoxia in Silver Sillago (Sillago sihama). J. Ocean Univ. China 2021, 20, 949–958. [Google Scholar] [CrossRef]
- Shi, W.; Wang, P.; Hu, R.; Wan, X.; Shen, H.; Li, H.; Wang, L.; Qiao, Y.; Jiang, G.; Cheng, J.; et al. Transcriptome analysis reveals hub genes in the hepatopancreas of Exopalaemon carinicauda in response to hypoxia and reoxygenation. Aqu. Int. 2021, 1–27. [Google Scholar] [CrossRef]
- Xu, L.; Fu, Y.; Fu, H.; Zhang, W.; Qiao, H.; Jiang, S.; Xiong, Y.; Jin, S.; Gong, Y.; Wang, Y.; et al. Transcriptome analysis of hepatopancreas from different living states oriental river prawn (Macrobrachium nipponense) in response to hypoxia. Comp. Biochem. Physiol. Part D 2021, 40, 100902. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Miao, Z.; Li, X.; Lin, H.; Cheng, Y.; Pan, J.; Xu, Z. Hypoxia-reoxygenation stress modulates the hepatopancreas transcriptome of Chinese mitten crab Eriocheir sinensis. Gene 2021, 771, 145361. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Tu, K.; Li, L.; Wu, B.; Wu, L.; Liu, Z.; Zhou, L.; Tian, J.; Yang, A. Integrated transcriptome and metabolome analysis reveals molecular responses of the clams to acute hypoxia. Mar. Environ. Res. 2021, 168, 105317. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Yuan, G.; Zhang, Q.; Xuan, L.; Li, J.; Zhou, L.; Shi, H.; Wang, X.; Wang, C. Transcriptome and metabolome analyses reveal the pivotal role of hydrogen sulfide in promoting submergence tolerance in Arabidopsis. Environ. Exp. Bot. 2021, 183, 104365. [Google Scholar] [CrossRef]
- Su, L.; Yang, C.; Meng, J.; Zhou, L.; Zhang, C. Comparative transcriptome and metabolome analysis of Ostrinia furnacalis female adults under UV-A exposure. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, A.; Freitas, R.; Ferreira, D.; Relvas-Santos, M.; Paulo, P.; Cardoso, M.; Soares, J.; Gaiteiro, C.; Palmeira, C.; Teixeira, F.; et al. Metabolomics, transcriptomics and functional glycomics reveals bladder cancer cells plasticity and enhanced aggressiveness facing hypoxia and glucose deprivation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Yang, M.; Sun, S.; Fu, H.; Qiao, H.; Zhang, W.; Gong, Y.; Jiang, S.; Xiong, Y.; Xu, L.; Zhao, C.; et al. Hypoxia and reoxygenation on antioxidant enzyme activities and histological structure of Macrobrachium nipponense. J. Fish Sci. China 2019, 26, 493–503. [Google Scholar] [CrossRef]
- Sun, S.; Xuan, F.; Fu, H.; Zhu, J.; Ge, X.; Gu, Z. Transciptomic and histological analysis of hepatopancreas, muscle and gill tissues of oriental river prawn (Macrobrachium nipponense) in response to chronic hypoxia. BMC Genom. 2015, 16, 491. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Xuan, F.; Ge, X.; Fu, H.; Zhu, J.; Zhang, S. Identification of differentially expressed genes in hepatopancreas of oriental river prawn, Macrobrachium nipponense exposed to environmental hypoxia. Gene 2014, 534, 298–306. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, A.; Barbas, C. Gas chromatography-mass spectrometry (GC-MS)-based metabolomics. Metab. Profiling Hum. Press 2011, 191–204. [Google Scholar] [CrossRef]
