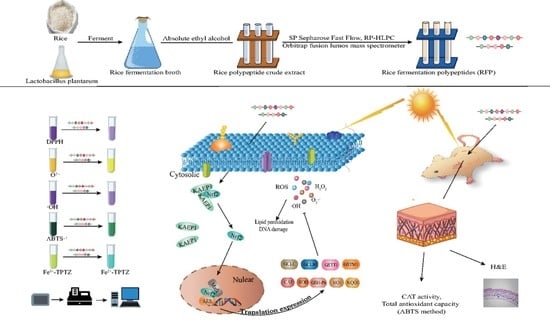
Purification and Identification of Antioxidant Peptides from Rice Fermentation of Lactobacillus plantarum and Their Protective Effects on UVA−Induced Oxidative Stress in Skin
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Rice Fermentation Broth
2.3. Purification and Identification of Antioxidant Peptides from Fermentation Broth
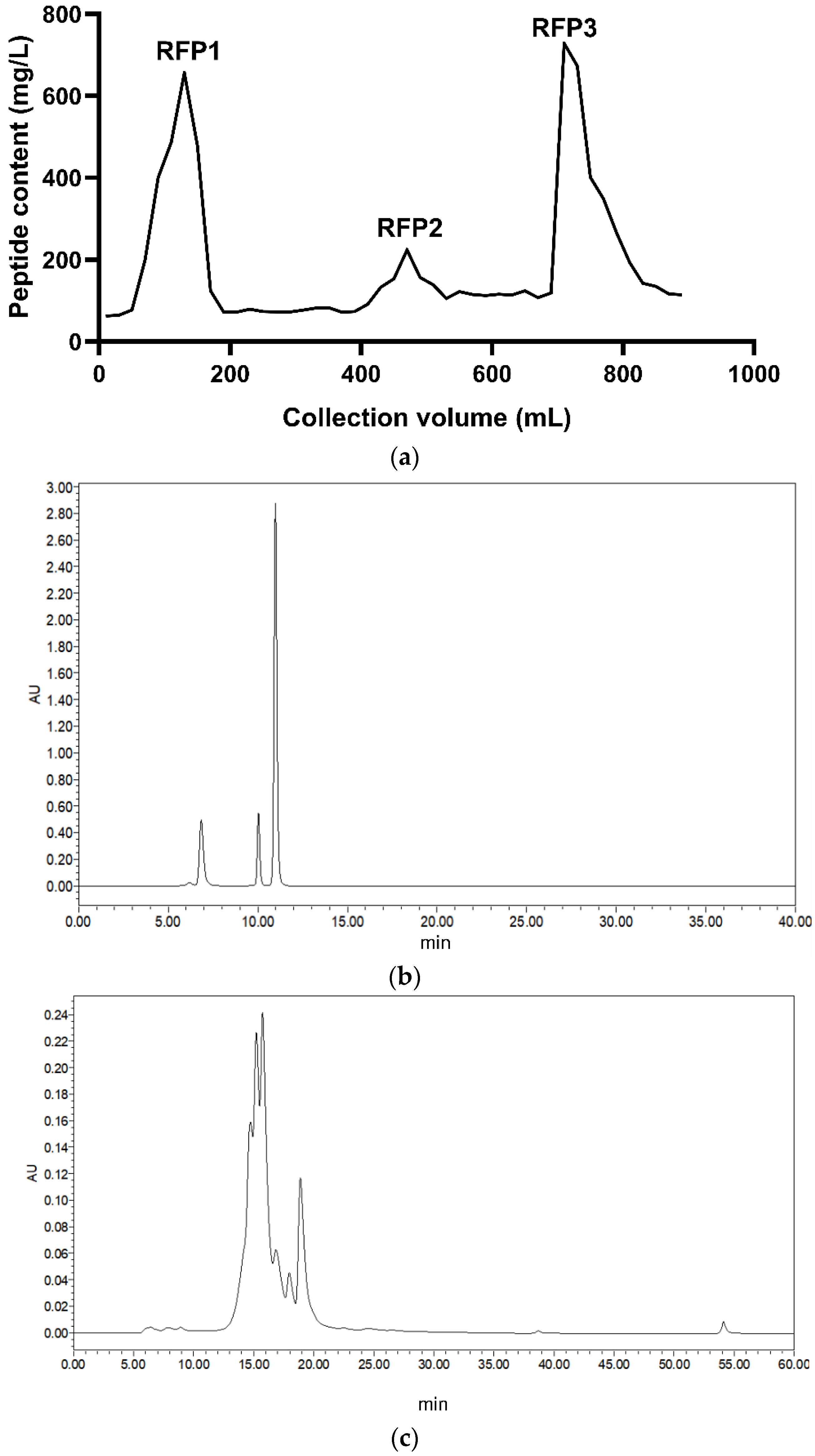
2.3.1. Separation and Purification
2.3.2. Molecular Weight Detection
2.3.3. Peptide Sequencing
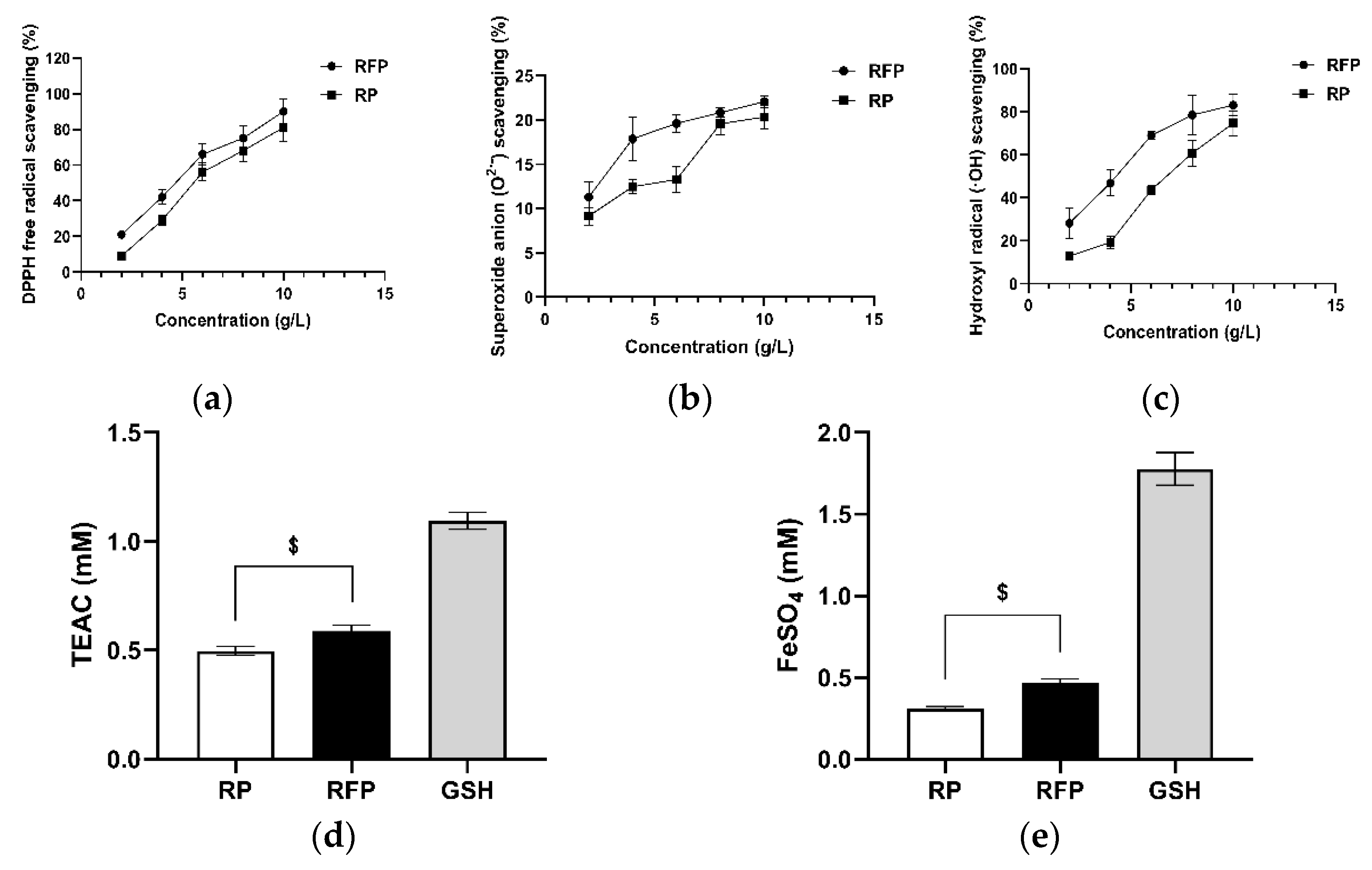
2.4. Determination of Antioxidant Capacity of RFP In Vitro
2.5. Protective Effects of RFP on UVA Damaged Cells
2.5.1. Cell Resuscitation and Culture
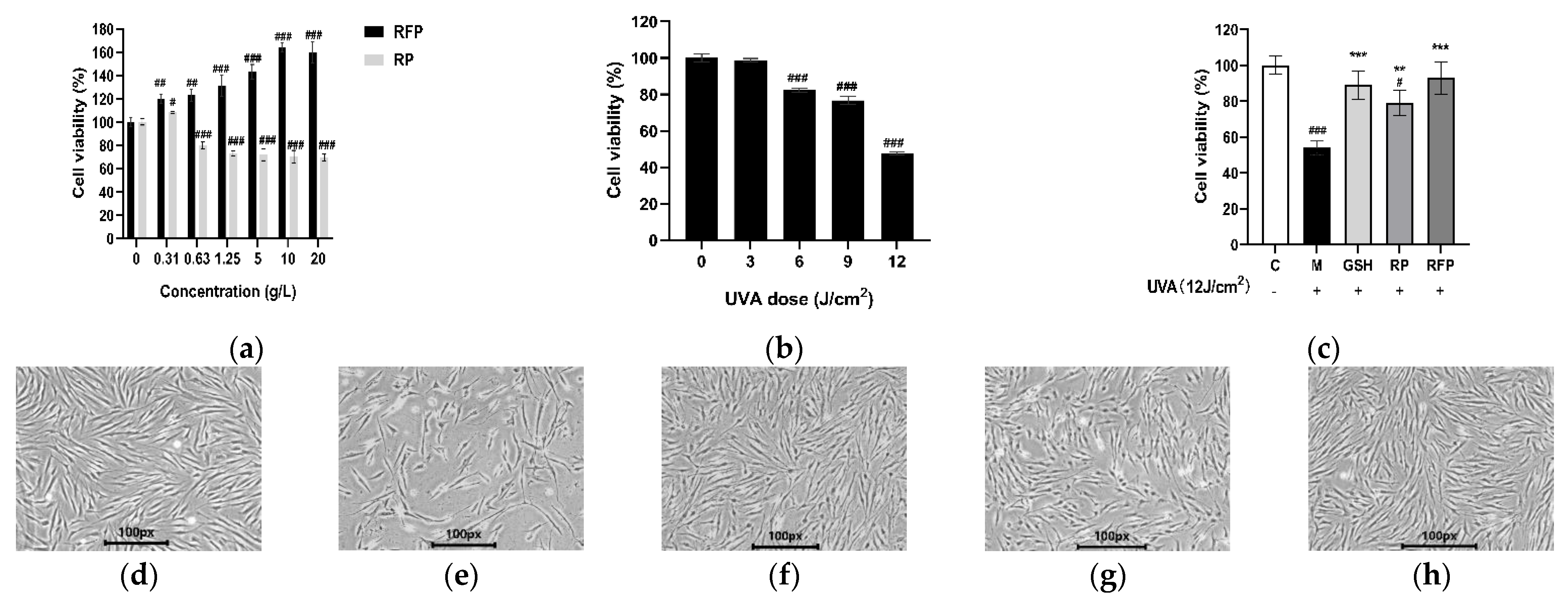
2.5.2. Cell Viability Assay
2.5.3. Determination of ROS
2.5.4. Determination of MDA, Total Antioxidant Capacity, and Three Antioxidant Enzymes
2.5.5. Transcriptome Sequencing Analysis
2.5.6. RT−qPCR
2.5.7. ELISA
2.6. Experiments on Animals
2.6.1. Establishment of UVA Oxidative Damage Mouse Skin Model
2.6.2. Tissue Sample Collection and Preparation
2.7. Statistical Analysis
3. Results
3.1. Purification and Identification of Antioxidant Peptides from Rice Fermentation Broth
3.2. In Vitro Antioxidant Capacity
3.3. Protective Effects of Antioxidant Peptides on UVA−Induced Oxidative Stress of HSF Cells
3.3.1. Cell Viability
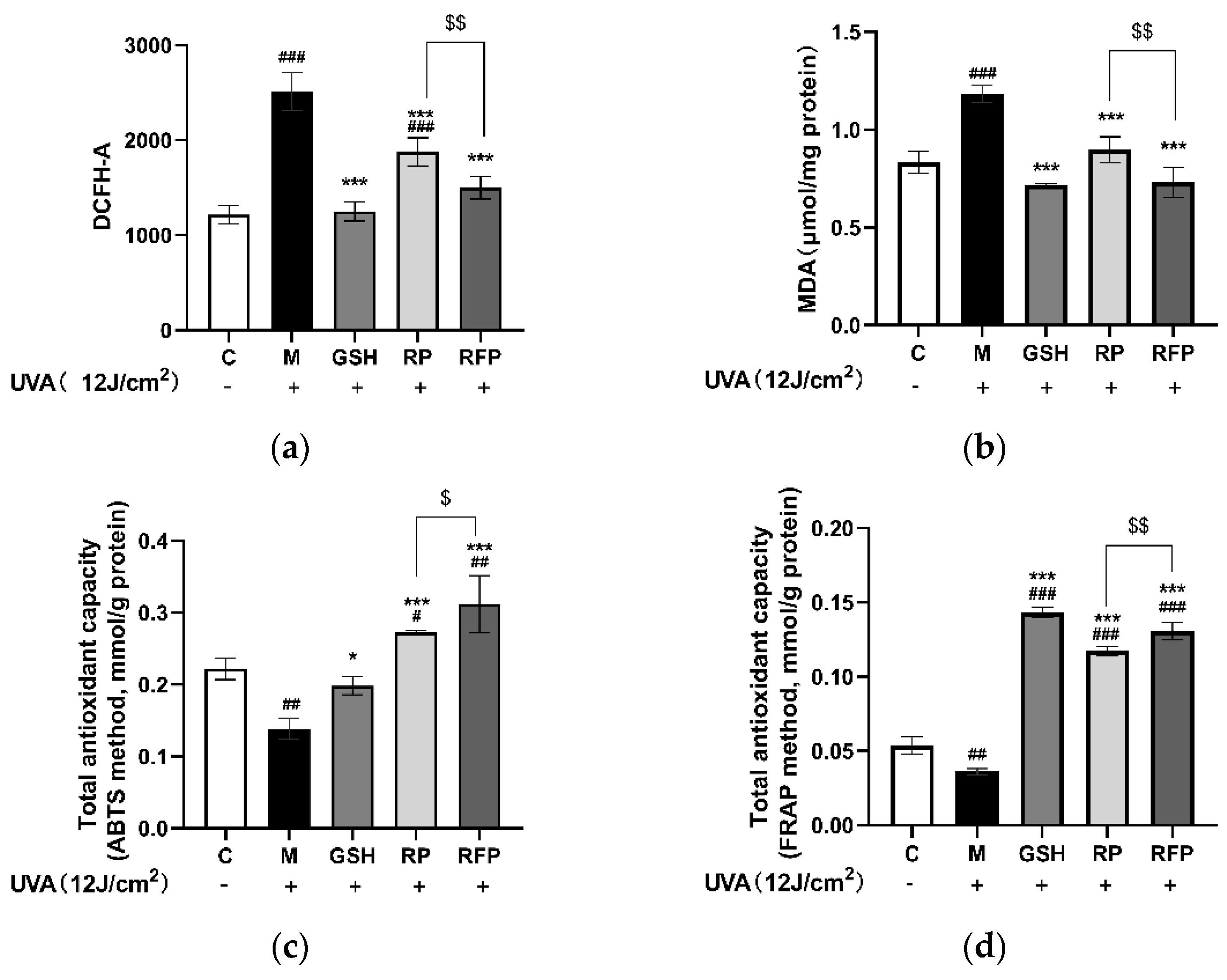
3.3.2. Antioxidation
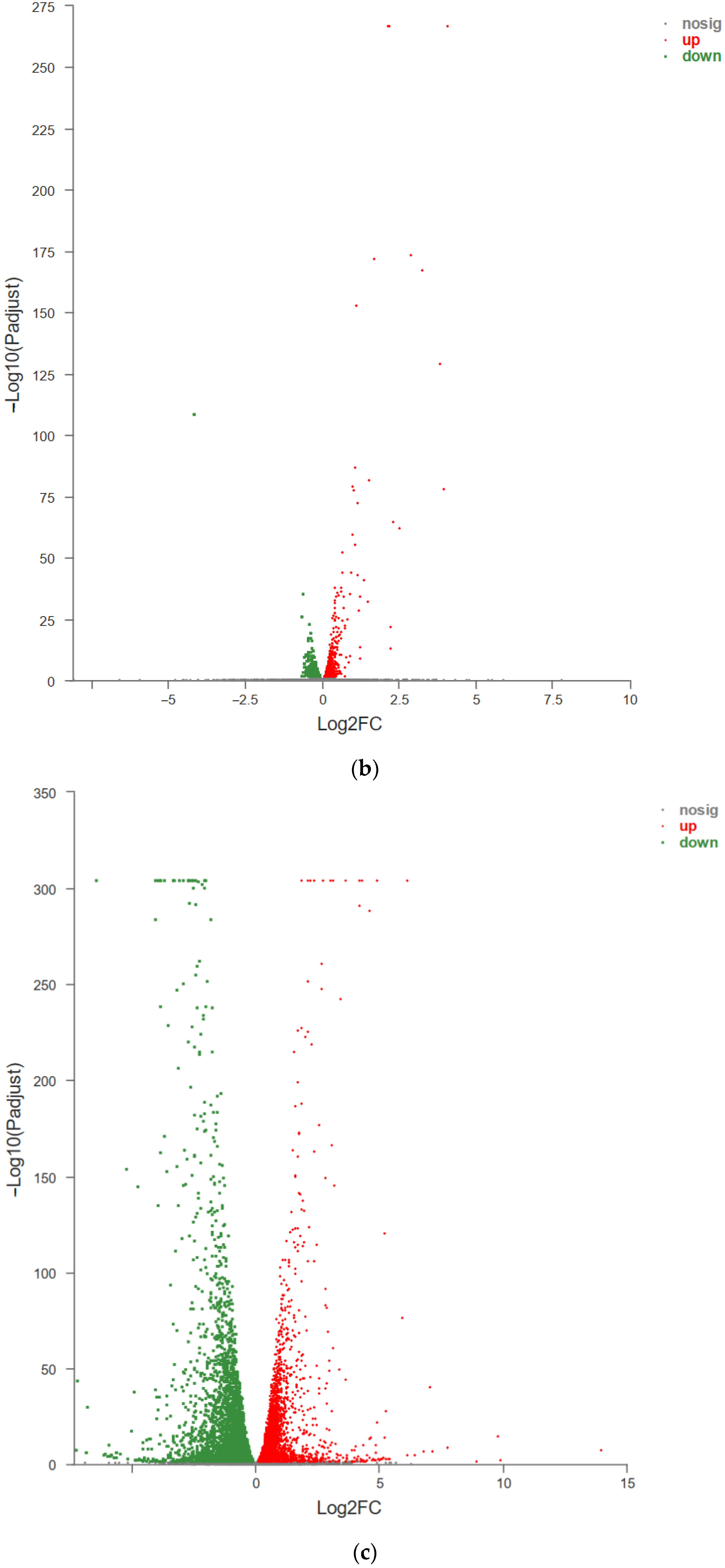
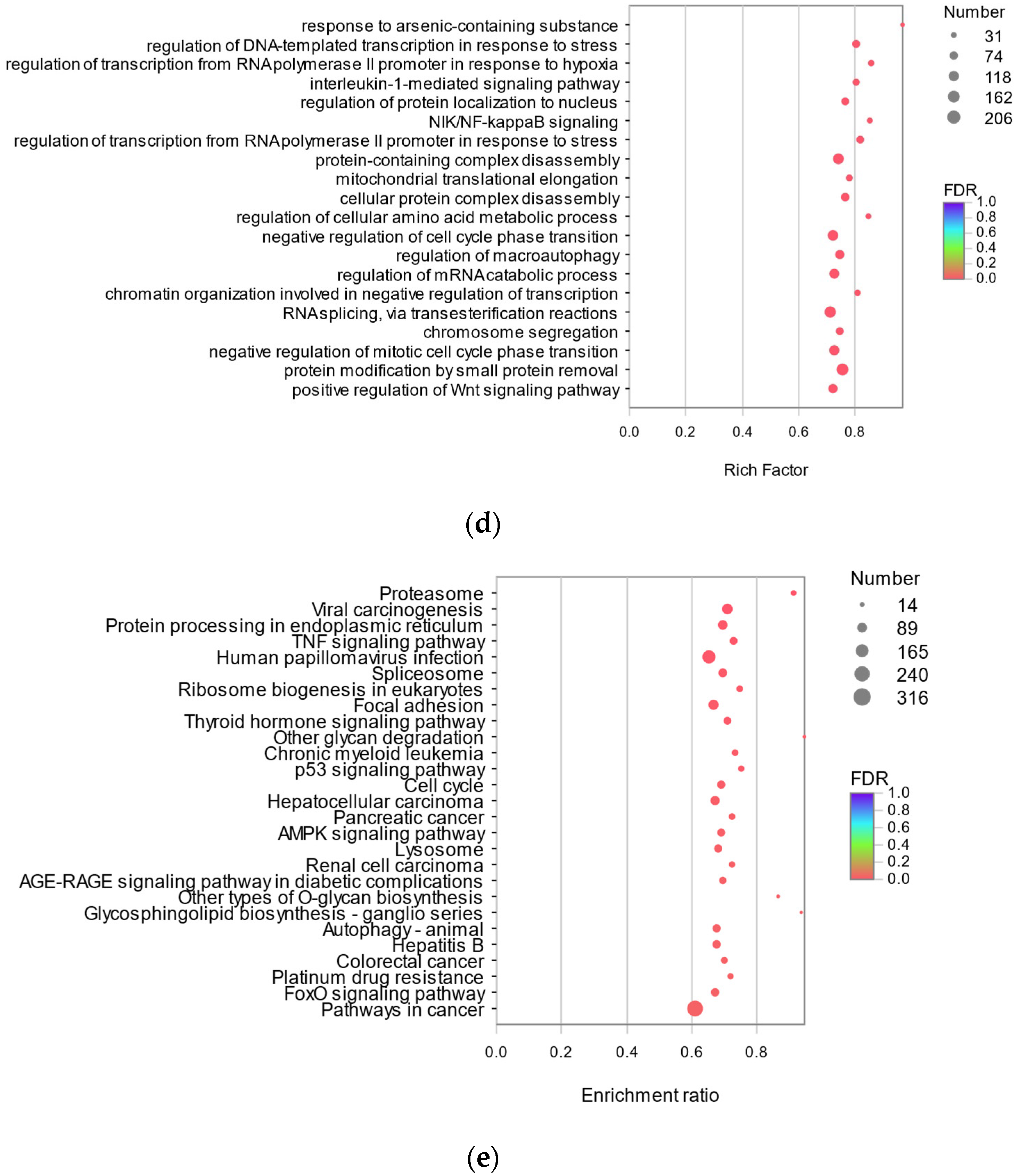
3.3.3. Analysis of Protective Mechanism of RFP against UVA Oxidative Stress in HSF Cells Based on Transcriptome
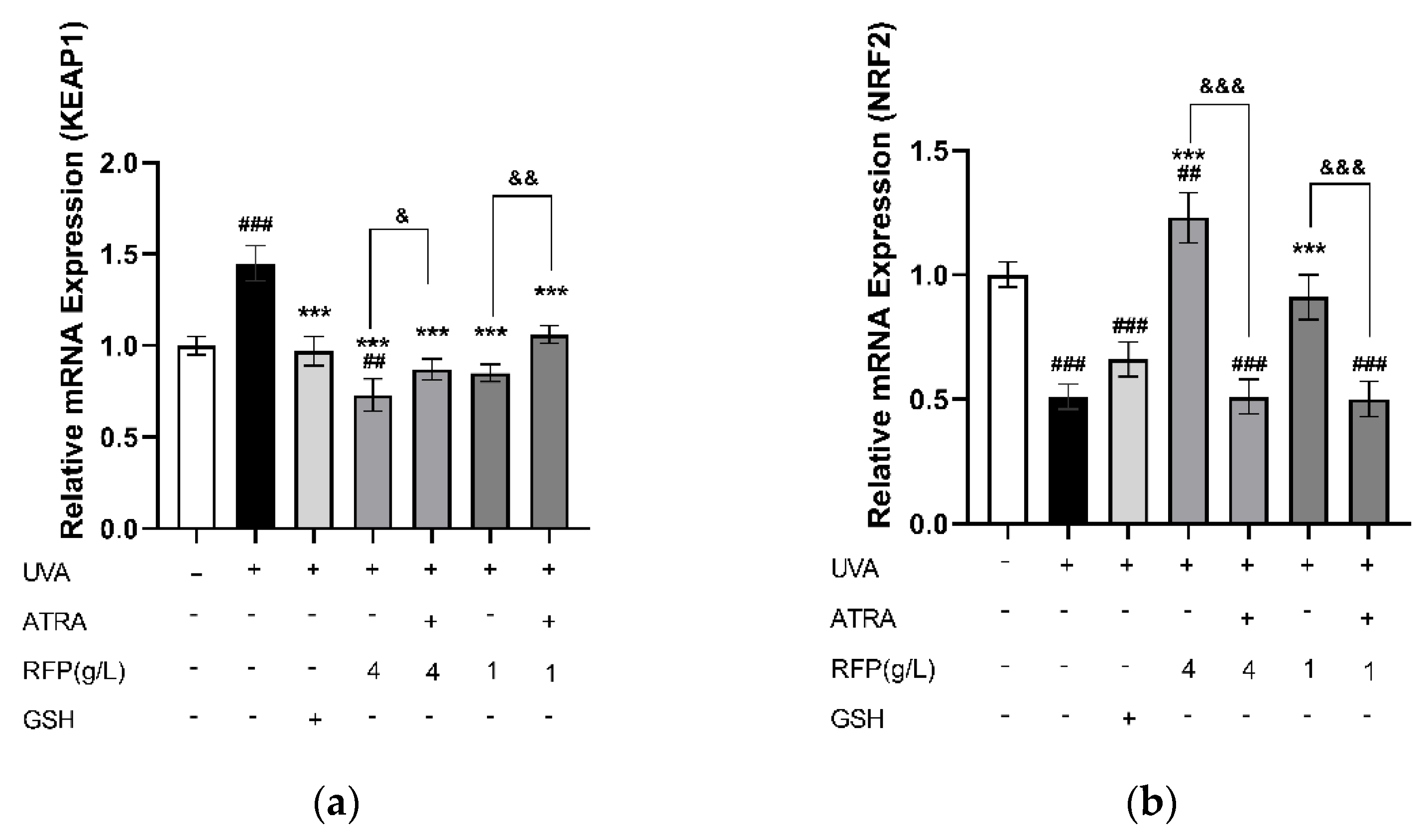
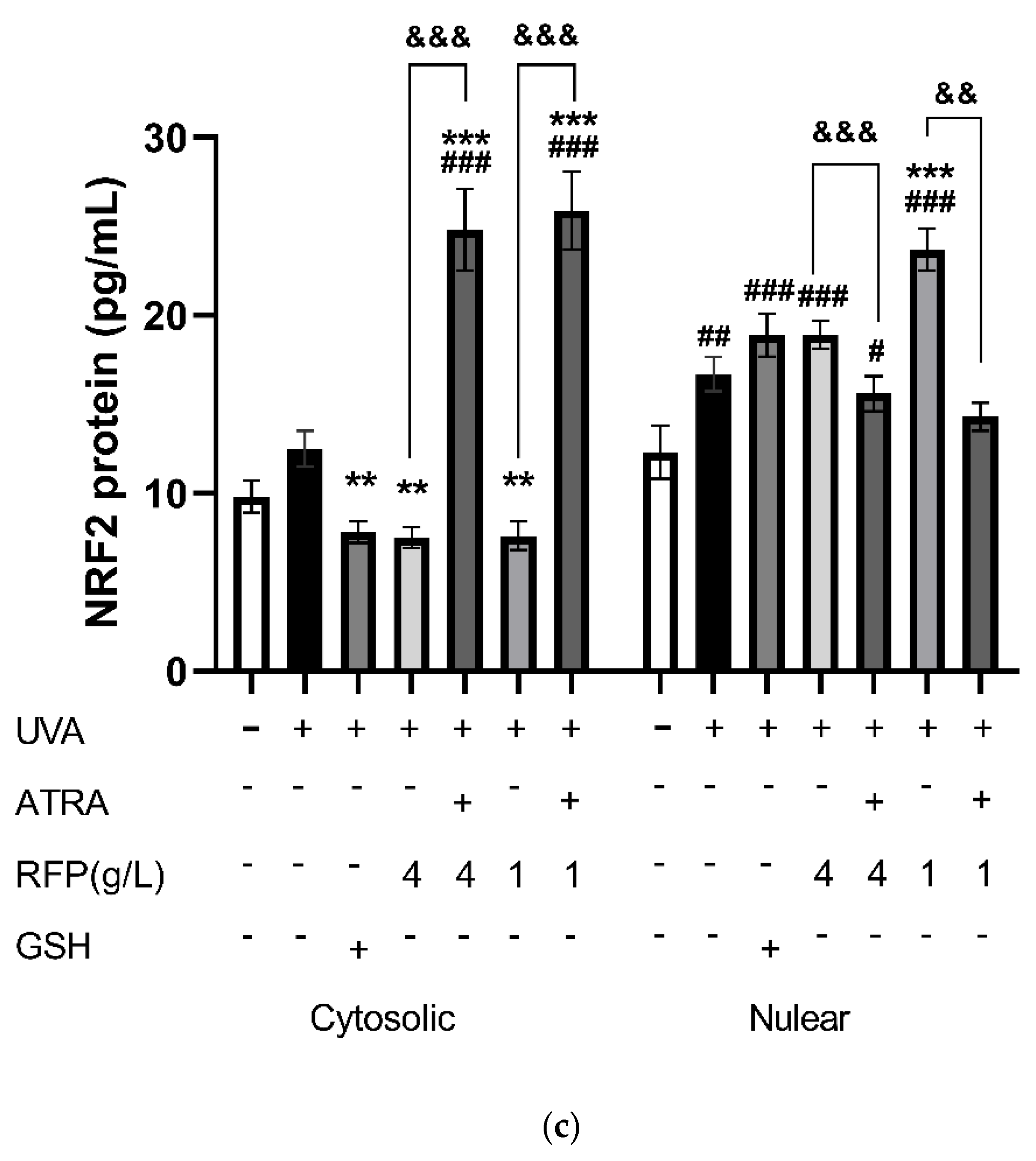
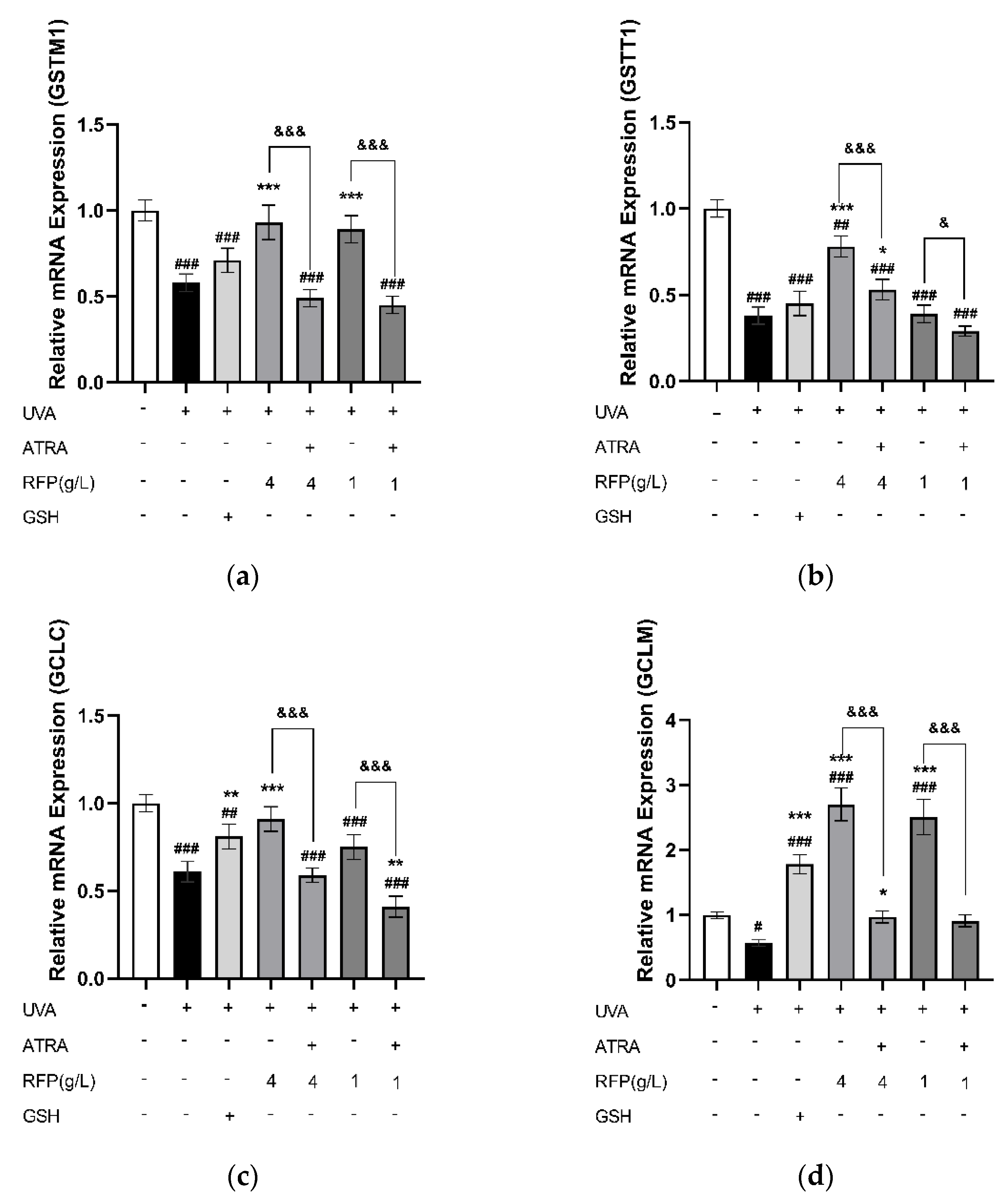
