Molecular Regulation and Evolution of Redox Homeostasis in Photosynthetic Machinery
Abstract
1. Introduction
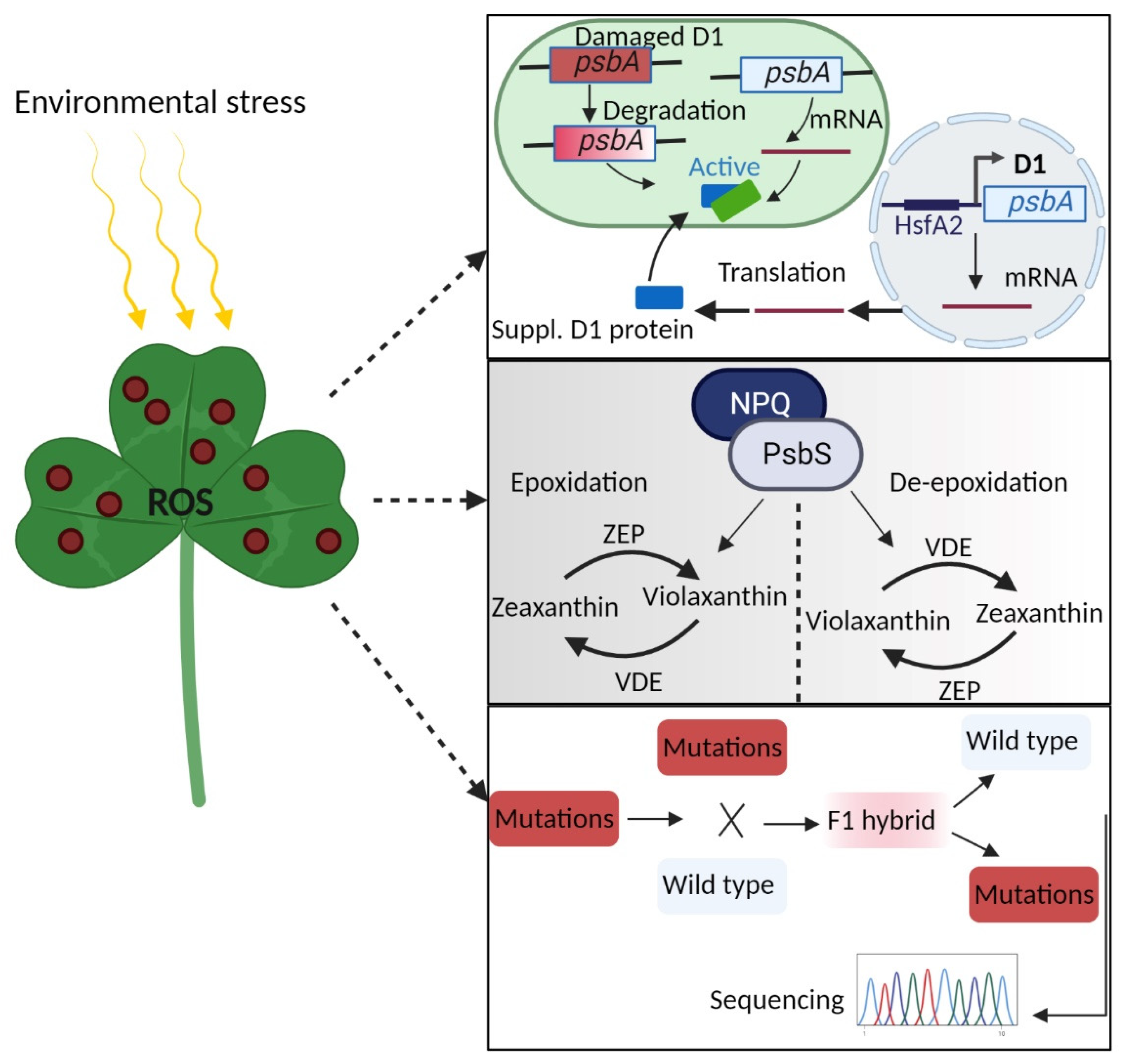
2. ROS Signaling Is a Double-Edged Sword in Plant Photosynthesis
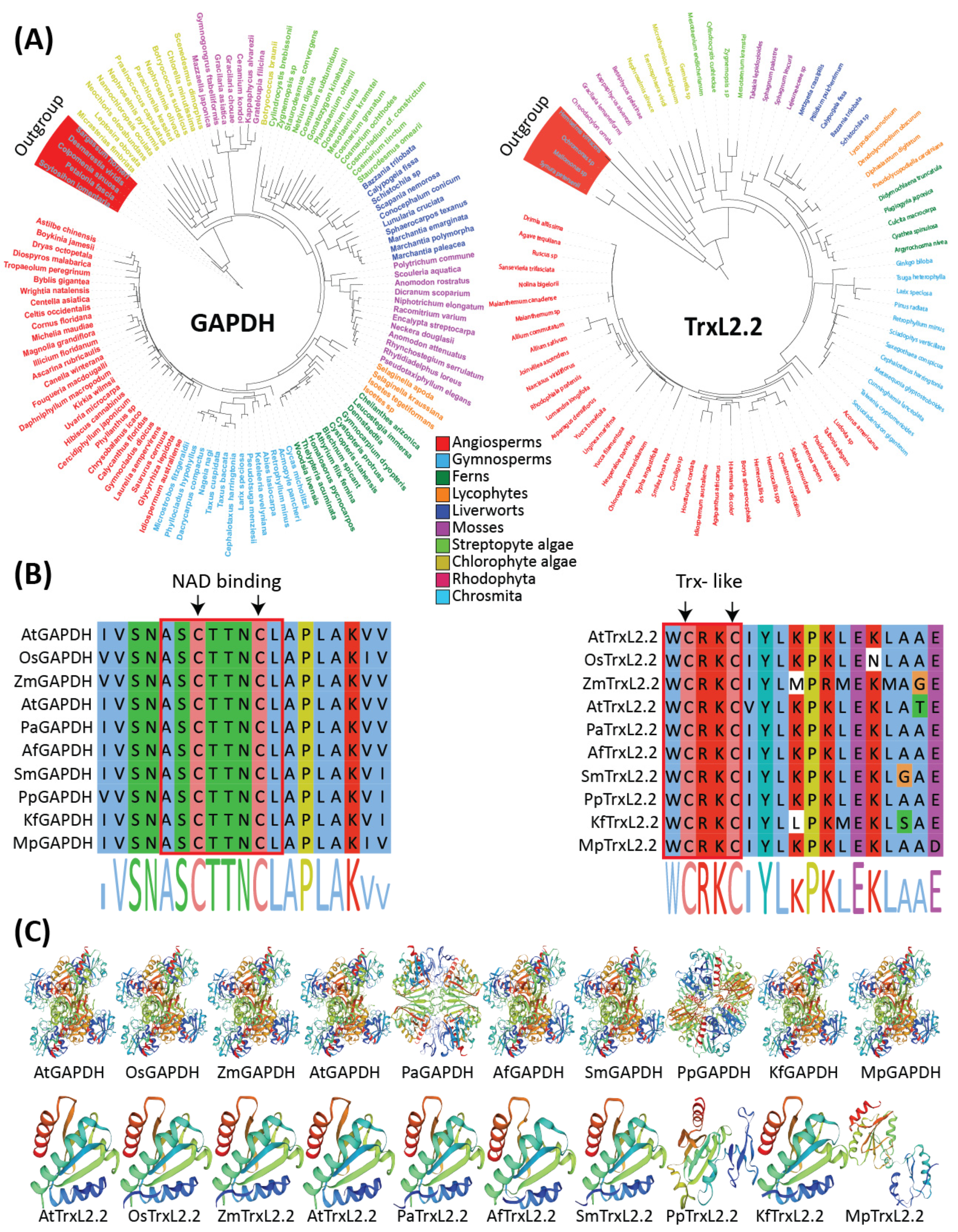
3. Molecular Evolution of Redox Regulatory Network
4. ROS Related Gene Families Are Highly Conserved across Land Plants and Green Algae
5. Key Photosynthesis Related Gene Families Are Evolved from Streptophyta Algae
6. Antioxidant Related Gene Families Are Conserved across Land Plants and Green Algae
7. Redox Regulation during Photosynthesis
8. Regulation in Calvin Benson Cycle
9. Regulation in Kranz Anatomy
10. Regulation in Crassulacean Acid Metabolism (CAM)
11. Regulation of Redox in Early Divergent Plants and Green Algae
12. Metabolism of Redox Regulators in Maintaining Redox Homeostasis for Photosynthesis
13. Manipulating ROS Signaling to Enhance Plant Photosynthetic Efficiency and Crop Yield
14. Concluding Remarks and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Olson, J.M. Evolution of photosynthetic reaction centers. Biosystems 1981, 14, 89–94. [Google Scholar] [CrossRef]
- Hohmann-Marriott, M.F.; Blankenship, R.E. Evolution of Photosynthesis. Annu. Rev. Plant Biol. 2011, 62, 515–548. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.W.; Hemp, J.; Johnson, J.E. Evolution of Oxygenic Photosynthesis. Annu. Rev. Earth Planet. Sci. 2016, 44, 647–683. [Google Scholar] [CrossRef]
- Zhu, X.G.; Long, S.P.; Ort, D.R. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef]
- Lu, Z.; Chang, Y.C.; Yin, Q.-Z.; Ng, C.; Jackson, W.M.J.S. Evidence for direct molecular oxygen production in CO2 photodissociation. Science 2014, 346, 61–64. [Google Scholar] [CrossRef]
- Meadows, V.S.J.A. Reflections on O2 as a biosignature in exoplanetary atmospheres. Astrobiology 2017, 17, 1022–1052. [Google Scholar] [CrossRef] [PubMed]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Rosing, M.T.; Frei, R.J.E.; Letters, P.S. U-rich Archaean sea-floor sediments from Greenland–indications of >3700 Ma oxygenic photosynthesis. Earth Planet. Sci. Lett. 2004, 217, 237–244. [Google Scholar] [CrossRef]
- Anbar, A.D.J.S. Elements and evolution. Science 2008, 322, 1481–1483. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.S.; Thomas, K.M.; Dai, Y.; Boyd, J.M.; Outten, F.W.J.B. Interplay between oxygen and Fe–S cluster biogenesis: Insights from the Suf pathway. Biochemistry 2014, 53, 5834–5847. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Mehla, N.; Sindhi, V.; Josula, D.; Bisht, P.; Wani, S.H. An Introduction to Antioxidants and Their Roles in Plant Stress Tolerance. In Reactive Oxygen Species and Antioxidant Systems in Plants: Role and Regulation under Abiotic Stress; Khan, M.I.R., Khan, N.A., Eds.; Springer: Singapore, 2017; pp. 1–23. [Google Scholar]
- Hasanuzzaman, M.; Bhuyan, M.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed]
- Raja, V.; Majeed, U.; Kang, H.; Andrabi, K.I.; John, R. Abiotic stress: Interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox signaling in plants. Antioxid. Redox Signal. 2013, 18, 2087–2090. [Google Scholar] [CrossRef]
- Dietz, K.-J.; Turkan, I.; Krieger-Liszkay, A. Redox- and Reactive Oxygen Species-Dependent Signaling into and out of the Photosynthesizing Chloroplast. Plant Physiol. 2016, 171, 1541–1550. [Google Scholar] [CrossRef]
- Huang, S.; Van Aken, O.; Schwarzländer, M.; Belt, K.; Millar, A.H. The Roles of Mitochondrial Reactive Oxygen Species in Cellular Signaling and Stress Response in Plants. Plant Physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Sandalio, L.M.; Romero-Puertas, M.C. Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann. Bot. 2015, 116, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kumar, A.; Yadav, S.; Singh, I.K. Reactive oxygen species-mediated signaling during abiotic stress. Plant Gene 2019, 18, 100173. [Google Scholar] [CrossRef]
- König, J.; Muthuramalingam, M.; Dietz, K.-J. Mechanisms and dynamics in the thiol/disulfide redox regulatory network: Transmitters. Sens. Targets 2012, 15, 261–268. [Google Scholar]
- Mignolet-Spruyt, L.; Xu, E.; Idänheimo, N.; Hoeberichts, F.A.; Mühlenbock, P.; Brosché, M.; Van Breusegem, F.; Kangasjärvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef] [PubMed]
- Vaahtera, L.; Brosché, M.; Wrzaczek, M.; Kangasjärvi, J. Specificity in ROS signaling and transcript signatures. Antioxid. Redox Signal. 2014, 21, 1422–1441. [Google Scholar] [CrossRef] [PubMed]
- Fichman, Y.; Mittler, R. Rapid systemic signaling during abiotic and biotic stresses: Is the ROS wave master of all trades? Plant J. 2020, 102, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Willems, P.; Mhamdi, A.; Stael, S.; Storme, V.; Kerchev, P.; Noctor, G.; Gevaert, K.; Van Breusegem, F.J.P.P. The ROS wheel: Refining ROS transcriptional footprints. Plant Physiol. 2016, 171, 1720–1733. [Google Scholar] [CrossRef]
- Kerchev, P.I.; Van Breusegem, F. Improving oxidative stress resilience in plants. Plant J. 2022, 109, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Reichheld, J.P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.-F. Superoxide dismutases: Ancient enzymes and new insights. FEBS Lett. 2012, 586, 585–595. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef]