- Hu, Y.; Fu, H.; Qiao, H.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Validation and evaluation of reference genes for quantitative real-time PCR in Macrobrachium nipponense. Int. J. Mol. Sci. 2018, 19, 2258. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Qiao, H.; Fu, H.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y.; et al. Identification and characterization of opsin gene and its role in ovarian maturation in the oriental river prawn Macrobrachium nipponense. Comp. Biochem. Physiol. Part B 2018, 218, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Webster, K.A. Evolution of the coordinate regulation of glycolytic enzyme genes by hypoxia. J. Exp. Biol. 2003, 206, 2911–2922. [Google Scholar] [CrossRef] [Green Version]
- Nie, H.; Wang, H.; Jiang, K.; Yan, X. Transcriptome analysis reveals differential immune related genes expression in Ruditapes philippinarum under hypoxia stress: Potential HIF and NF-κB crosstalk in immune responses in clam. BMC Genom. 2020, 21, 1–16. [Google Scholar] [CrossRef] [Green Version]
- McClelland, G.B.; Brooks, G.A. Changes in MCT 1, MCT 4, and LDH expression are tissue specific in rats after long-term hypobaric hypoxia. J. Appl. Physiol. 2002, 92, 1573–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, J.; Li, X.; Yu, H.; Jiang, H. Respiratory metabolism responses of Chinese mitten crab, Eriocheir sinensis and Chinese grass shrimp, Palaemonetes sinensis, subjected to environmental hypoxia stress. Front. Physiol. 2018, 9, 1559. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, V. Multifunctional alpha-enolase: Its role in diseases. Cell Mol. Life Sci. 2001, 58, 902–920. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordet, J.; Maire, P.; Giallongo, A. Hypoxia response elements in the aldolase A.; enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [Green Version]
- Sedoris, K.C.; Thomas, S.D.; Miller, D.M. Hypoxia induces differential translation of enolase/MBP-1. BMC Cancer 2010, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Riddle, S.R.; Ahmad, A.; Ahmad, S.; Deeb, S.S.; Malkki, M.; Schneider, B.K.; Allen, C.B.; White, C.W. Hypoxia induces hexokinase II gene expression in human lung cell line A549. Am. J. Physiol. Lung Cell Mol. Physiol. 2000, 278, L407–L416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Xuan, F.; Fu, H.; Zhu, J.; Ge, X. Molecular cloning and functional characterization of a hexokinase from the oriental river prawn Macrobrachium nipponense in response to hypoxia. Int. J. Mol. Sci. 2017, 18, 1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosas, C.; Cuzon, G.; Gaxiola, G.; Le Priol, Y.; Pascual, C.; Rossignyol, J.; Contreras, F.; Sa’nchez, A.; van Wormhoudt, A. Metabolism and growth of juveniles of Litopenaeus vannamei: Effect of salinity and dietary carbohydrates level. J. Exp. Mar. Biol. Ecol. 2001, 259, 1–22. [Google Scholar] [CrossRef]
