Review of the Mechanisms by Which Transcription Factors and Exogenous Substances Regulate ROS Metabolism under Abiotic Stress
Abstract
:1. ROS Metabolism under Abiotic Stress
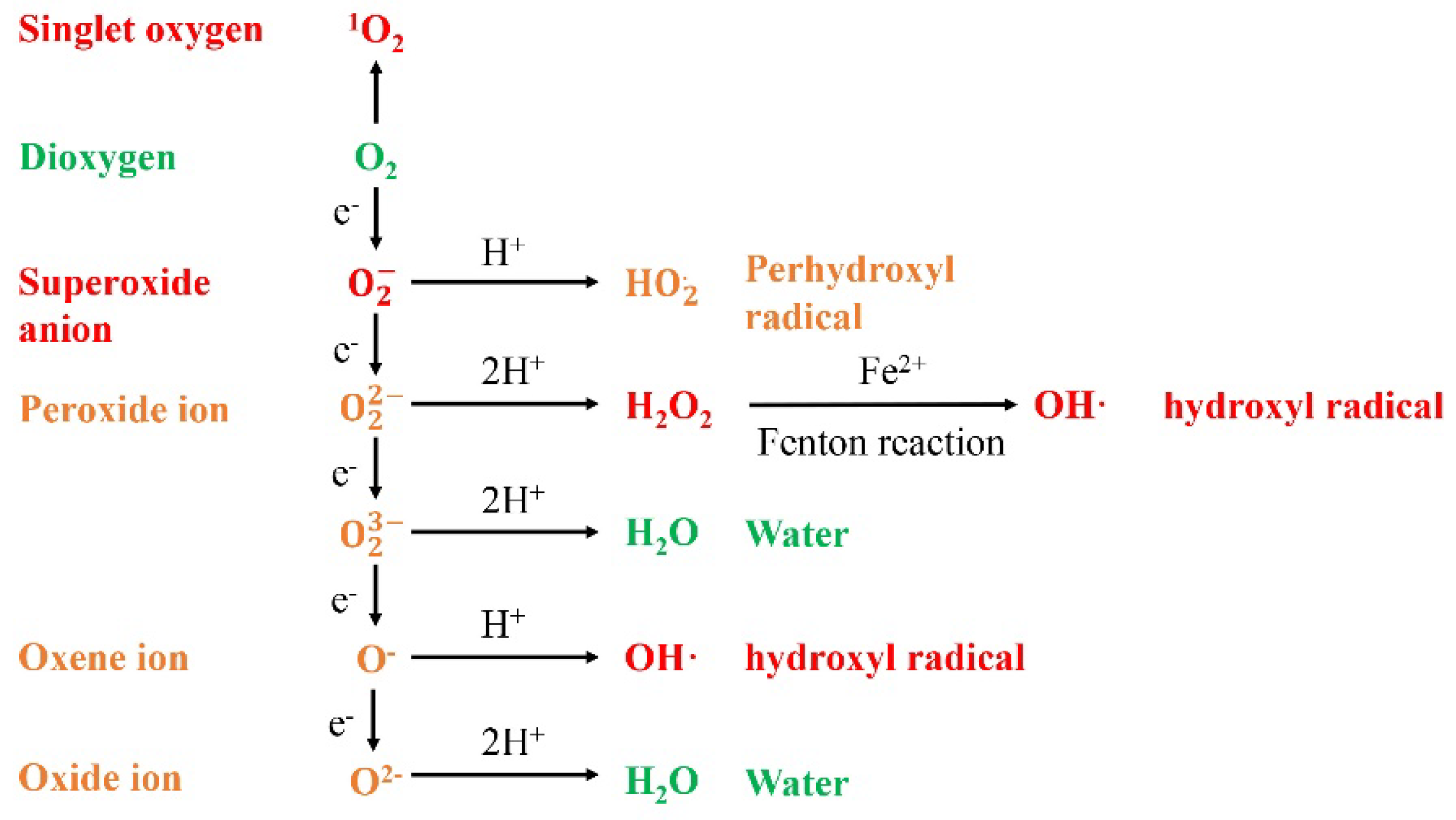
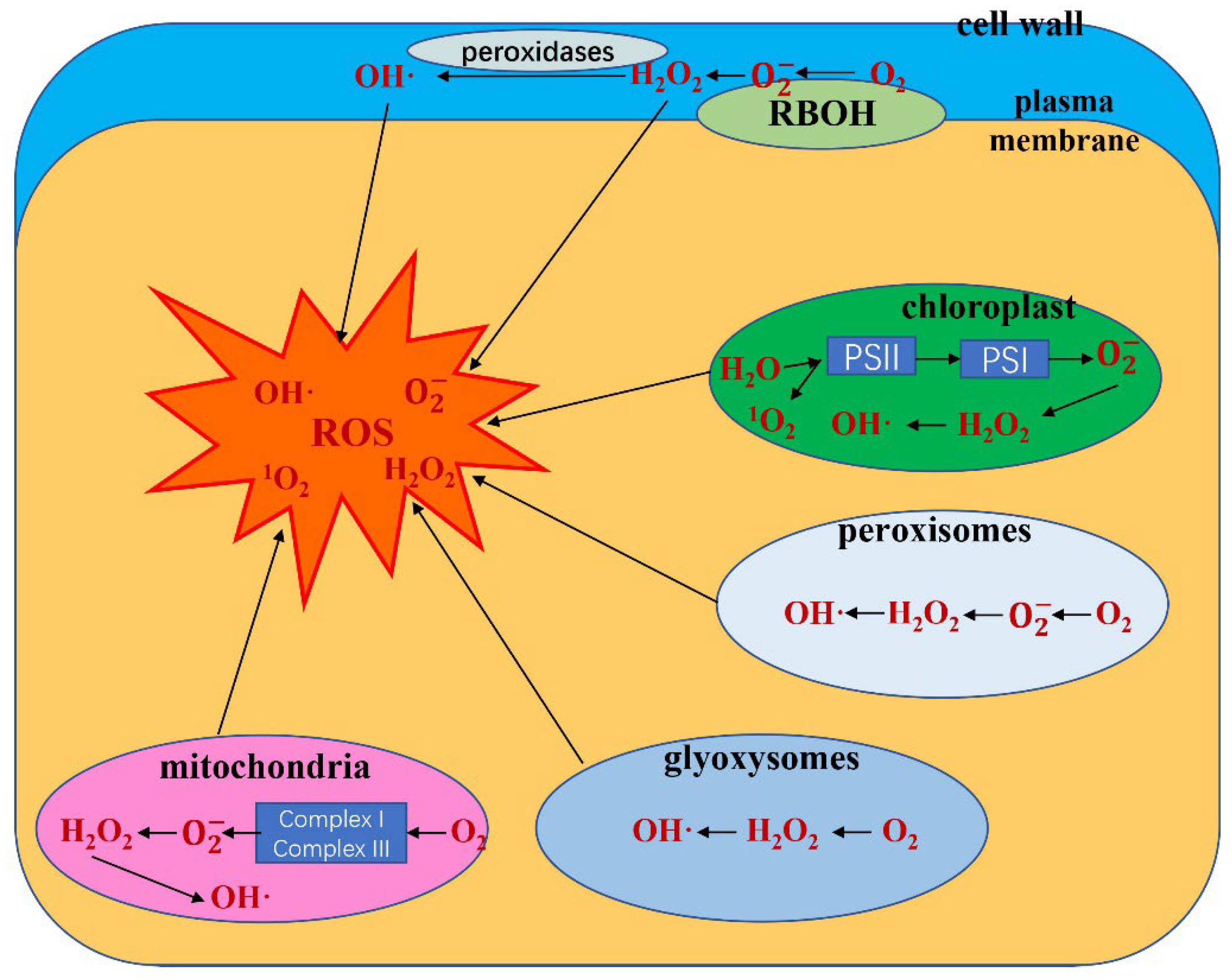
1.1. ROS Generation under Abiotic Stress
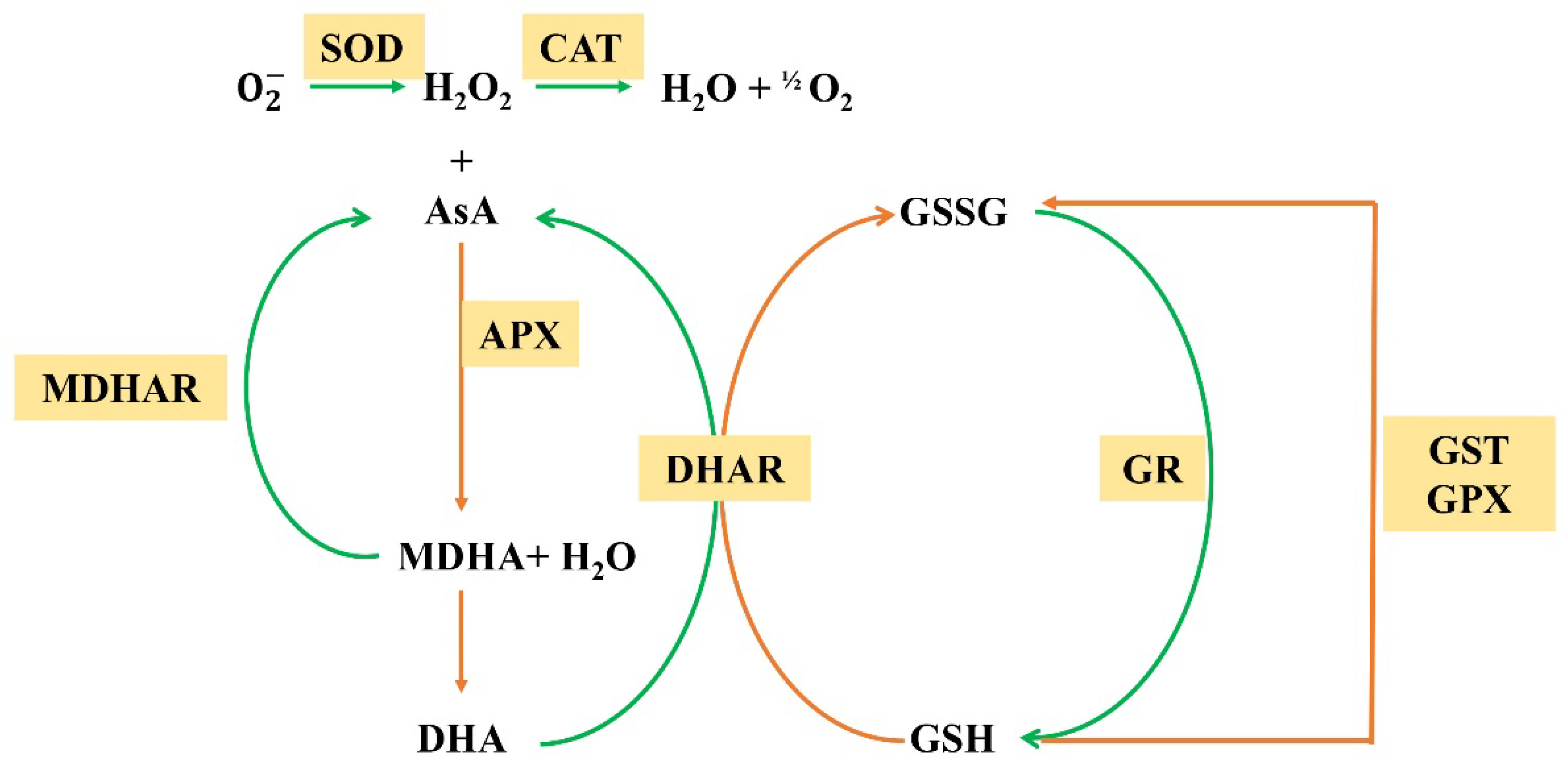
1.2. ROS Scavenging under Abiotic Stress
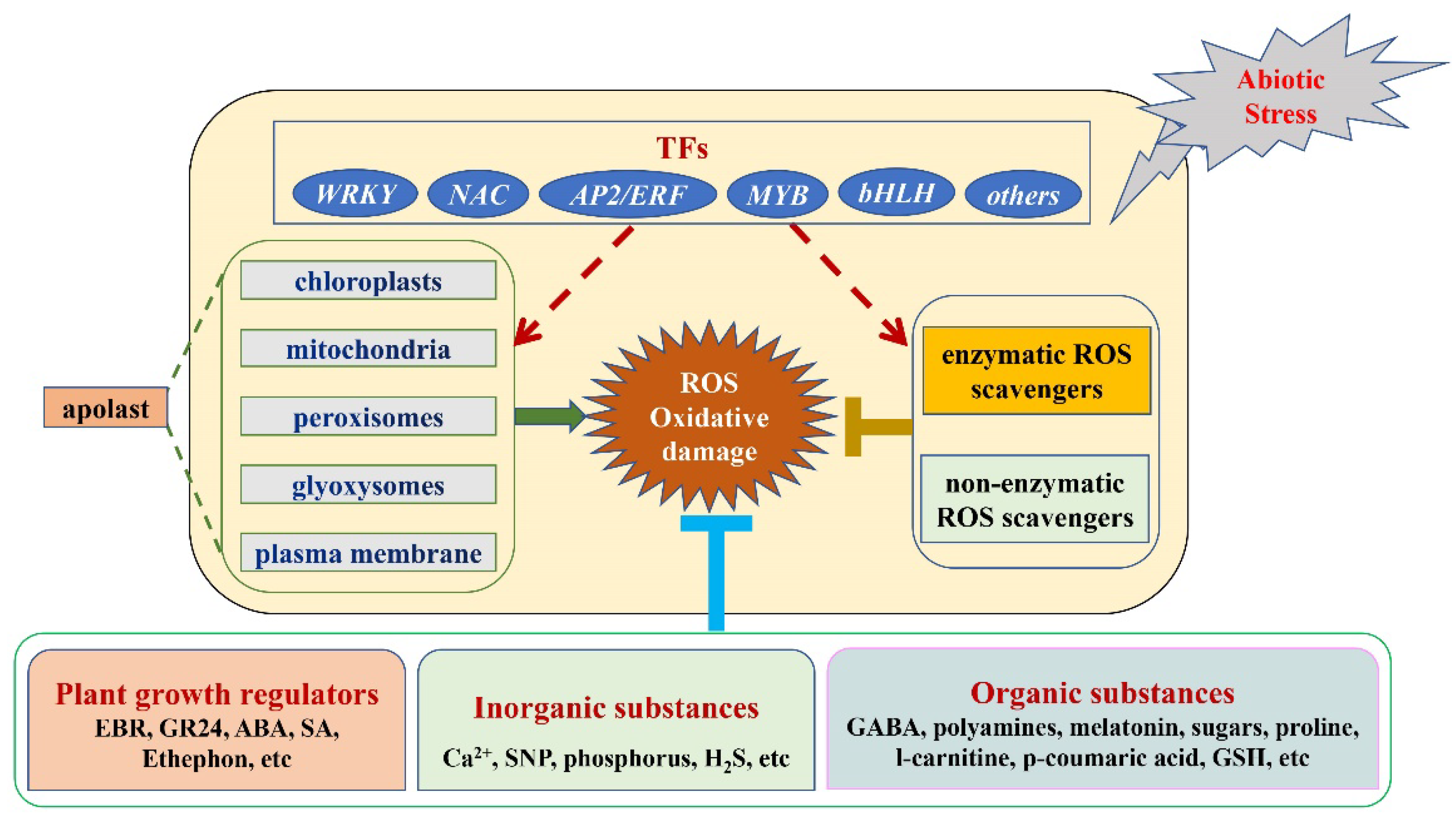
2. The Function of Transcription Factors in ROS Metabolism under Abiotic Stress
3. The Mechanism by Which Exogenous Substances Regulate ROS Metabolism
3.1. Plant Growth Regulators
3.1.1. Epibrassinolide (EBR)
3.1.2. GR24
3.1.3. Abscisic Acid (ABA)
3.1.4. Salicylic Acid (SA)
3.1.5. Ethephon
3.2. Inorganic Substances
3.2.1. Ca2+
3.2.2. Sodium Nitroprusside (SNP)
3.2.3. Other Inorganic Substances
3.3. Organic Substances
3.3.1. Gamma-Aminobutyric Acid (GABA)
3.3.2. Polyamines
3.3.3. Melatonin
3.3.4. Sugars
3.3.5. Other Organic Substances
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2010, 138, 414–429. [Google Scholar] [CrossRef] [PubMed]
- Zurbriggen, M.D.; Carrillo, N.; Hajirezaei, M.-R. ROS signaling in the hypersensitive response: When, where and what for? Plant Signal. Behav. 2010, 5, 393–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oracz, K.; Karpiński, S. Phytohormones Signaling Pathways and ROS Involvement in Seed Germination. Front. Plant Sci. 2016, 7, 864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, H.-W.; Koh, E.-J.; Kim, S.-H.; Kim, K.-I.; Lee, H.; Hong, S.-W. Glucosamine causes overproduction of reactive oxygen species, leading to repression of hypocotyl elongation through a hexokinase-mediated mechanism in Arabidopsis. J. Plant Physiol. 