- Llorente, B.; Segretin, M.E.; Giannini, E.; Lobais, C.; Juárez, M.E.; Paulsen, I.T.; Blanco, N.E. Homecoming: Rewinding the reductive evolution of the chloroplast genome for increasing crop yields. Nat. Commun. 2021, 12, 6734. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Danila, F.R.; Furbank, R.T.; von Caemmerer, S. On the road to C4 rice: Advances and perspectives. Plant J. 2020, 101, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, H.; Ouyang, B.; Shi, C.; Demidchik, V.; Hao, Z.; Yu, M.; Shabala, S. NADPH oxidases and the evolution of plant salinity tolerance. Plant Cell Environ. 2020, 43, 2957–2968. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Smirnoff, N.; Van Breusegem, F.; Dietz, K.-J.; Noctor, G. Plant redox biology—On the move. Plant Physiol. 2021, 186, 1–3. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011, 14, 691–699. [Google Scholar] [CrossRef]
- Laurindo, F.R.; Araujo, T.L.; Abrahao, T.B. Nox NADPH oxidases and the endoplasmic reticulum. Antioxid. Redox Signal. 2014, 20, 2755–2775. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Vanderauwera, S.; Suzuki, N.; Miller, G.; van de Cotte, B.; Morsa, S.; Ravanat, J.L.; Hegie, A.; Triantaphylidès, C.; Shulaev, V.; Van Montagu, M.C.; et al. Extranuclear protection of chromosomal DNA from oxidative stress. Proc. Natl. Acad. Sci. USA 2011, 108, 1711–1716. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Van Breusegem, F.; Mhamdi, A. Redox-dependent control of nuclear transcription in plants. J. Exp. Bot. 2018, 69, 3359–3372. [Google Scholar] [CrossRef] [PubMed]
- Willems, P.; Van Breusegem, F.; Huang, J. Contemporary proteomic strategies for cysteine redoxome profiling. Plant Physiol. 2021, 186, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Mullineaux, P.M.; Exposito-Rodriguez, M.; Laissue, P.P.; Smirnoff, N.; Park, E. Spatial chloroplast-to-nucleus signalling involving plastid-nuclear complexes and stromules. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190405. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to Disease Immunity Pathways. Plant Physiol. 2016, 171, 1635–1650. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, J.M. Hydrogen peroxide concentrations in leaves under natural conditions. J. Exp. Bot. 2006, 57, 2435–2444. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Stress-triggered redox signalling: What’s in pROSpect? Plant Cell Environ. 2016, 39, 951–964. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and Glutathione: The Heart of the Redox Hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Dvořák, P.; Krasylenko, Y.; Ovečka, M.; Basheer, J.; Zapletalová, V.; Šamaj, J.; Takac, T. FSD1: Developmentally-regulated plastidial, nuclear and cytoplasmic enzyme with anti-oxidative and osmoprotective role. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Fischer, B.B.; Hideg, É.; Krieger-Liszkay, A. Production, Detection, and Signaling of Singlet Oxygen in Photosynthetic Organisms. Antioxid. Redox Signal. 2013, 18, 2145–2162. [Google Scholar] [CrossRef] [PubMed]
- Gollan, P.J.; Tikkanen, M.; Aro, E.M. Photosynthetic light reactions: Integral to chloroplast retrograde signalling. Curr. Opin. Plant Biol. 2015, 27, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Krieger-Liszkay, A.; Fufezan, C.; Trebst, A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 2008, 98, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Bobik, K.; Burch-Smith, T.M. Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 2015, 6, 781. [Google Scholar] [CrossRef]
- Galvez-Valdivieso, G.; Fryer, M.J.; Lawson, T.; Slattery, K.; Truman, W.; Smirnoff, N.; Asami, T.; Davies, W.J.; Jones, A.M.; Baker, N.R.; et al. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 2009, 21, 2143–2162. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. The Water-Water Cycle In Chloroplasts: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Zaffagnini, M.; Morisse, S.; Sparla, F.; Pérez-Pérez, M.E.; Francia, F.; Danon, A.; Marchand, C.H.; Fermani, S.; Trost, P.; et al. Redox regulation of the Calvin-Benson cycle: Something old, something new. Front. Plant Sci. 2013, 4, 470. [Google Scholar] [CrossRef]
- Awad, J.; Stotz, H.U.; Fekete, A.; Krischke, M.; Engert, C.; Havaux, M.; Berger, S.; Mueller, M.J. 2-Cysteine Peroxiredoxins and Thylakoid Ascorbate Peroxidase Create a Water-Water Cycle That Is Essential to Protect the Photosynthetic Apparatus under High Light Stress Conditions. Plant Physiol. 2015, 167, 1592–1603. [Google Scholar] [CrossRef]
- Vogel, M.O.; Moore, M.; König, K.; Pecher, P.; Alsharafa, K.; Lee, J.; Dietz, K.-J. Fast Retrograde Signaling in Response to High Light Involves Metabolite Export, Mitogen-Activated Protein Kinase6, and AP2/ERF Transcription Factors in Arabidopsis. Plant Cell 2014, 26, 1151–1165. [Google Scholar] [CrossRef]
- Exposito-Rodriguez, M.; Laissue, P.P.; Yvon-Durocher, G.; Smirnoff, N.; Mullineaux, P.M. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 2017, 8, 49. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, Y.; Chan, K.X.; Marchant, D.B.; Franks, P.J.; Randall, D.; Tee, E.E.; Chen, G.; Ramesh, S.; Phua, S.Y.; et al. Evolution of chloroplast retrograde signaling facilitates green plant adaptation to land. Proc. Natl. Acad. Sci. USA 2019, 116, 5015–5020. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Haigh, A.M.; Holford, P.; Chen, Z.-H. Roles of Chloroplast Retrograde Signals and Ion Transport in Plant Drought Tolerance. Int. J. Mol. Sci. 2018, 19, 963. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; Van Aken, O.; De Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef] [PubMed]
- Shapiguzov, A.; Vainonen, J.P.; Hunter, K.; Tossavainen, H.; Tiwari, A.; Järvi, S.; Hellman, M.; Aarabi, F.; Alseekh, S.; Wybouw, B.; et al. Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. eLife 2019, 8, e43284. [Google Scholar] [CrossRef]