- Rossignol, F.; Solares, M.; Balanza, E.; Coudert, J.; Clottes, E. Expression of lactate dehydrogenase A and B genes in different tissues of rats adapted to chronic hypobaric hypoxia. J. Cell. Biochem. 2003, 89, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Gaxiola, G.; Cuzon, G.; Garcia, T.; Taboada, G.; Brito, R.; Chimal, M.E.; Paredes, A.; Soto, L.; Rosas, C.; van Wormhoudt, A. Factorial effects of salinity, dietary carbohydrate and moult cycle on digestive carbohydrases and hexokinases in Litopenaeus vannamei (Boone, 1931). Comp. Biochem. Physiol. A 2005, 140, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Leegood, R.C.; Walker, R.P. Regulation and roles of phosphoenolpyruvate carboxykinase in plants. Arch. Biochem. Biophys. 2003, 414, 204–210. [Google Scholar] [CrossRef]
- Westermeier, F.; Holyoak, T.; Gatica, R.; Martínez, F.; Negron, Ḿ.; Yánez, Ã.J.; Nahmias, D.; Nualart, F.; Burbulis, I.; Bertinat, R. Cytosolic phosphoenolpyruvate carboxykinase is expressed in a-cells from human and murine pancreas. J. Cell. Physiol. 2020, 235, 166–175. [Google Scholar] [CrossRef]
- Hara, Y.; Watanabe, N. Changes in expression of genes related to glucose metabolism in liver and skeletal muscle of rats exposed to acute hypoxia. Heliyon 2020, 6, e04334. [Google Scholar] [CrossRef] [PubMed]
- Martin-Requero, A.; Ayuso, M.S.; Parrilla, R. Rate-limiting steps for hepatic gluconeogenesis mechanism of oxamate inhibition of mitochondrial pyruvate metabolism. J. Biol. Chem. 1986, 261, 13973–13978. [Google Scholar] [CrossRef]
- Gabbay, R.A.; Sutherland, C.; Gnudi, L.; Kahn, B.B.; O’brien, R.M.; Granner, D.K.; Flier, J.S. Insulin regulation of phosphoenolpyruvate carboxykinase gene expression does not require activation of the Ras/mitogen-activated protein kinase signaling pathway. J. Biol. Chem. 1996, 271, 1890–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owczarek, A.; Gieczewska, K.; Jarzyna, R.; Jagielski, A.K.; Kiersztan, A.; Gruza, A.; Winiarska, K. Hypoxia increases the rate of renal gluconeogenesis via hypoxia-inducible factor-1-dependent activation of phosphoenolpyruvate carboxykinase expression. Biochimie 2020, 171, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Ramos, C.A.; Peregrino-Uriarte, A.B.; Cota-Ruiz, K.; Valenzuela-Soto, E.M.; Leyva Carrillo, L.; Yepiz-Plascencia, G. Phosphoenolpyruvate carboxykinasecytosolic and mitochondrial isoforms are expressed and active during hypoxia in thewhite shrimp Litopenaeus vannamei. Comp. Biochem. Physiol. B Biochem. Mol.Biol. 2018, 226, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wierenga, R.K.; Kapetaniou, E.G.; Venkatesan, R. Triosephosphate isomerase: A highly evolved biocatalyst. Cell Mol Life Sci. 2010, 67, 3961–9682. [Google Scholar] [CrossRef]