2009, 166, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Tsukagoshi, H.; Busch, W.; Benfey, P.N. Transcriptional Regulation of ROS Controls Transition from Proliferation to Differentiation in the Root. Cell 2010, 143, 606–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mhamdi, A.; Van Breusegem, F. Reactive oxygen species in plant development. Development 2018, 145, dev164376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailly, C. The signalling role of ROS in the regulation of seed germination and dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Prakash, V.; Vishwakarma, K.; Singh, V.; Rai, P.; Ramawat, N.; Tripathi, D.K.; Sharma, S. NO and ROS implications in organization of root system architecture. Physiol. Plant. 2019, 168, 473–489. [Google Scholar] [CrossRef] [PubMed]
- Schippers, J.H.; Foyer, C.H.; van Dongen, J.T. Redox regulation in shoot growth, SAM maintenance and flowering. Curr. Opin. Plant Biol. 2016, 29, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Singh, S.; Parihar, P.; Mishra, R.K.; Tripathi, D.K.; Singh, V.P.; Chauhan, D.K.; Prasad, S.M. Reactive Oxygen Species (ROS): Beneficial Companions of Plant’ Developmental Processes. Front. Plant Sci. 2016, 7, 1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Li, G.; Manzoor, M.A.; Wang, H.; Abdullah, M.; Su, X.; Zhang, J.; Jiang, T.; Jin, Q.; Cai, Y.; et al. In Silico Genome-Wide Analysis of Respiratory Burst Oxidase Homolog (RBOH) Family Genes in Five Fruit-Producing Trees, and Potential Functional Analysis on Lignification of Stone Cells in Chinese White Pear. Cells 2019, 8, 520. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Cao, X.; Wang, X.; Zhang, W.; Li, W.; Wang, X.; Liu, S.; Lyu, D. RBOH-dependent hydrogen peroxide signaling mediates melatonin-induced anthocyanin biosynthesis in red pear fruit. Plant Sci. 2021, 313, 111093. [Google Scholar] [CrossRef]