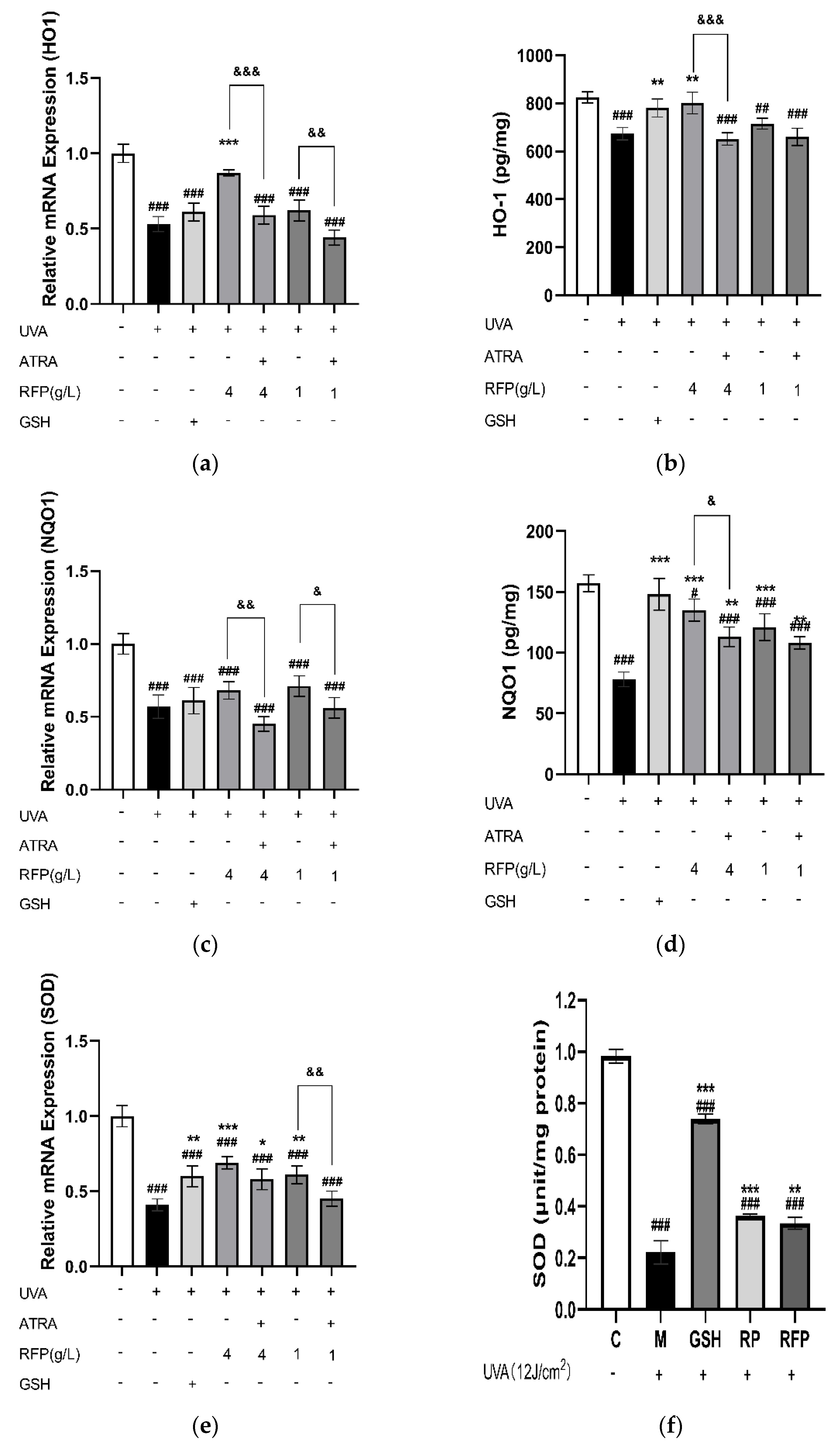
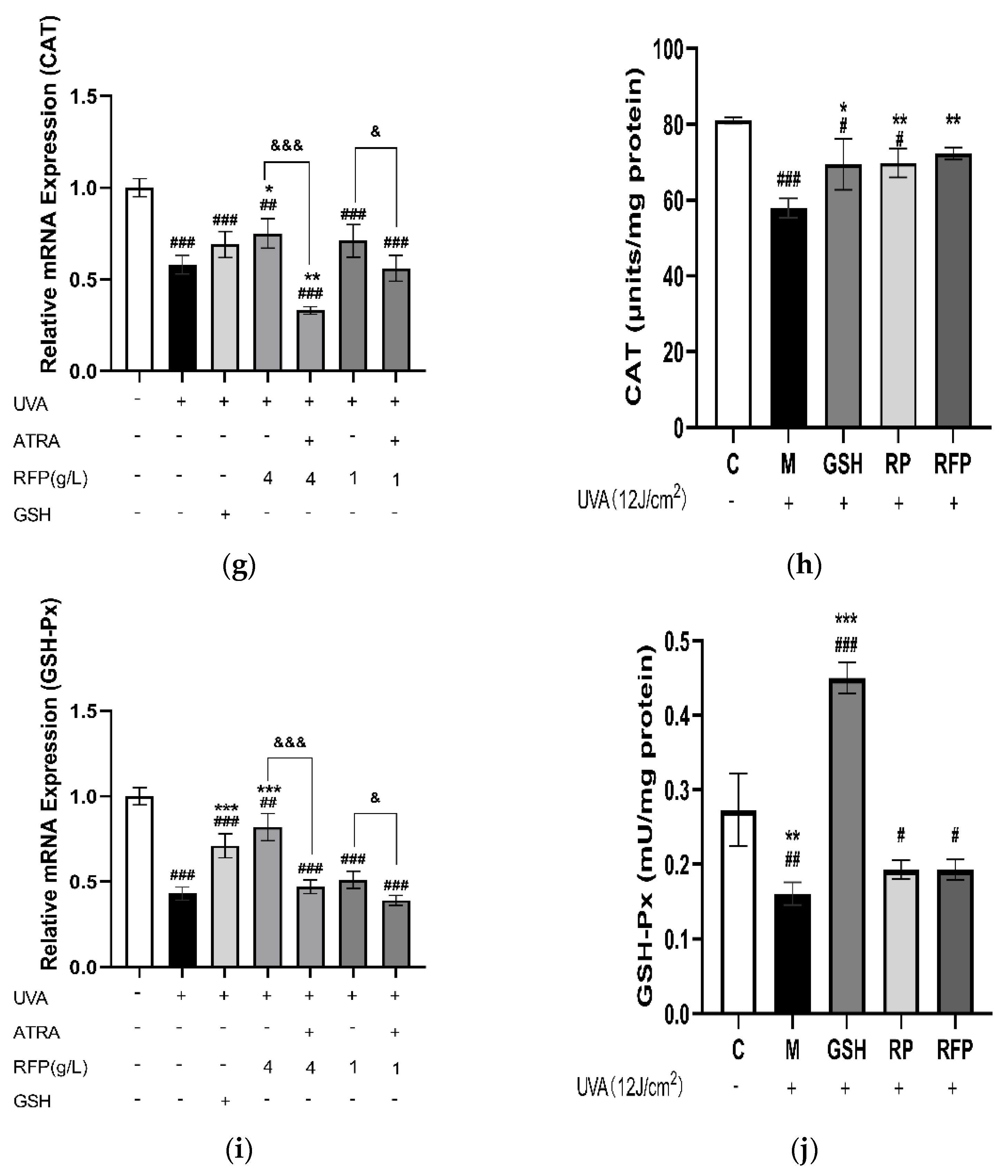
3.3.4. Quantitative Expression Analysis
3.4. Protective Mechanism of RFP against Oxidative Stress in Mice
3.4.1. Blood Routine Test Results
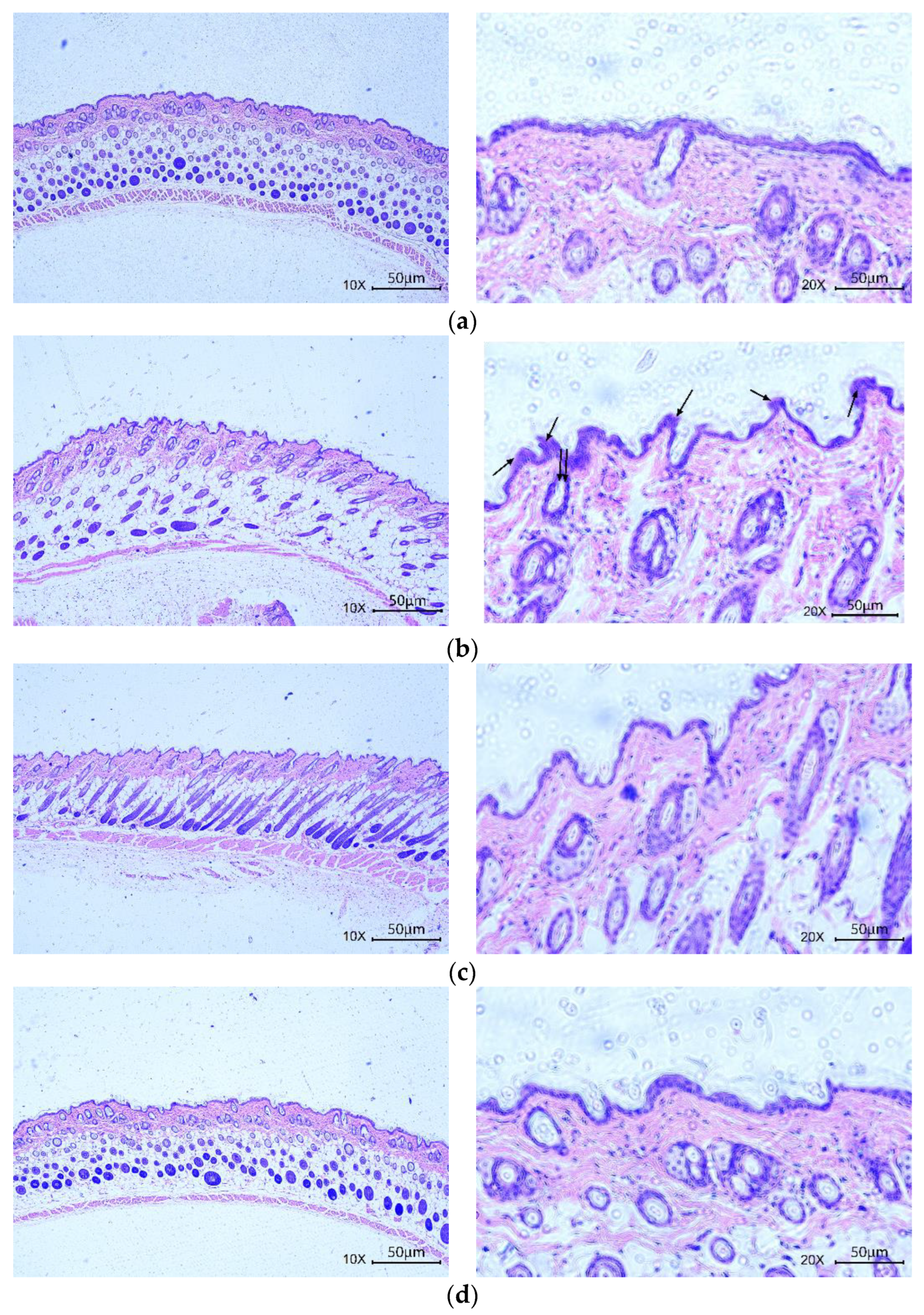
3.4.2. Skin Histological Observation
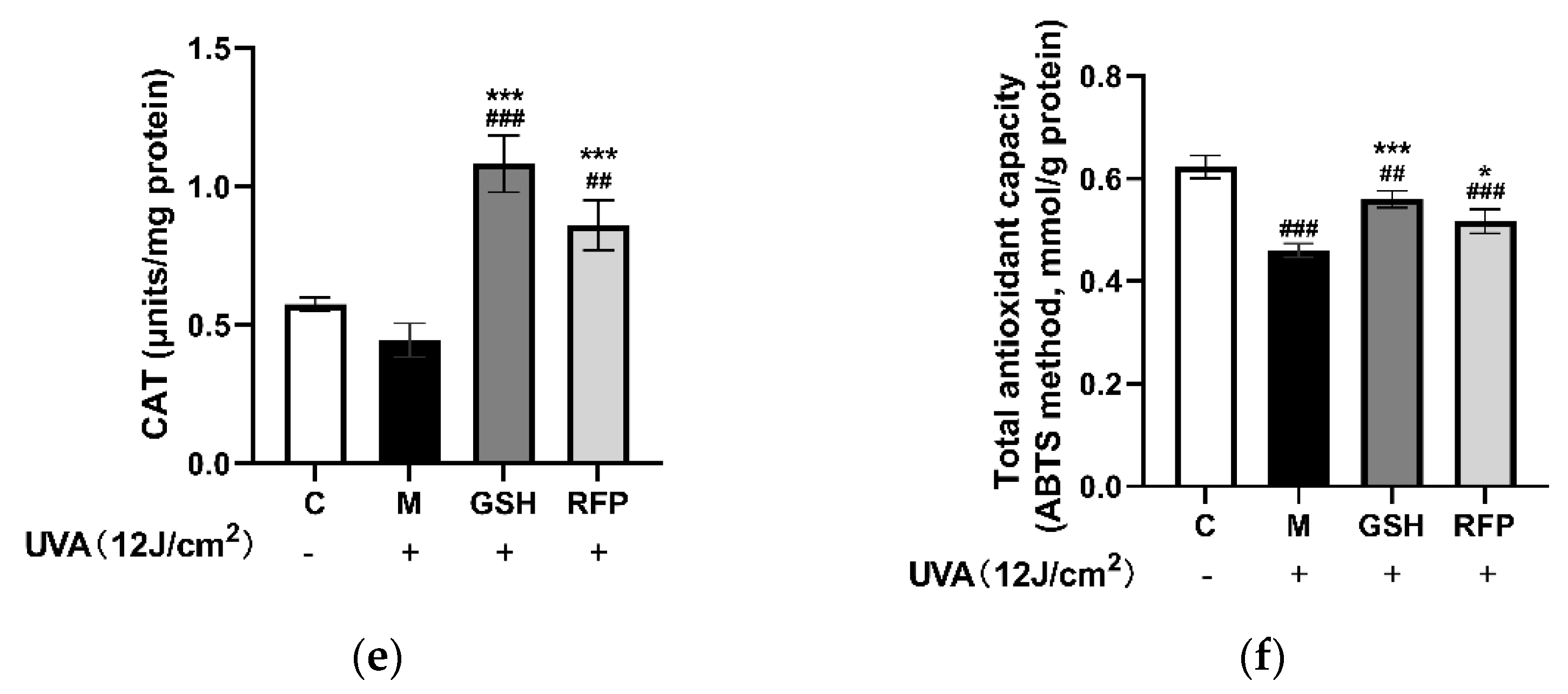
3.4.3. Total Antioxidant Capacity and CAT Enzyme Activity in Mice Tissue
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | Clean Reads | Q20 (%) | Q30 (%) | GC Content (%) | Total Mapped |
|---|---|---|---|---|---|
| RFP1 | 172,381,762 | 98.71 | 95.55 | 47.3 | 119,055,944 (69.07%) |
| RFP2 | 210,034,384 | 98.69 | 95.62 | 48.4 | 145,725,178 (69.38%) |
| RFP3 | 193,501,058 | 98.65 | 95.59 | 48.06 | 134,169,921 (69.34%) |
| Control1 | 201,140,460 | 98.79 | 95.72 | 47.29 | 117,966,520 (58.65%) |
| Control2 | 212,140,016 | 98.67 | 95.57 | 47.35 | 124,435,711 (58.66%) |
| Control3 | 199,240,206 | 98.66 | 95.53 | 46.74 | 111,945,599 (56.19%) |
| Model1 | 180,873,264 | 98.73 | 95.57 | 48.12 | 128,051,630 (70.8%) |
| Model2 | 216,959,018 | 98.73 | 95.73 | 48.43 | 149,826,586 (69.06%) |
| Model3 | 204,370,646 | 98.59 | 95.39 | 48.6 | 143,448,434 (70.19%) |
| Average | 198,960,090 | 98.69 | 95.59 | 47.81 | 65.70% |
| Gene ID | Gene Name | Gene Description |
|---|---|---|
| ENSG00000079999 | KEAP1 | Kelch−like ECH associated protein 1 |
| ENSG00000116044 | NFR2 | Nuclear factor erythroid 2−related factor 2 |
| ENSG00000181019 | NQO1 | NAD(P)H quinone dehydrogenase 1 |
| ENSG00000100292 | HMOX1 | Heme oxygenase 1 |
| ENSG00000065621 | GSTO2 | Glutathione S−transferase omega 2 |
| ENSG00000112096 | SOD2 | Superoxide dismutase 2 |
| ENSG00000285441 | SOD2 | Superoxide dismutase 2 |