- op den Camp, R.G.; Przybyla, D.; Ochsenbein, C.; Laloi, C.; Kim, C.; Danon, A.; Wagner, D.; Hideg, E.; Göbel, C.; Feussner, I.; et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 2003, 15, 2320–2332. [Google Scholar] [CrossRef]
- Vass, I.; Aro, E.-M. Photoinhibition of photosynthetic electron transport, in Primary Processes of Photosynthesis: Priniciples and Apparatus. Part I. Prim. Process. Photosynth. Basic Princ. Appar. 2008, 1, 393–425. [Google Scholar]
- Queval, G.; Issakidis-Bourguet, E.; Hoeberichts, F.A.; Vandorpe, M.; Gakière, B.; Vanacker, H.; Miginiac-Maslow, M.; Van Breusegem, F.; Noctor, G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007, 52, 640–657. [Google Scholar] [CrossRef]
- Devireddy, A.R.; Zandalinas, S.I.; Fichman, Y.; Mittler, R. Integration of reactive oxygen species and hormone signaling during abiotic stress. Plant J. 2021, 105, 459–476. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. The roles of reactive oxygen metabolism in drought: Not so cut and dried. Plant Physiol. 2014, 164, 1636–1648. [Google Scholar] [CrossRef]
- Balsera, M.; Uberegui, E.; Schürmann, P.; Buchanan, B.B. Evolutionary development of redox regulation in chloroplasts. Antioxid. Redox Signal. 2014, 21, 1327–1355. [Google Scholar] [CrossRef]
- Gütle, D.D.; Roret, T.; Hecker, A.; Reski, R.; Jacquot, J.-P. Dithiol disulphide exchange in redox regulation of chloroplast enzymes in response to evolutionary and structural constraints. Plant Sci. 2017, 255, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Groen, S.C.; Ćalić, I.; Joly-Lopez, Z.; Platts, A.E.; Choi, J.Y.; Natividad, M.; Dorph, K.; Mauck, W.M., 3rd; Bracken, B.; Cabral, C.L.U.; et al. The strength and pattern of natural selection on gene expression in rice. Nature 2020, 578, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Kingsolver, J.G.; Hoekstra, H.E.; Hoekstra, J.M.; Berrigan, D.; Vignieri, S.N.; Hill, C.E.; Hoang, A.; Gibert, P.; Beerli, P. The strength of phenotypic selection in natural populations. Am. Nat. 2001, 157, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Chen, G.; Dai, F.; Wang, Y.; Hills, A.; Ruan, Y.L.; Zhang, G.; Franks, P.J.; Nevo, E.; Blatt, M.R. Molecular Evolution of Grass Stomata. Trends Plant Sci. 2017, 22, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Zeng, F.; Chen, G.; Feng, X.; Riaz, A.; Wu, X.; Gao, W.; Wu, F.-b.; Holford, P.; Chen, Z.-H. Metalloid hazards: From plant molecular evolution to mitigation strategies. J. Hazard. Mater. 2020, 409, 124495. [Google Scholar] [CrossRef]
- Jiang, W.; Tong, T.; Li, W.; Huang, Z.; Chen, G.; Zeng, F.; Riaz, A.; Amoanimaa-Dede, H.; Pan, R.; Zhang, W.; et al. Molecular Evolution of Plant 14-3-3 Proteins and Function of Hv14-3-3A in Stomatal Regulation and Drought Tolerance. Plant Cell Physiol. 2022. [Google Scholar] [CrossRef]
- Adem, G.D.; Chen, G.; Shabala, L.; Chen, Z.-H.; Shabala, S. GORK Channel: A Master Switch of Plant Metabolism? Trends Plant Sci. 2020, 25, 434–445. [Google Scholar] [CrossRef]
- Deng, F.; Zeng, F.; Shen, Q.; Abbas, A.; Cheng, J.; Jiang, W.; Chen, G.; Shah, A.N.; Holford, P.; Tanveer, M.; et al. Molecular evolution and functional modification of plant miRNAs with CRISPR. Trends Plant Sci. 2022, 27, 890–907. [Google Scholar] [CrossRef]
- Li, Q.; Tong, T.; Jiang, W.; Cheng, J.; Deng, F.; Wu, X.; Chen, Z.H.; Ouyang, Y.; Zeng, F. Highly Conserved Evolution of Aquaporin PIPs and TIPs Confers Their Crucial Contribution to Flowering Process in Plants. Front. Plant Sci. 2021, 12, 761713. [Google Scholar] [CrossRef]
- Cai, S.; Huang, Y.; Chen, F.; Zhang, X.; Sessa, E.; Zhao, C.; Marchant, D.B.; Xue, D.; Chen, G.; Dai, F.; et al. Evolution of rapid blue-light response linked to explosive diversification of ferns in angiosperm forests. New Phytol. 2021, 230, 1201–1213. [Google Scholar] [CrossRef]
- Tong, T.; Li, Q.; Jiang, W.; Chen, G.; Xue, D.; Deng, F.; Zeng, F.; Chen, Z.H. Molecular Evolution of Calcium Signaling and Transport in Plant Adaptation to Abiotic Stress. Int. J. Mol. Sci. 2021, 22, 12308. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, W.; Tong, T.; Chen, G.; Zeng, F.; Jang, S.; Gao, W.; Li, Z.; Mak, M.; Deng, F.; et al. Molecular Interaction and Evolution of Jasmonate Signaling With Transport and Detoxification of Heavy Metals and Metalloids in Plants. Front. Plant Sci. 2021, 12, 665842. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Chibani, K.; Wingsle, G.; Jacquot, J.-P.; Gelhaye, E.; Rouhier, N. Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa. Mol. Plant 2009, 2, 308–322. [Google Scholar] [CrossRef]
- Eliyahu, E.; Rog, I.; Inbal, D.; Danon, A. ACHT4-driven oxidation of APS1 attenuates starch synthesis under low light intensity in Arabidopsis plants. Proc. Natl. Acad. Sci. USA 2015, 112, 12876–12881. [Google Scholar] [CrossRef]
- Yoshida, K.; Hara, A.; Sugiura, K.; Fukaya, Y.; Hisabori, T. Thioredoxin-like2/2-Cys peroxiredoxin redox cascade supports oxidative thiol modulation in chloroplasts. Proc. Natl. Acad. Sci. USA 2018, 115, E8296–E8304. [Google Scholar] [CrossRef]
- Cejudo, F.J.; Ojeda, V.; Delgado-Requerey, V.; González, M.; Pérez-Ruiz, J.M.P. Chloroplast redox regulatory mechanisms in plant adaptation to light and darkness. Front. Plant Sci. 2019, 10, 380. [Google Scholar] [CrossRef]
- Yoshida, K.; Uchikoshi, E.; Hara, S.; Hisabori, T. Thioredoxin-like2/2-Cys peroxiredoxin redox cascade acts as oxidative activator of glucose-6-phosphate dehydrogenase in chloroplasts. Biochem. J. 2019, 476, 1781–1790. [Google Scholar] [CrossRef]