- Wang, B.; Li, F.; Dong, B.; Zhang, X.; Zhang, C.; Xiang, J. Discovery of the genes in response to white spot syndrome virus (WSSV) infection in Fenneropenaeus chinensis through cDNA microarray. Mar. Biotechnol. 2006, 8, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Zavala, A.A.; Carrasco-Miranda, J.S.; Ramirez-Aguirre, C.D.; López-Hidalgo, M.; Benitez-Cardoza, C.G.; Ochoa-Leyva, A.; Cardona-Felix, C.S.; Diaz-Quezada, C.; Rudiño-Piñera, E.; SoteloMundo, R.R.; et al. Structural insights from a novel invertebrate triosephosphate isomerase from Litopenaeus Vannamei. Biochim. Biophys. Acta BBA-Proteins Proteom. 2016, 1864, 1696–1706. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Zhou, J.; Wang, H.; Mu, C.; Shi, C.; Liu, L.; Wang, C. Transcriptome analysis of genes and pathways associated with metabolism in Scylla paramamosain under different light intensities during indoor overwintering. BMC Genom. 2020, 21, 1–15. [Google Scholar] [CrossRef]
- Prathibha, Y.; Senthilkumaran, B. Expression of wnt4/5 during reproductive cycle of catfish and wnt5 promoter analysis. J. Endocrinol. 2017, 232, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Schleiffarth, J.; Jessurun, J.; Sumanas, S.; Petryk, A.; Lin, S.; Ekker, S. Wnt5 signaling in vertebrate pancreas development. BMC Biol. 2005, 3, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Xiao, A.; Choi, S.Y.; Kan, Q.; Zhou, W.; Chacon-Heszele, M.F.; Ryu, Y.K.; McKenna, S.; Zuo, X.; Kuruvilla, R.; et al. Wnt5a is necessary for normal kidney development in zebrafish and mice. Nephron Exp. Nephrol. 2014, 128, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Kilian, B.; Mansukoski, H.; Barbosa, F.; Ulrich, F.; Tada, M.; Heisenberg, C. The role of Ppt/Wnt5 in regulating cell shape and movement during zebrafish gastrulation. Mech. Dev. 2003, 120, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, C.Z.; Yang, Q.H.; Dong, X.; Chi, S.; Liu, H.; Shi, L.; Tan, B. Molecular cloning, characterization and expression analysis of Wnt4, Wnt5, Wnt6, Wnt7, Wnt10 and Wnt16 from Litopenaeus Vannamei. Fish Shellfish. Immunol. 2016, 54, 445–455. [Google Scholar] [CrossRef]
- Du, J.; Zhang, X.; Yuan, J.; Zhang, X.; Li, F.; Xiang, J. Wnt gene family members and their expression profiling in Litopenaeus Vannamei. Fish Shellfish. Immunol. 2018, 77, 233–243. [Google Scholar] [CrossRef]
- Baril, M.; Es-Saad, S.; Chatel-Chaix, L.; Fink, K.; Pham, T.; Raymond, V.A.; Audette, K.; Guenier, A.; Duchaine, J.; Servant, M.; et al. Genome-wide RNAi screen reveals a new role of a WNT/CTNNB1 signalingpathway as negative regulator of virus-induced innate immune responses. PLoS Pathog. 2013, 9, e1003416. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Pan, L.; Wei, C.; Tong, R.; Li, Y.; Ding, M.; Wang, H. Crustacean hyperglycemic hormone (CHH) regulates the ammonia excretion and metabolism in white shrimp, Litopenaeus vannamei under ammonia-N stress. Sci. Total Environ. 2020, 723, 138128. [Google Scholar] [CrossRef]