| ENSG00000079999 | KEAP1 | Kelch−like ECH associated protein 1 |
| Control Group | MODEL GROUP | GSH Group | RFP Group | |
|---|---|---|---|---|
| WBC (109/L) | 6.85 ± 1.24 | 7.94 ± 0.83 | 6.12 ± 1.42 | 6.11 ± 0.68 |
| RBC (1012/L) | 10.2 ± 1.21 | 9.88 ± 0.89 | 9.75 ± 0.95 | 10.11 ± 1.12 |
| Hb (g/L) | 165.63 ± 9.54 | 155.32 ± 11.45 | 160.32 ± 12.98 | 160.55 ± 16.21 |
| HCT (%) | 50.14 ± 5.03 | 48.33 ± 4.34 | 49.32 ± 3.93 | 47.49 ± 4.55 |
| PLT (109/L) | 1056.33 ± 208.48 | 725.56 ± 104.38 | 701.67 ± 125.03 | 602.37 ± 136.78 |
| LY% (%) | 75.12 ± 3.87 | 76.22 ± 3.54 | 65.23 ± 3.86 | 66.71 ± 4.77 |
| NEUT% (%) | 30.8 ± 2.87 | 33.16 ± 1.99 | 32.17 ± 2.46 | 31.22 ± 2.67 |
| BAS (109/L) | 0.101 ± 0.01 | 0.104 ± 0.005 | 0.102 ± 0.007 | 0.101 ± 0.008 |
References
- Liochev, S.I. Reactive oxygen species and the free radical theory of aging. Free Radic. Biol. Med. 2013, 60, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Blume−Peytavi, U.; Kottner, J.; Sterry, W.; Hodin, M.W.; Griffiths, T.W.; Watson, R.E.B.; Hay, R.J.; Griffiths, C.E.M. Age−Associated Skin Conditions and Diseases: Current Perspectives and Future Options. Gerontologist 2016, 56, S230–S242. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Maity, N.; Nema, N.K.; Sarkar, B.K. Bioactive compounds from natural resources against skin aging. Phytomedicine 2011, 19, 64–73. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP−UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- Amagliani, L.; O’Regan, J.; Kelly, A.L.; O’Mahony, J.A. The composition, extraction, functionality and applications of rice proteins: A review. Trends Food Sci. Technol. 2017, 64, 1–12. [Google Scholar] [CrossRef]