- Chu-Puga, A.; González-Gordo, S.; Rodríguez-Ruiz, M.; Palma, J.M.; Corpas, F.J. NADPH Oxidase (Rboh) Activity is Up Regulated during Sweet Pepper (Capsicum annuum L.) Fruit Ripening. Antioxidants 2019, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Orman-Ligeza, B.; Parizot, B.; de Rycke, R.; Fernandez, A.; Himschoot, E.; Van Breusegem, F.; Bennett, M.J.; Périlleux, C.; Beeckman, T.; Draye, X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 2016, 143, 3328–3339. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Sun, L.; Ma, L.; Hao, F.-S. Both AtrbohD and AtrbohF are essential for mediating responses to oxygen deficiency in Arabidopsis. Plant Cell Rep. 2017, 36, 947–957. [Google Scholar] [CrossRef]
- Ren, X.; Wang, M.; Wang, Y.; Huang, A. Superoxide anion generation response to wound in Arabidopsis hypocotyl cutting. Plant Signal. Behav. 2020, 16, 1848086. [Google Scholar] [CrossRef] [PubMed]
- Kurusu, T.; Kuchitsu, K.; Tada, Y. Plant signaling networks involving Ca2+ and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Sci. 2015, 6, 427. [Google Scholar] [CrossRef] [Green Version]
- Asai, S.; Ichikawa, T.; Nomura, H.; Kobayashi, M.; Kamiyoshihara, Y.; Mori, H.; Kadota, Y.; Zipfel, C.; Jones, J.D.G.; Yoshioka, H. The Variable Domain of a Plant Calcium-dependent Protein Kinase (CDPK) Confers Subcellular Localization and Substrate Recognition for NADPH Oxidase. J. Biol. Chem. 2013, 288, 14332–14340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, M.; Ohura, I.; Kawakita, K.; Yokota, N.; Fujiwara, M.; Shimamoto, K.; Doke, N.; Yoshioka, H. Calcium-Dependent Protein