- Yokochi, Y.; Sugiura, K.; Takemura, K.; Yoshida, K.; Hara, S.; Wakabayashi, K.-I.; Kitao, A.; Hisabori, T. Impact of key residues within chloroplast thioredoxin-f on recognition for reduction and oxidation of target proteins. J. Biol. Chem. 2019, 294, 17437–17450. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Yokochi, Y.; Hisabori, T. New light on chloroplast redox regulation: Molecular mechanism of protein thiol oxidation. Front. Plant Sci. 2019, 10, 1534. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hisabori, T.J.P.; Physiology, C. Biochemical basis for redox regulation of chloroplast-localized phosphofructokinase from Arabidopsis thaliana. Plant Cell Physiol. 2021, 62, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Mestres-Ortega, D.; Meyer, Y. The Arabidopsis thaliana genome encodes at least four thioredoxins m and a new prokaryotic-like thioredoxin. Gene 1999, 240, 307–316. [Google Scholar] [CrossRef]
- Arsova, B.; Hoja, U.; Wimmelbacher, M.; Greiner, E.; Ustün, S.; Melzer, M.; Petersen, K.; Lein, W.; Börnke, F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: Evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 2010, 22, 1498–1515. [Google Scholar] [CrossRef]
- Schröter, Y.; Steiner, S.; Matthäi, K.; Pfannschmidt, T. Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics 2010, 10, 2191–2204. [Google Scholar] [CrossRef]
- Gelhaye, E.; Rouhier, N.; Navrot, N.; Jacquot, J.-P. The plant thioredoxin system. Experientia 2005, 62, 24–35. [Google Scholar] [CrossRef]
- Ojeda, V.; Pérez-Ruiz, J.M.; Cejudo, F.J. 2-Cys peroxiredoxins participate in the oxidation of chloroplast enzymes in the dark. Mol. Plant 2018, 11, 1377–1388. [Google Scholar] [CrossRef]
- Dangoor, I.; Peled-Zehavi, H.; Levitan, A.; Pasand, O.; Danon, A. A small family of chloroplast atypical thioredoxins. Plant Physiol. 2009, 149, 1240–1250. [Google Scholar] [CrossRef]
- Yokochi, Y.; Fukushi, Y.; Wakabayashi, K.I.; Yoshida, K.; Hisabori, T. Oxidative regulation of chloroplast enzymes by thioredoxin and thioredoxin-like proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2114952118. [Google Scholar] [CrossRef]
- Belin, C.; Bashandy, T.; Cela, J.; Delorme-Hinoux, V.; Riondet, C.; Reichheld, J. A comprehensive study of thiol reduction gene expression under stress conditions in A rabidopsis thaliana. Plant Cell Environ. 2015, 38, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Portis, A.R.; Li, C.; Wang, D.; Salvucci, M.E. Regulation of Rubisco activase and its interaction with Rubisco. J. Exp. Bot. 2007, 59, 1597–1604. [Google Scholar] [CrossRef]
- Yin, Z.; Meng, F.; Song, H.; Wang, X.; Xu, X.; Yu, D. Expression Quantitative Trait Loci Analysis of Two Genes Encoding Rubisco Activase in Soybean. Plant Physiol. 2009, 152, 1625–1637. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, M.E.; Werneke, J.M.; Ogren, W.L.; Portis, A.R., Jr. Purification and Species Distribution of Rubisco Activase. Plant Physiol. 1987, 84, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Teich, R.; Becker, B.; Cerff, R.; Brinkmann, H. The GapA/B Gene Duplication Marks the Origin of Streptophyta (Charophytes and Land Plants). Mol. Biol. Evol. 2006, 23, 1109–1118. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Robbens, S.; Petersen, J.; Brinkmann, H.; Rouzé, P.; Van de Peer, Y. Unique regulation of the Calvin cycle in the ultrasmall green alga Ostreococcus. J. Mol. Evol. 2007, 64, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Liaud, M.F.; Brandt, U.; Scherzinger, M.; Cerff, R. Evolutionary origin of cryptomonad microalgae: Two novel chloroplast/cytosol-specific GAPDH genes as potential markers of ancestral endosymbiont and host cell components. J. Mol. Evol. 1997, 44 (Suppl. S1), S28–S37. [Google Scholar] [CrossRef]
- Pedersen, T.A.; Kirk, M.; Bassham, J.A. Light-Dark Transients in Levels of Intermediate Compounds during Photosynthesis in Air-Adapted Chlorella. Physiol. Plant. 1966, 19, 219–231. [Google Scholar] [CrossRef]
- Champigny, M.-L.; Bismuth, E. Role of Photosynthetic Electron Transfer in Light Activation of Calvin Cycle Enzymes. Physiol. Plant. 1976, 36, 95–100. [Google Scholar] [CrossRef]
- Avron, M.; Gibbs, M. Properties of phosphoribulokinase of whole chloroplasts. Plant Physiol. 1974, 53, 136–139. [Google Scholar] [CrossRef]
- Harrison, D.H.T.; Runquist, J.A.; Holub, A.; Miziorko, H.M. The Crystal Structure of Phosphoribulokinase from Rhodobacter sphaeroides Reveals a Fold Similar to That of Adenylate Kinase. Biochemistry 1998, 37, 5074–5085. [Google Scholar] [CrossRef] [PubMed]
- Marri, L.; Zaffagnini, M.; Collin, V.; Issakidis-Bourguet, E.; Lemaire, S.D.; Pupillo, P.; Sparla, F.; Miginiac-Maslow, M.; Trost, P. Prompt and easy activation by specific thioredoxins of calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol. Plant 2009, 2, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Tamoi, M.; Ishikawa, T.; Takeda, T.; Shigeoka, S. Molecular characterization and resistance to hydrogen peroxide of two fructose-1,6-bisphosphatases from Synechococcus PCC 7942. Arch. Biochem. Biophys. 1996, 334, 27–36. [Google Scholar] [CrossRef]
- Raines, C.A.; Lloyd, J.C.; Willingham, N.M.; Potts, S.; Dyer, T.A. cDNA and gene sequences of wheat chloroplast sedoheptulose-1,7-bisphosphatase reveal homology with fructose-1,6-bisphosphatases. Eur. J. Biochem. 1992, 205, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Bloom, A.J.; Queval, G.; Noctor, G. Photorespiratory Metabolism: Genes, Mutants, Energetics, and Redox Signaling. Annu. Rev. Plant Biol. 2009, 60, 455–484. [Google Scholar] [CrossRef]