- Venegas−Ortega, M.G.; Flores−Gallegos, A.C.; Martinez−Hernandez, J.L.; Aguilar, C.N.; Nevarez−Moorillon, G.V. Production of Bioactive Peptides from Lactic Acid Bacteria: A Sustainable Approach for Healthier Foods. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein−derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef]
- Chai, T.-T.; Law, Y.-C.; Wong, F.-C.; Kim, S.-K. Enzyme−Assisted Discovery of Antioxidant Peptides from Edible Marine Invertebrates: A Review. Mar. Drugs 2017, 15, 42. [Google Scholar] [CrossRef] [PubMed]
- He, S.H.; Wei, J.F. A Method of Separating L−Amino Acid Oxidase from Snake Venom. China National Intellectual Property Administration, China. CN1614012, 19 November 2004. [Google Scholar]
- Chen, A.Y.; Jia, J.P.; Ye, X.D.; Cheng, M.; Liao, Y.P.; Zhou, Y.G.; Zheng, G.L. Determination of the Molecular Weight Distribution of Small Molecule Oligopeptide in Fish Skin Collagen Hydrolyzate. Chin. J. Mod. Appl. Pharm. 2011, 28, 160–162. [Google Scholar]
- Wang, Z.X.; Liu, X.Y.; Xie, H.K.; Liu, Z.Q.; Rakariyatham, K.; Yu, C.X.; Shahidi, F.; Zhou, D.Y. Antioxidant activity and functional properties of Alcalase−hydrolyzed scallop protein hydrolysate and its role in the inhibition of cytotoxicity in vitro. Food Chem. 2021, 344, 128566. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.W.; Li, Y.N.; Chen, W.J. Antioxidant Activity of Polygonatum sibricum Instant Powder in vitro and its Protective Effect to Ethanol−induced Oxidative Damage in Mice. Food Res. Dev. 2018, 39, 182–186. [Google Scholar] [CrossRef]
- Wu, L.; Shen, H.S.; Tang, B.S.; Chen, J.C.; Zheng, H.G. Determination of Antioxidant Activity of Agrocybe chaxingu Fermentation Broth by a Modified Fenton Reaction Method. Fujian J. Agric. Sci. 2018, 33, 638–643. [Google Scholar] [CrossRef]