Kinases Regulate the Production of Reactive Oxygen Species by Potato NADPH Oxidase. Plant Cell 2007, 19, 1065–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakubowicz, M.; Gałgańska, H.; Nowak, W.; Sadowski, J. Exogenously induced expression of ethylene biosynthesis, ethylene perception, phospholipase D, and Rboh-oxidase genes in broccoli seedlings. J. Exp. Bot. 2010, 61, 3475–3491. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Gu, Y.; Lee, Y.; Yang, Z.; Lee, Y. Phosphatidic Acid Induces Leaf Cell Death in Arabidopsis by Activating the Rho-Related Small G Protein GTPase-Mediated Pathway of Reactive Oxygen Species Generation. Plant Physiol. 2004, 134, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Gilroy, S.; Białasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- Kerchev, P.; Waszczak, C.; Lewandowska, A.; Willems, P.; Shapiguzov, A.; Li, Z.; Alseekh, S.; Mühlenbock, P.; Hoeberichts, F.A.; Huang, J.; et al. Lack of GLYCOLATE OXIDASE1, but Not GLYCOLATE OXIDASE2, Attenuates the Photorespiratory Phenotype of CATALASE2-Deficient Arabidopsis. Plant Physiol. 2016, 171, 1704–1719. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, M.; Romero-Puertas, M.C.; Sanz-Fernández, M.; Hu, J.; Sandalio, L.M. Peroxisomes Extend Peroxules in a Fast Response to Stress via a Reactive Oxygen Species-Mediated Induction of the Peroxin PEX11a. Plant Physiol. 2016, 171, 1665–1674. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Xia, X.J.; Zhou, Y.H.; Shi, K.; Chen, Z.X.; Yu, J.Q. RBOH1-dependent H2O2 production and subsequent activation of MPK1/2 play an important role in acclimation-induced cross-tolerance in tomato. J. Exp. Bot. 2014, 65, 595–607. [Google Scholar]