- Miginiac-Maslow, M.; Lancelin, J.M. Intrasteric inhibition in redox signalling: Light activation of NADP-malate dehydrogenase. Photosynth. Res. 2002, 72, 1–12. [Google Scholar] [CrossRef]
- Lemaire, S.D.; Quesada, A.; Merchan, F.; Corral, J.M.; Igeno, M.I.; Keryer, E.; Issakidis-Bourguet, E.; Hirasawa, M.; Knaff, D.B.; Miginiac-Maslow, M. NADP-Malate Dehydrogenase from Unicellular Green Alga Chlamydomonas reinhardtii. A First Step toward Redox Regulation? Plant Physiol. 2005, 137, 514–521. [Google Scholar] [CrossRef]
- Latifi, A.; Ruiz, M.; Zhang, C.-C. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 2009, 33, 258–278. [Google Scholar] [CrossRef]
- Diaz, J.M.; Plummer, S. Production of extracellular reactive oxygen species by phytoplankton: Past and future directions. J. Plankton Res. 2018, 40, 655–666. [Google Scholar] [CrossRef]
- Bernroitner, M.; Zamocky, M.; Furtmüller, P.G.; Peschek, G.A.; Obinger, C. Occurrence, phylogeny, structure, and function of catalases and peroxidases in cyanobacteria. J. Exp. Bot. 2009, 60, 423–440. [Google Scholar] [CrossRef]
- Rose, A.J. The influence of extracellular superoxide on iron redox chemistry and bioavailability to aquatic microorganisms. Front. Microbiol. 2012, 3, 124. [Google Scholar] [CrossRef]
- Vassiliev, I.R.; Ronan, M.T.; Hauska, G.; Golbeck, J.H. The bound electron acceptors in green sulfur bacteria: Resolution of the g-tensor for the FX iron-sulfur cluster in Chlorobium tepidum. Biophys. J. 2000, 78, 3160–3169. [Google Scholar] [CrossRef]
- Wallace, M.A.; Liou, L.-L.; Martins, J.; Clement, M.H.; Bailey, S.; Longo, V.D.; Valentine, J.S.; Gralla, E.B. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis: Cross-compartment protection by CuZn-superoxide dismutase. J. Biol. Chem. 2004, 279, 32055–32062. [Google Scholar] [CrossRef]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.-F.; Teixeira, M.; Valentine, J.S. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef]
- Yokochi, Y.; Yoshida, K.; Hahn, F.; Miyagi, A.; Wakabayashi, K.-I.; Kawai-Yamada, M.; Weber, A.P.M.; Hisabori, T. Redox regulation of NADP-malate dehydrogenase is vital for land plants under fluctuating light environment. Proc. Natl. Acad. Sci. USA 2021, 118, e2016903118. [Google Scholar] [CrossRef] [PubMed]
- Genty, B.; Harbinson, J. Regulation of Light Utilization for Photosynthetic Electron Transport. In Photosynthesis and the Environment; Baker, N.R., Ed.; Springer: Dordrecht, The Netherlands, 1996; pp. 67–99. [Google Scholar]
- Buchanan, B.B. Role of light in the regulation of chloroplast enzymes. Annu. Rev. Plant Physiol. 1980, 31, 341–374. [Google Scholar] [CrossRef]
- Chaux, F.; Peltier, G.; Johnson, X. A security network in PSI photoprotection: Regulation of photosynthetic control, NPQ and O2 photoreduction by cyclic electron flow. Front. Plant Sci. 2015, 6, 875. [Google Scholar] [CrossRef]
- Sonoike, K. Photoinhibition of photosystem I. Physiol. Plant 2011, 142, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Kirilovsky, D.; Kerfeld, C.A. The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim. Biophys. Acta 2012, 1817, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Ruban, A.V. Nonphotochemical Chlorophyll Fluorescence Quenching: Mechanism and Effectiveness in Protecting Plants from Photodamage. Plant Physiol. 2016, 170, 1903–1916. [Google Scholar] [CrossRef]
- Li, Z.; Wakao, S.; Fischer, B.B.; Niyogi, K.K. Sensing and Responding to Excess Light. Annu. Rev. Plant Biol. 2009, 60, 239–260. [Google Scholar] [CrossRef] [PubMed]
- Pinnola, A.; Bassi, R. Molecular mechanisms involved in plant photoprotection. Biochem. Soc. Trans. 2018, 46, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Munekage, Y.N.; Genty, B.; Peltier, G. Effect of PGR5 Impairment on Photosynthesis and Growth in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Rumeau, D.; Peltier, G.; Cournac, L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007, 30, 1041–1051. [Google Scholar] [CrossRef]
- Konno, H.; Nakane, T.; Yoshida, M.; Ueoka-Nakanishi, H.; Hara, S.; Hisabori, T.J.P.; Physiology, C. Thiol modulation of the chloroplast ATP synthase is dependent on the energization of thylakoid membranes. Plant Cell Physiol. 2012, 53, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Matsuoka, Y.; Hara, S.; Konno, H.; Hisabori, T. Distinct redox behaviors of chloroplast thiol enzymes and their relationships with photosynthetic electron transport in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Wolosiuk, R.A.; Buchanan, B.B. Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature 1977, 266, 565–567. [Google Scholar] [CrossRef]
- Serrato, A.J.; Pérez-Ruiz, J.M.; Spínola, M.a.C.; Cejudo, F.J. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 43821–43827. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, B.; Mignée, C.; Krieger-Liszkay, A.; Hornero-Méndez, D.; Gallardo-Guerrero, L.; Cejudo, F.J.; Lindahl, M. The chloroplast NADPH thioredoxin reductase C, NTRC, controls non-photochemical quenching of light energy and photosynthetic electron transport in Arabidopsis. Plant Cell Environ. 2016, 39, 804–822. [Google Scholar] [CrossRef]
- Yoshida, K.; Hisabori, T. Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl. Acad. Sci. USA 2016, 113, E3967–E3976. [Google Scholar] [CrossRef]
- Pérez-Ruiz, J.M.; Naranjo, B.; Ojeda, V.; Guinea, M.; Cejudo, F.J. NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc. Natl. Acad. Sci. USA 2017, 114, 12069–12074. [Google Scholar] [CrossRef] [PubMed]