- Li, Z.L.; Ouyang, W.Q.; Zhang, L.; Liu, Y.M.; Yang, G.M. Protective effects of EMP on oxidative damage of mice skin irradiated by ultraviolet A. J. Northwest AF Univ. (Nat. Sci. Ed.) 2007, 35, 19–22. [Google Scholar] [CrossRef]
- Tian, M.H. Study on Antioxidative Mechanism of Rice Active Peptide In Vitro. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2015. [Google Scholar]
- Wang, X.J.; Hayes, J.D.; Henderson, C.J.; Wolf, C.R. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc. Natl. Acad. Sci. USA 2007, 104, 19589–19594. [Google Scholar] [CrossRef]
- Ke, Y.; Wang, X.J. TGF beta Signaling in Photoaging and UV−Induced Skin Cancer. J. Investig. Dermatol. 2021, 141, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Douki, T. Formation of UV−induced DNA damage contributing to skin cancer development. Photochem. Photobiol. Sci. 2018, 17, 1816–1841. [Google Scholar] [CrossRef]
- Marionnet, C.; Pierrard, C.; Golebiewski, C.; Bernerd, F. Diversity of Biological Effects Induced by Longwave UVA Rays (UVA1) in Reconstructed Skin. PLoS ONE 2014, 9, e105263. [Google Scholar] [CrossRef]
- Elfawy, H.A.; Das, B. Crosstalk between mitochondrial dysfunction, oxidative stress, and age related neurodegenerative disease: Etiologies and therapeutic strategies. Life Sci. 2019, 218, 165–184. [Google Scholar] [CrossRef]
- Lyakhovich, V.V.; Vavilin, V.A.; Zenkov, N.K.; Menshchikova, E.B. Active defense under oxidative stress. The antioxidant responsive element. Biochemistry 2006, 71, 962–974. [Google Scholar] [CrossRef]
- Tovar−Perez, E.G.; Guerrero−Becerra, L.; Lugo−Cervantes, E. Antioxidant activity of hydrolysates and peptide fractions of glutelin from cocoa (Theobroma cacao L.) seed. CyTA−J. Food 2017, 15, 489–496. [Google Scholar] [CrossRef]