- Torres, M.A.; Dangl, J.L.; Jones, J.D.G. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mori, I.C.; Pei, Z.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Kabała, K.; Reda, M.; Wdowikowska, A.; Janicka, M. Role of Plasma Membrane NADPH Oxidase in Response to Salt Stress in Cucumber Seedlings. Antioxidants 2022, 11, 1534. [Google Scholar] [CrossRef] [PubMed]
- Gémes, K.; Kim, Y.J.; Park, K.Y.; Moschou, P.N.; Andronis, E.; Valassaki, C.; Roussis, A.; Roubelakis-Angelakis, K.A. An NADPH-Oxidase/Polyamine Oxidase Feedback Loop Controls Oxidative Burst Under Salinity. Plant Physiol. 2016, 172, 1418–1431. [Google Scholar] [CrossRef] [Green Version]
- Di, Q.; Li, Y.; Li, S.; Shi, A.; Zhou, M.; Ren, H.; Yan, Y.; He, C.; Wang, J.; Sun, M.; et al. Photosynthesis Mediated by RBOH-Dependent Signaling Is Essential for Cold Stress Memory. Antioxidants 2022, 11, 969. [Google Scholar] [CrossRef]
- Zhang, X.; Bian, A.; Li, T.; Ren, L.; Li, L.; Su, Y.; Zhang, Q. ROS and calcium oscillations are required for polarized root hair growth. Plant Signal. Behav. 2022, 17, 2106410. [Google Scholar] [CrossRef]
- Voothuluru, P.; Thompson, H.J.; Flint-Garcia, S.A.; Sharp, R.E. Genetic variability of oxalate oxidase activity and elongation in water-stressed primary roots of diverse maize and rice lines. Plant Signal. Behav. 2013, 8, e23454. [Google Scholar] [CrossRef] [Green Version]
- Berna, A.; Bernier, F. Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Mol. Biol. 1999, 39, 539–549. [Google Scholar] [CrossRef]
- Li, Z.; Wakao, S.; Fischer, B.B.; Niyogi, K.K. Sensing and Responding to Excess Light. Annu. Rev. Plant Biol. 2009, 60, 239–260. [Google Scholar] [CrossRef]