- Hu, X.; Zhou, C.; He, G.; Cheng, Y.; Pan, M.; Gao, Y. Inhibition of Frizzled-2 by small interfering RNA protects rat hepatic BRL-3A cells against cytotoxicity and apoptosis induced by Hypoxia/Reoxygenation. Gastroenterol. Y Hepatol. 2020, 43, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Tang, J.; Mu, D.Z. Bioinformatic Analysis of Differentially Expressed Genes Involved in the Post-hypoxic Ischemic Brain Damage of Newborn Rats. J. Sichuan Univ. 2016, 47, 722–726. [Google Scholar]
- Hu, C.A.A.; Khalil, S.; Zhaorigetu, S.; Liu, Z.; Tyler, M.; Wan, G.; Valle, D. Human Δ 1-pyrroline-5-carboxylate synthase: Function and regulation. Amino Acids 2008, 35, 665–672. [Google Scholar] [CrossRef] [Green Version]
- Baumgartner, M.R.; Rabier, D.; Nassogne, M.-C.; Dufier, J.-L.; Padovani, J.-P.; Kamoun, P.; Valle, D.; Saudubray, J.-M. Δ1-pyrroline-5-carboxylate synthase deficiency: Neurodegeneration, cataracts and connective tissue manifestations combined with hyperammonaemia and reduced ornithine, citrulline, arginine and proline. Eur. J. Pediatrics 2005, 164, 31–36. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, X.; Shang, W.; Liu, Y.; Ji, J.F.; Liu, J.P.; Tong, C. Pyrroline-5-carboxylate synthase senses cellular stress and modulates metabolism by regulating mitochondrial respiration. Cell Death Differ. 2021, 28, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Goharrizi, K.J.; Baghizadeh, A.; Afroushteh, M.; Amirmahani, F.; Kermani, S.G. Effects of salinity stress on proline content and expression of Δ1-pyrroline-5-carboxylate synthase and vacuolar-type H+ subunit E genes in wheat. Plant Genet. Resour. 2020, 18, 334–342. [Google Scholar] [CrossRef]
- Liang, Q.J.; Wu, X.J.; Yang, P.; Kong, J.; Wei, W.; Qiao, X.; Liu, Y.; Wang, W. The role of delta-1-pyrroline-5-carboxylate dehydrogenase (P5CDh) in the Pacific white shrimp (Litopenaeus vannamei) during biotic and abiotic stress. Aquat. Toxicol. 2019, 208, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.A.; Hou, Y. Mammalian P5CR and P5CDH protein structure and disease associationc. Am. For. 2014, 1, 1935–1937. [Google Scholar] [CrossRef]
- Palmer, R.; Ashton, A.; Moncada, S. Vascular endothelial cells synthesize nitric oxide from l-arginine. Nature 1998, 333, 664–666. [Google Scholar] [CrossRef]
- Ignarro, L.F.; Buga, G.M.; Wei, L.H.; Bauer, P.M.; Wu, G.; del Soldato, P. Role of the arginine-nitric oxide pathway in the regulation of vascular smooth muscle cell proliferation. Proc. Natl. Acad. Sci. USA 2001, 98, 4202–4208. [Google Scholar] [CrossRef] [Green Version]