- Chi, C.F.; Hu, F.Y.; Wang, B.; Li, T.; Ding, G.F. Antioxidant and anticancer peptides from the protein hydrolysate of blood clam (Tegillarca granosa) muscle. J. Funct. Foods 2015, 15, 301–313. [Google Scholar] [CrossRef]
- Jiang, X.; Cui, Z.Q.; Wang, L.H.; Xu, H.J.; Zhang, Y.G. Production of bioactive peptides from corn gluten meal by solid−state fermentation with Bacillus subtilis MTCC5480 and evaluation of its antioxidant capacity in vivo. LWT−Food Sci. Technol. 2020, 131, 109767. [Google Scholar] [CrossRef]
- Sun, T.; Xie, W.M.; Xu, P.X. Superoxide anion scavenging activity of graft chitosan derivatives. Carbohydr. Polym. 2004, 58, 379–382. [Google Scholar] [CrossRef]
- Raha, S.; Robinson, B.H. Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem. Sci. 2000, 25, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Ling, W.H.; Cheng, Q.X.; Ma, J.; Wang, T. Red and black rice decrease atherosclerotic plaque formation and increase antioxidant status in rabbits. Eur. J. Nutr. 2001, 131, 1421–1426. [Google Scholar] [CrossRef]
- Hu, X.M.; Wang, Y.M.; Zhao, Y.Q.; Chi, C.F.; Wang, B. Antioxidant Peptides from the Protein Hydrolysate of Monkfish (Lophius litulon) Muscle: Purification, Identification, and Cytoprotective Function on HepG2 Cells Damage by H2O2. Mar. Drugs 2020, 18, 153. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.C.; Xiao, J.B.; Wang, S.Y.; Ee, K.Y.; Chai, T.T. Advances on the antioxidant peptides from edible plant sources. Trends Food Sci. Technol. 2020, 99, 44–57. [Google Scholar] [CrossRef]