- Thormählen, I.; Meitzel, T.; Groysman, J.; Öchsner, A.B.; von Roepenack-Lahaye, E.; Naranjo, B.; Cejudo, F.J.; Geigenberger, P. Thioredoxin f 1 and NADPH-dependent thioredoxin reductase C have overlapping functions in regulating photosynthetic metabolism and plant growth in response to varying light conditions. Plant Physiol. 2015, 169, 1766–1786. [Google Scholar] [CrossRef] [PubMed]
- Nikkanen, L.; Toivola, J.; Rintamäki, E. Crosstalk between chloroplast thioredoxin systems in regulation of photosynthesis. Plant Cell Environ. 2016, 39, 1691–1705. [Google Scholar] [CrossRef]
- Nikkanen, L.; Rintamäki, E. Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochem. J. 2019, 476, 1159–1172. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, H.; Nagasaki, J.; Yoshikawa, N.; Yamamoto, A.; Takito, S.; Kawasaki, M.; Sugiyama, T.; Miyake, H.; Weber, A.P.; Taniguchi, M. The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 2011, 65, 15–26. [Google Scholar] [CrossRef]
- Selinski, J.; Scheibe, R. Malate valves: Old shuttles with new perspectives. Plant Biol. 2019, 21 (Suppl. 1), 21–30. [Google Scholar] [CrossRef] [PubMed]
- Scheibe, R. NADP+-malate dehydrogenase in C3-plants: Regulation and role of a light-activated enzyme. Physiol. Plant. 1987, 71, 393–400. [Google Scholar] [CrossRef]
- Berkemeyer, M.; Scheibe, R.; Ocheretina, O. A Novel, Non-redox-regulated NAD-dependent Malate Dehydrogenase from Chloroplasts of Arabidopsis thalianaL. J. Biol. Chem. 1998, 273, 27927–27933. [Google Scholar] [CrossRef]
- Turner, W.L.; Waller, J.C.; Vanderbeld, B.; Snedden, W.A. Cloning and characterization of two NAD kinases from Arabidopsis. Identification of a calmodulin binding isoform. Plant Physiol. 2004, 135, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Correll, C.C.; Ludwig, M.L.; Bruns, C.M.; Karplus, P.A. Structural prototypes for an extended family of flavoprotein reductases: Comparison of phthalate dioxygenase reductase with ferredoxin reductase and ferredoxin. Protein Sci. A Publ. Protein Soc. 1993, 2, 2112–2133. [Google Scholar] [CrossRef]
- Piubelli, L.; Aliverti, A.; Arakaki, A.K.; Carrillo, N.; Ceccarelli, E.A.; Karplus, P.A.; Zanetti, G. Competition between C-terminal tyrosine and nicotinamide modulates pyridine nucleotide affinity and specificity in plant ferredoxin-NADP(+) reductase. J. Biol. Chem. 2000, 275, 10472–10476. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.; Luquita, A.; Tejero, J.; Hermoso, J.; Mayoral, T.; Sanz-Aparicio, J.; Grever, K.; Gomez-Moreno, C. Probing the determinants of coenzyme specificity in ferredoxin-NADP+ reductase by site-directed mutagenesis. J. Biol. Chem. 2001, 276, 11902–11912. [Google Scholar] [CrossRef]
- Barber, J. The intact chloroplast. FEBS Lett. 1976, 97, 171–214. [Google Scholar]
- Balsera, M.; Uberegui, E.; Susanti, D.; Schmitz, R.A.; Mukhopadhyay, B.; Schürmann, P.; Buchanan, B.B. Ferredoxin:thioredoxin reductase (FTR) links the regulation of oxygenic photosynthesis to deeply rooted bacteria. Planta 2013, 237, 619–635. [Google Scholar] [CrossRef] [PubMed]
- Gütle, D.D.; Roret, T.; Müller, S.J.; Couturier, J.; Lemaire, S.D.; Hecker, A.; Dhalleine, T.; Buchanan, B.B.; Reski, R.; Einsle, O.; et al. Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc. Natl. Acad. Sci. USA 2016, 113, 6779–6784. [Google Scholar] [CrossRef]
- Ross, S.A.; Zhang, M.X.; Selman, B.R. Role of the Chlamydomonas reinhardtii Coupling Factor 1 γ-Subunit Cysteine Bridge in the Regulation of ATP Synthase(∗). J. Biol. Chem. 1995, 270, 9813–9818. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Ort, D.R. Mutation in the cysteine bridge domain of the gamma-subunit affects light regulation of the ATP synthase but not photosynthesis or growth in Arabidopsis. Photosynth. Res. 2008, 97, 185–193. [Google Scholar] [CrossRef]
- Miginiac-Maslow, M.; Issakidis, E.; Lemaire, M.; Ruelland, E.; Jacquot, J.-P.; Decottignies, P. Light-dependent Activation of NADP-Malate Dehydrogenase: A Complex Process. Funct. Plant Biol. 1997, 24, 529–542. [Google Scholar] [CrossRef]
- Hebbelmann, I.; Selinski, J.; Wehmeyer, C.; Goss, T.; Voss, I.; Mulo, P.; Kangasjärvi, S.; Aro, E.M.; Oelze, M.L.; Dietz, K.J.; et al. Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J. Exp. Bot. 2012, 63, 1445–1459. [Google Scholar] [CrossRef]
- Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant 2004, 120, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Knuesting, J.; Scheibe, R. Small Molecules Govern Thiol Redox Switches. Trends Plant Sci. 2018, 23, 769–782. [Google Scholar] [CrossRef]
- Heckmann, D.; Schulze, S.; Denton, A.; Gowik, U.; Westhoff, P.; Weber, A.P.; Lercher, M.J. Predicting C4 photosynthesis evolution: Modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 2013, 153, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, U.; Bräutigam, A.; Gowik, U.; Melzer, M.; Christin, P.-A.; Kurz, S.; Mettler-Altmann, T.; Weber, A.P. Photosynthesis in C3–C4 intermediate Moricandia species. J. Exp. Bot. 2016, 68, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Bräutigam, A.; Schliesky, S.; Külahoglu, C.; Osborne, C.P.; Weber, A.P. Towards an integrative model of C4 photosynthetic subtypes: Insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. J. Exp. Bot. 2014, 65, 3579–3593. [Google Scholar] [CrossRef]
- Bräutigam, A.; Kajala, K.; Wullenweber, J.; Sommer, M.; Gagneul, D.; Weber, K.L.; Carr, K.M.; Gowik, U.; Maß, J.; Lercher, M.J.; et al. An mRNA Blueprint for C4 Photosynthesis Derived from Comparative Transcriptomics of Closely Related C3 and C4 Species. Plant Physiol. 2010, 155, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Iwano, M.; Havaux, M.; Yokota, A.; Munekage, Y.N. Promotion of cyclic electron transport around photosystem I during the evolution of NADP-malic enzyme-type C4 photosynthesis in the genus Flaveria. New Phytol. 2013, 199, 832–842. [Google Scholar] [CrossRef]