- Murchie, E.H.; Niyogi, K.K. Manipulation of Photoprotection to Improve Plant Photosynthesis. Plant Physiol. 2011, 155, 86–92. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.; Li, X.-P.; Niyogi, K.K. Non-Photochemical Quenching. A Response to Excess Light Energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruban, A.V. Nonphotochemical Chlorophyll Fluorescence Quenching: Mechanism and Effectiveness in Protecting Plants from Photodamage. Plant Physiol. 2016, 170, 1903–1916. [Google Scholar] [CrossRef] [Green Version]
- Marchi, S.; Giorgi, C.; Suski, J.M.; Agnoletto, C.; Bononi, A.; Bonora, M.; De Marchi, E.; Missiroli, S.; Patergnani, S.; Poletti, F.; et al. Mitochondria-Ros Crosstalk in the Control of Cell Death and Aging. J. Signal Transduct. 2011, 2012, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffens, B.; Steffens, B. The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 2014, 5, 685. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.M.; Umbach, A.L.; Subbaiah, C.C.; Siedow, J.N. Mitochondrial Reactive Oxygen Species. Contribution to Oxidative Stress and Interorganellar Signaling. Plant Physiol. 2006, 141, 357–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dröse, S.; Brandt, U. Molecular Mechanisms of Superoxide Production by the Mitochondrial Respiratory Chain. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 748, pp. 145–169. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, S.; Murata, N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 2008, 13, 178–182. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of Photosynthesis during Abiotic Stress-Induced Photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [Green Version]