- Sah, B.N.P.; Vasiljevic, T.; McKechnie, S.; Donkor, O.N. Antioxidative and antibacterial peptides derived from bovine milk proteins. Crit. Rev. Food Sci. Nutr. 2018, 58, 726–740. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Taguchi, K.; Yamamoto, M. The KEAP1−NRF2 System as a Molecular Target of Cancer Treatment. Cancers 2021, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Espinosa−Diez, C.; Miguel, V.; Mennerich, D.; Kietzmann, T.; Sanchez−Perez, P.; Cadenas, S.; Lamas, S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015, 6, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Das, B.C.; Sehgal, A.; Mehrotra, R.; Kar, P.; Sardana, S.; Phukan, R.; Mahanta, J.; Purkayastha, J.; Saxena, S.; et al. GSTM1 and GSTT1 polymorphism and susceptibility to esophageal cancer in high− and low−risk regions of India. Tumour Biol. 2013, 34, 3249–3257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.W.; Chen, H.Y.; Li, J.; Bian, Y.Y.; Song, Y.L.; Li, Z.H.; He, F.Y.; Liu, S.Y.; Tsai, Y. Hirudin protects against isoproternol−induced myocardial infraction by alleviating oxidative via an Nrf2 dependent manner. Int. J. Biol. Macromol. 2020, 162, 425–435. [Google Scholar] [CrossRef]

| Gene | Direction | Primer Pair Sequence (5′→3′) |
|---|---|---|
| β−actin | F | TGGCACCCAGCACAATGAA |
| R | CTAAGTCATAGTCCGCCTAGAAGCA | |
| NRF2 | F | CAACTCAGCACCTTGTATC |
| R | TTCTTAGTATCTGGCTTCTT | |
| KEAP1 | F | GGAGGCGGAGCCCGA |
| R | GATGCCCTCAATGGACACCA | |
| HO−1 | F | CAAGCGCTATGTTCAGCGAC |
| R | GCTTGAACTTGGTGGCACTG | |
| NQO1 | F | CAGCCAATCAGCGTTCGGTA |
| R | CTTCATGGCGTAGTTGAATGATGTC | |
| GCLC | F | CAGTCAAGGACCGGCACAAG |
| R | CAAGAACATCGCCTCCATTCAG | |
| GCLM | F | TAAATCCCGATGAAAGAG |
| R | AACAGGAGGTGAAGCAAT | |
| GSTM1 | F | GAACTCCCTGAAAAGCTAAAGC |
| R | GTTGGGCTCAAATATACGGTGG | |
| GSTT1 | F | TTCCTTACTGGTCCTCACATCTC |
| R | TCACCGGATCATGGCCAGGCGCA | |
| SOD | F | TGGAGATAATACAGCAGGCT |
| R | AGTCACATTGCCCAAGTCTC | |