- Shikanai, T. Central role of cyclic electron transport around photosystem I in the regulation of photosynthesis. Curr. Opin. Biotechnol. 2014, 26, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Suorsa, M.; Rossi, F.; Tadini, L.; Labs, M.; Colombo, M.; Jahns, P.; Kater, M.M.; Leister, D.; Finazzi, G.; Aro, E.M.; et al. PGR5-PGRL1-Dependent Cyclic Electron Transport Modulates Linear Electron Transport Rate in Arabidopsis thaliana. Mol. Plant 2016, 9, 271–288. [Google Scholar] [CrossRef]
- Krieger-Liszkay, A.; Feilke, K. The Dual Role of the Plastid Terminal Oxidase PTOX: Between a Protective and a Pro-oxidant Function. Front. Plant Sci. 2015, 6, 1147. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. Production and Scavenging of Reactive Oxygen Species in Chloroplasts and Their Functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Dietz, K.J. Thiol-Based Peroxidases and Ascorbate Peroxidases: Why Plants Rely on Multiple Peroxidase Systems in the Photosynthesizing Chloroplast? Mol. Cells 2016, 39, 20–25. [Google Scholar] [CrossRef]
- Lee, S.D.; Kim, S.J.; Jung, S.I.; Son, K.-C.; Kays, S.J. Diurnal CO2 assimilation patterns in nine species of CAM-type succulent plants. HortScience 2006, 41, 1373–1376. [Google Scholar] [CrossRef]
- Cushman, J. CAM Plants. In Encyclopedia of Applied Plant Sciences, 2nd ed.; Thomas, B., Murray, B.G., Murphy, D.J., Eds.; Academic Press: Oxford, UK, 2017; pp. 60–77. [Google Scholar]
- Turkan, I.; Uzilday, B.; Dietz, K.J.; Bräutigam, A.; Ozgur, R. Reactive oxygen species and redox regulation in mesophyll and bundle sheath cells of C4 plants. J. Exp. Bot. 2018, 69, 3321–3331. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-J.; Zhang, S.-B.; Huang, W. Photosynthetic regulation under fluctuating light in young and mature leaves of the CAM plant Bryophyllum pinnatum. Biochim. Biophys. Acta 2019, 1860, 469–477. [Google Scholar] [CrossRef]
- Yang, Y.-J.; Tan, S.-L.; Huang, J.-L.; Zhang, S.-B.; Huang, W.J.E.; Botany, E. The water-water cycle facilitates photosynthetic regulation under fluctuating light in the epiphytic orchid Dendrobium officinale. Environ. Exp. Bot. 2020, 180, 104238. [Google Scholar] [CrossRef]
- Sun, H.; Shi, Q.; Zhang, S.-B.; Huang, W.J.P. Coordination of cyclic electron flow and water–water cycle facilitates photoprotection under fluctuating light and temperature stress in the epiphytic orchid Dendrobium officinale. Plants 2021, 10, 606. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.-Q.; Zeng, Z.-L.; Yu, H.; Huang, W. Photosynthesis under fluctuating light in the CAM plant Vanilla planifolia. Plant Sci. 2022, 317, 111207. [Google Scholar] [CrossRef] [PubMed]
- Jacquot, J.-P.; Eklund, H.; Rouhier, N.; Schürmann, P. Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci. 2009, 14, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Schürmann, P.; Buchanan, B.B. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 2008, 10, 1235–1274. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H. Redox homeostasis: Opening up ascorbate transport. Nat. Plants 2015, 1, 14012. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J.; Jacob, S.; Oelze, M.-L.; Laxa, M.; Tognetti, V.; de Miranda, S.M.N.; Baier, M.; Finkemeier, I. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 2006, 57, 1697–1709. [Google Scholar] [CrossRef] [PubMed]
- Horemans, N.; Foyer, C.H.; Potters, G.; Asard, H. Ascorbate function and associated transport systems in plants. Plant Physiol. Biochem. 2000, 38, 531–540. [Google Scholar] [CrossRef]
- Beck, E.; Burkert, A.; Hofmann, M. Uptake of L-ascorbate by intact spinach chloroplasts. Plant Physiol. 1983, 73, 41–45. [Google Scholar] [CrossRef] [PubMed]
- de Koning, H.; Diallinas, G. Nucleobase transporters. Mol. Membr. Biol. 2000, 17, 75–94. [Google Scholar] [PubMed]
- Bürzle, M.; Suzuki, Y.; Ackermann, D.; Miyazaki, H.; Maeda, N.; Clémençon, B.; Burrier, R.; Hediger, M.A. The sodium-dependent ascorbic acid transporter family SLC23. Mol. Asp. Med. 2013, 34, 436–454. [Google Scholar] [CrossRef]
- Diallinas, G.; Valdez, J.; Sophianopoulou, V.; Rosa, A.; Scazzocchio, C. Chimeric purine transporters of Aspergillus nidulans define a domain critical for function and specificity conserved in bacterial, plant and metazoan homologues. EMBO J. 1998, 17, 3827–3837. [Google Scholar] [CrossRef] [PubMed]
- Argyrou, E.; Sophianopoulou, V.; Schultes, N.; Diallinas, G. Functional characterization of a maize purine transporter by expression in Aspergillus nidulans. Plant Cell 2001, 13, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Schultes, N.P.; Brutnell, T.P.; Allen, A.; Dellaporta, S.L.; Nelson, T.; Chen, J. Leaf permease1 gene of maize is required for chloroplast development. Plant Cell 1996, 8, 463–475. [Google Scholar] [PubMed]
- Miyaji, T.; Kuromori, T.; Takeuchi, Y.; Yamaji, N.; Yokosho, K.; Shimazawa, A.; Sugimoto, E.; Omote, H.; Ma, J.F.; Shinozaki, K.; et al. AtPHT4;4 is a chloroplast-localized ascorbate transporter in Arabidopsis. Nat. Commun. 2015, 6, 5928. [Google Scholar] [CrossRef]
- Zhang, J.; Kirkham, M.B. Antioxidant responses to drought in sunflower and sorghum seedlings. New Phytol. 1996, 132, 361–373. [Google Scholar] [CrossRef]
- Stepien, P.; Klobus, G. Antioxidant defense in the leaves of C3 and C4 plants under salinity stress. Physiol. Plant. 2005, 125, 31–40. [Google Scholar] [CrossRef]