- Maslenkova, L.T.; Zanev, Y.; Popova, L.P. Effect of abscisic acid on the photosynthetic oxygen evolution in barley chloroplasts. Photosyn. Res. 1989, 21, 45–50. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Harb, A.; Krishnan, A.; Ambavaram, M.M.; Pereira, A. Molecular and Physiological Analysis of Drought Stress in Arabidopsis Reveals Early Responses Leading to Acclimation in Plant Growth. Plant Physiol. 2010, 154, 1254–1271. [Google Scholar] [CrossRef] [Green Version]
- Elsawy, H.I.; Mekawy, A.; Elhity, M.A.; Abdel-Dayem, S.M.; Abdelaziz, M.N.; Assaha, D.V.; Ueda, A.; Saneoka, H. Differential responses of two Egyptian barley (Hordeum vulgare L.) cultivars to salt stress. Plant Physiol. Biochem. 2018, 127, 425–435. [Google Scholar] [CrossRef]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant Flavonoids—Biosynthesis, Transport and Involvement in Stress Responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef] [Green Version]
- Kaling, M.; Kanawati, B.; Ghirardo, A.; Albert, A.; Winkler, J.B.; Heller, W.; Barta, C.; Loreto, F.; Schmitt-Kopplin, P.; Schnitzler, J.-P. UV-B mediated metabolic rearrangements in poplar revealed by non-targeted metabolomics. Plant Cell Environ. 2014, 38, 892–904. [Google Scholar] [CrossRef]
- Abouzeid, S.; Beutling, U.; Selmar, D. Stress-induced modification of indole alkaloids:Phytomodificines as a new category of specialized metabolites. Phytochemistry 2018, 159, 102–107. [Google Scholar] [CrossRef]
- Nomura, M.; Ma, W.Y.; Huang, C.; Yang, C.S.; Bowden, G.T.; Miyamoto, K.I.; Dong, Z. Inhibition of ultraviolet B-induced AP-1 activation by theaflavins from black tea. Mol Carcinog. 2000, 28, 148–155. [Google Scholar] [CrossRef]
- Alseekh, S.; Ofner, I.; Liu, Z.; Osorio, S.; Vallarino, J.; Last, R.L.; Zamir, D.; Tohge, T.; Fernie, A.R. Quantitative trait loci analysis of seed-specialized metabolites reveals seed-specific flavonols and differential regulation of glycoalkaloid content in tomato. Plant J. 2020, 103, 2007–2024. [Google Scholar] [CrossRef]