| CAT | F | CCTTCGACCCAAGCAA |
| R | CGATGGCGGTGAGTGT | |
| GSH−Px | F | AGAAGTGCGAGGTGAACGGT |
| R | CCCACCAGGAACTTCTCAAA |
| Sequence | Length | Mass (Da) |
|---|---|---|
| PLL | 3 | 341.23146 |
| YNDGDAPIVA | 10 | 1033.4716 |
| LLLP | 4 | 454.31552 |
| YNDGDAPIV | 9 | 962.43453 |
| VRVF | 5 | 575.3319 |
| LLSPF | 4 | 519.31692 |
| FYNDGDAPIV | 10 | 1109.5029 |
| YNEGDAPVVA | 10 | 1033.4716 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, Q.; You, S.; Fu, H.; Wang, D.; Zhang, J.; Wang, C.; Li, M. Purification and Identification of Antioxidant Peptides from Rice Fermentation of Lactobacillus plantarum and Their Protective Effects on UVA−Induced Oxidative Stress in Skin. Antioxidants 2022, 11, 2333. https://doi.org/10.3390/antiox11122333
Mo Q, You S, Fu H, Wang D, Zhang J, Wang C, Li M. Purification and Identification of Antioxidant Peptides from Rice Fermentation of Lactobacillus plantarum and Their Protective Effects on UVA−Induced Oxidative Stress in Skin. Antioxidants. 2022; 11(12):2333. https://doi.org/10.3390/antiox11122333
Chicago/Turabian StyleMo, Qiuting, Shiquan You, Hao Fu, Dongdong Wang, Jiachan Zhang, Changtao Wang, and Meng Li. 2022. "Purification and Identification of Antioxidant Peptides from Rice Fermentation of Lactobacillus plantarum and Their Protective Effects on UVA−Induced Oxidative Stress in Skin" Antioxidants 11, no. 12: 2333. https://doi.org/10.3390/antiox11122333
APA StyleMo, Q., You, S., Fu, H., Wang, D., Zhang, J., Wang, C., & Li, M. (2022). Purification and Identification of Antioxidant Peptides from Rice Fermentation of Lactobacillus plantarum and Their Protective Effects on UVA−Induced Oxidative Stress in Skin. Antioxidants, 11(12), 2333. https://doi.org/10.3390/antiox11122333