- Nayyar, H.; Gupta, D. Differential sensitivity of C3 and C4 plants to water deficit stress: Association with oxidative stress and antioxidants. Environ. Exp. Bot. 2006, 58, 106–113. [Google Scholar] [CrossRef]
- Uzilday, B.; Turkan, I.; Sekmen, A.H.; Ozgur, R.; Karakaya, H.C. Comparison of ROS formation and antioxidant enzymes in Cleome gynandra (C4) and Cleome spinosa (C3) under drought stress. Plant Sci. 2012, 182, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Uzilday, B.; Turkan, I.; Ozgur, R.; Sekmen, A.H. Strategies of ROS regulation and antioxidant defense during transition from C3 to C4 photosynthesis in the genus Flaveria under PEG-induced osmotic stress. J. Plant Physiol. 2014, 171, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Uzilday, B.; Ozgur, R.; Yalcinkaya, T.; Turkan, I.; Sekmen, A.H. Changes in redox regulation during transition from C3 to single cell C4 photosynthesis in Bienertia sinuspersici. J. Plant Physiol. 2018, 220, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lara, M.V.; Offermann, S.; Smith, M.; Okita, T.W.; Andreo, C.S.; Edwards, G.E. Leaf development in the single-cell C4 system in Bienertia sinuspersici: Expression of genes and peptide levels for C4 metabolism in relation to chlorenchyma structure under different light conditions. Plant Physiol. 2008, 148, 593–610. [Google Scholar] [CrossRef]
- Baier, M.; Noctor, G.; Foyer, C.H.; Dietz, K.J. Antisense suppression of 2-cysteine peroxiredoxin in Arabidopsis specifically enhances the activities and expression of enzymes associated with ascorbate metabolism but not glutathione metabolism. Plant Physiol. 2000, 124, 823–832. [Google Scholar] [CrossRef]
- Kangasjärvi, S.; Lepistö, A.; Hännikäinen, K.; Piippo, M.; Luomala, E.M.; Aro, E.M.; Rintamäki, E. Diverse roles for chloroplast stromal and thylakoid-bound ascorbate peroxidases in plant stress responses. Biochem. J. 2008, 412, 275–285. [Google Scholar] [CrossRef]
- Aro, E.M.; Virgin, I.; Andersson, B. Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1993, 1143, 113–134. [Google Scholar] [CrossRef]
- Chen, J.H.; Chen, S.T.; He, N.Y.; Wang, Q.L.; Zhao, Y.; Gao, W.; Guo, F.Q. Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nat. Plants 2020, 6, 570–580. [Google Scholar] [CrossRef]
- Theis, J.; Schroda, M. Revisiting the photosystem II repair cycle. Plant Signal. Behav. 2016, 11, e1218587. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Bueno, M.L.P.; Zia, A.; Horton, P.; Ruban, A.V. The Zeaxanthin-Independent and Zeaxanthin-Dependent qE Components of Nonphotochemical Quenching Involve Common Conformational Changes within the Photosystem II Antenna in Arabidopsis. Plant Physiol. 2008, 149, 1061–1075. [Google Scholar] [CrossRef]
- Jahns, P.; Holzwarth, A.R. The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim. Biophys. Acta 2012, 1817, 182–193. [Google Scholar] [CrossRef]
- Hieber, A.D.; Bugos, R.C.; Yamamoto, H.Y. Plant lipocalins: Violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim. Biophys. Acta 2000, 1482, 84–91. [Google Scholar] [CrossRef]
- Hoang, M.H.; Kim, H.-S.; Zulfugarov, I.S.; Lee, C.-H. Down-Regulation of Zeaxanthin Epoxidation in Vascular Plant Leaves Under Normal and Photooxidative Stress Conditions. J. Plant Biol. 2020, 63, 331–336. [Google Scholar] [CrossRef]
- Zhu, X.G.; Ort, D.R.; Whitmarsh, J.; Long, S.P. The slow reversibility of photosystem II thermal energy dissipation on transfer from high to low light may cause large losses in carbon gain by crop canopies: A theoretical analysis. J. Exp. Bot. 2004, 55, 1167–1175. [Google Scholar] [CrossRef]
- Murchie, E.H.; Niyogi, K.K. Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 2011, 155, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Kromdijk, J.; Głowacka, K.; Leonelli, L.; Gabilly, S.T.; Iwai, M.; Niyogi, K.K.; Long, S.P. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 2016, 354, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Molina, A.; Leister, D. Accelerated relaxation of photoprotection impairs biomass accumulation in Arabidopsis. Nat. Plants 2020, 6, 9–12. [Google Scholar] [CrossRef]
- Goss, R.; Lepetit, B. Biodiversity of NPQ. J. Plant Physiol. 2015, 172, 13–32. [Google Scholar] [CrossRef] [PubMed]
- Schierenbeck, L.; Ries, D.; Rogge, K.; Grewe, S.; Weisshaar, B.; Kruse, O. Fast forward genetics to identify mutations causing a high light tolerant phenotype in Chlamydomonas reinhardtii by whole-genome-sequencing. BMC Genom. 2015, 16, 57. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riaz, A.; Deng, F.; Chen, G.; Jiang, W.; Zheng, Q.; Riaz, B.; Mak, M.; Zeng, F.; Chen, Z.-H. Molecular Regulation and Evolution of Redox Homeostasis in Photosynthetic Machinery. Antioxidants 2022, 11, 2085. https://doi.org/10.3390/antiox11112085
Riaz A, Deng F, Chen G, Jiang W, Zheng Q, Riaz B, Mak M, Zeng F, Chen Z-H. Molecular Regulation and Evolution of Redox Homeostasis in Photosynthetic Machinery. Antioxidants. 2022; 11(11):2085. https://doi.org/10.3390/antiox11112085
Chicago/Turabian StyleRiaz, Adeel, Fenglin Deng, Guang Chen, Wei Jiang, Qingfeng Zheng, Bisma Riaz, Michelle Mak, Fanrong Zeng, and Zhong-Hua Chen. 2022. "Molecular Regulation and Evolution of Redox Homeostasis in Photosynthetic Machinery" Antioxidants 11, no. 11: 2085. https://doi.org/10.3390/antiox11112085
APA StyleRiaz, A., Deng, F., Chen, G., Jiang, W., Zheng, Q., Riaz, B., Mak, M., Zeng, F., & Chen, Z.-H. (2022). Molecular Regulation and Evolution of Redox Homeostasis in Photosynthetic Machinery. Antioxidants, 11(11), 2085. https://doi.org/10.3390/antiox11112085