- Kourembanas, S.; McQuillan, L.P.; Leung, G.K.; Faller, D.V. Nitric oxide regulates the expression of vasoconstrictors and growth factors by vascular endothelium under both normoxia and hypoxia. J. Clin. Investig. 1993, 92, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Pietra, G.G.; Capron, F.; Stewart, S.; Leone, O.; Humbert, M.; Robbins, I.M.; Reid, L.M.; Tuder, R. Pathologic assessment of vasculopathies in pulmonary hypertension. J. Am. Coll. Cardiol. 2004, 43, 25S–32S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Calvert, A.E.; Cui, H.; Nelin, L.D. Hypoxia promotes human pulmonary artery smooth muscle cell proliferation through induction of arginase. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2009, 297, L1151–L1159. [Google Scholar] [CrossRef] [Green Version]
- Nelin, L.D.; Chicoine, L.G.; Reber, K.M.; English, B.K.; Young, T.L.; Liu, Y. Cytokine-induced endothelial arginase expression is dependent on epidermal growth factor receptor. Am. J. Respir. Cell. Mol. Biol. 2005, 33, 394–401. [Google Scholar] [CrossRef] [Green Version]
- Ino, Y.; Yamazaki-Itoh, R.; Oguro, S.; Shimada, K.; Kosuge, T.; Zavada, J.; Kanai, Y.; Hiraoka, N. Arginase II expressed in cancer-associated fibroblasts indicates tissue hypoxia and predicts poor outcome in patients with pancreatic cancer. PLoS ONE 2013, 8, e55146. [Google Scholar] [CrossRef] [PubMed] [Green Version]

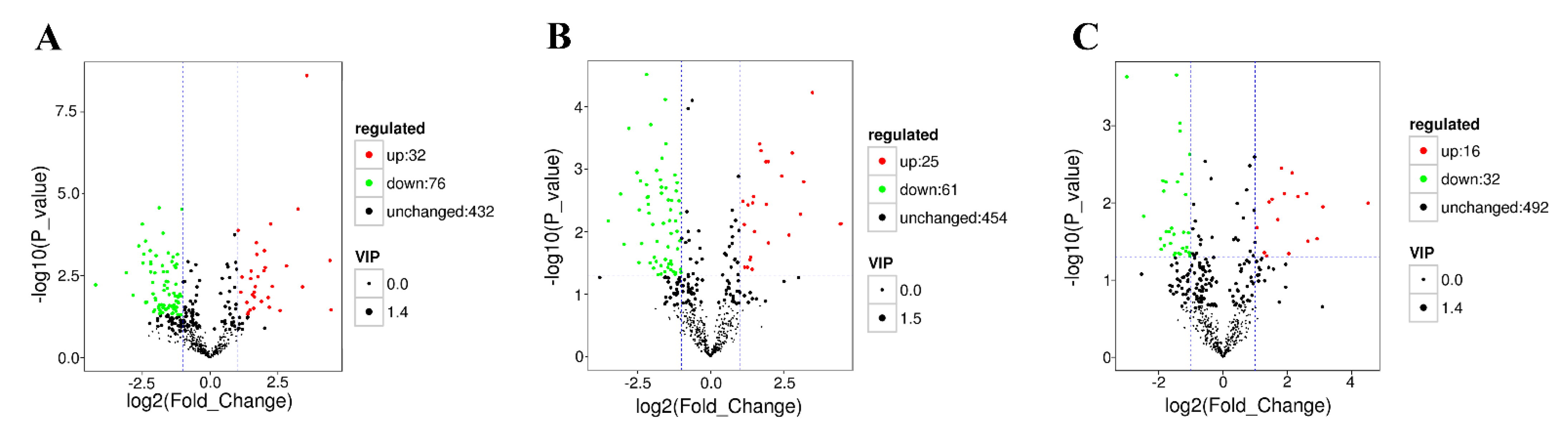
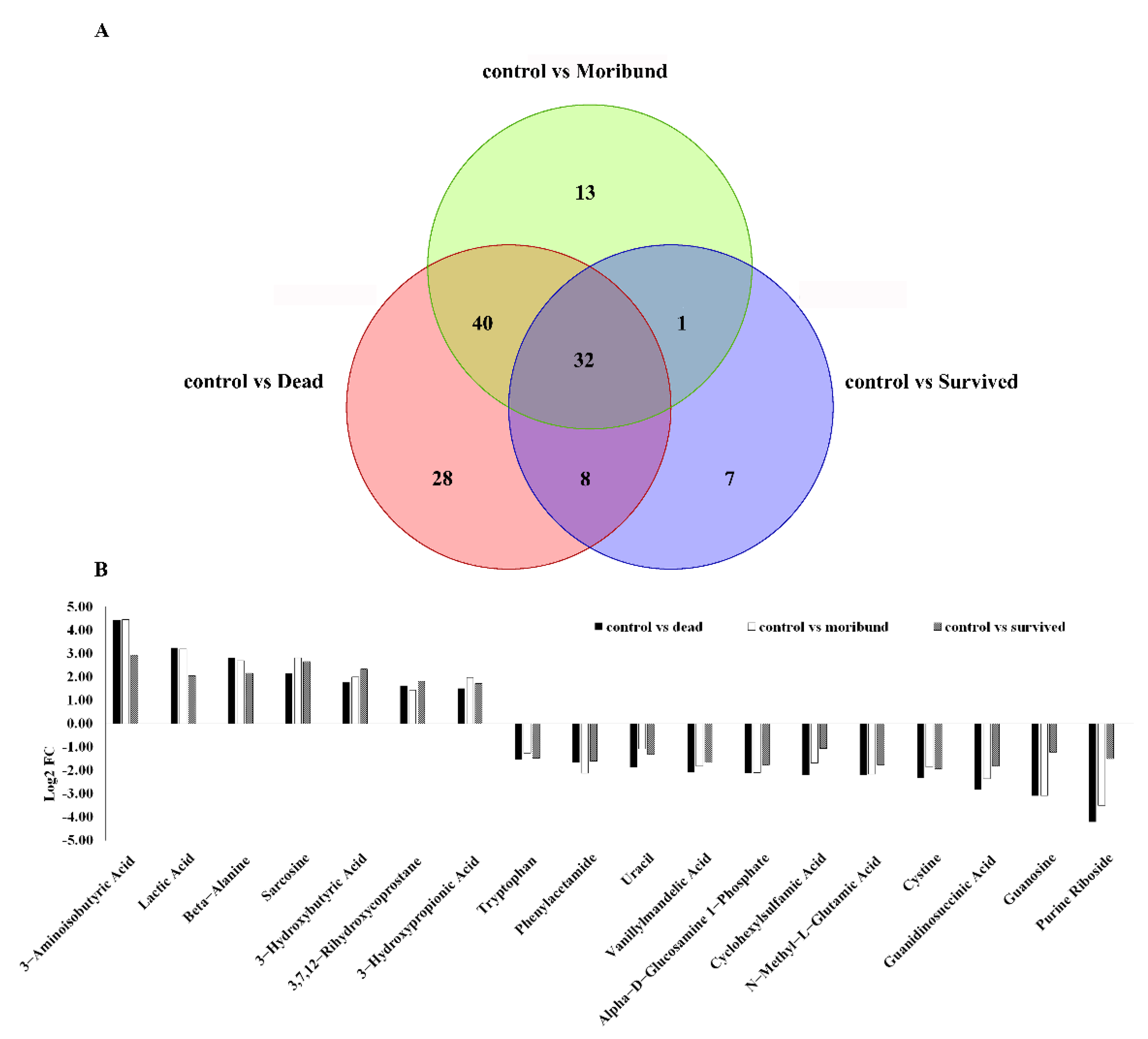

| All Different Metabolites | Down | Up | Number of Pathways | |
|---|---|---|---|---|
| Control vs. Dead | 108 | 76 | 32 | 81 |
| Control vs. Moribund | 86 | 61 | 25 | 37 |
| Control vs. Survived | 48 | 32 | 16 | 35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Zhang, W.; Qiao, H.; Jiang, S.; Xiong, Y.; Jin, S.; Gong, Y.; Fu, H. Integrated Metabolomics and Transcriptomic Analysis of Hepatopancreas in Different Living Status Macrobrachium nipponense in Response to Hypoxia. Antioxidants 2022, 11, 36. https://doi.org/10.3390/antiox11010036
Xu L, Zhang W, Qiao H, Jiang S, Xiong Y, Jin S, Gong Y, Fu H. Integrated Metabolomics and Transcriptomic Analysis of Hepatopancreas in Different Living Status Macrobrachium nipponense in Response to Hypoxia. Antioxidants. 2022; 11(1):36. https://doi.org/10.3390/antiox11010036
Chicago/Turabian StyleXu, Lei, Wenyi Zhang, Hui Qiao, Sufei Jiang, Yiwei Xiong, Shubo Jin, Yongsheng Gong, and Hongtuo Fu. 2022. "Integrated Metabolomics and Transcriptomic Analysis of Hepatopancreas in Different Living Status Macrobrachium nipponense in Response to Hypoxia" Antioxidants 11, no. 1: 36. https://doi.org/10.3390/antiox11010036
APA StyleXu, L., Zhang, W., Qiao, H., Jiang, S., Xiong, Y., Jin, S., Gong, Y., & Fu, H. (2022). Integrated Metabolomics and Transcriptomic Analysis of Hepatopancreas in Different Living Status Macrobrachium nipponense in Response to Hypoxia. Antioxidants, 11(1), 36. https://doi.org/10.3390/antiox11010036