- Liu, D.; Li, Y.-Y.; Zhou, Z.-C.; Xiang, X.; Liu, X.; Wang, J.; Hu, Z.-R.; Xiang, S.-P.; Li, W.; Xiao, Q.-Z.; et al. Tobacco transcription factor bHLH123 improves salt tolerance by activating NADPH oxidase NtRbohE expression. Plant Physiol. 2021, 186, 1706–1720. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; He, R.J.; Xie, Q.L.; Zhao, X.H.; Deng, X.M.; He, J.B.; Song, L.; He, J.; Marchant, A.; Chen, X.Y.; et al. ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 2017, 213, 1667–1681. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.A.; Al-Ghamdi, A.A.; Ali, H.M.; Ahmad, M. Overexpression of AtWRKY30 Transcription Factor Enhances Heat and Drought Stress Tolerance in Wheat (Triticum aestivum L.). Genes 2019, 10, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Hu, W.; Zhou, R.; Wang, L.; Wang, X.; Wang, Q.; Feng, Z.; Li, Y.; Qiu, D.; He, G.; et al. The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 2014, 34, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chi, X.; Guo, F.; Jin, X.; Luo, H.; Hawar, A.; Chen, Y.; Feng, K.; Wang, B.; Qi, J.; et al. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 2020, 246–247, 153142. [Google Scholar] [CrossRef]
- Li, J.-B.; Luan, Y.-S.; Liu, Z. Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 2015, 155, 248–266. [Google Scholar] [CrossRef]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Yang, J.-F.; Li, M.; Xu, Z.-S.; Fu, J.-D. The Maize WRKY Transcription Factor ZmWRKY40 Confers Drought Resistance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Li, M.; Zhao, D.; Yang, J.-F.; Fu, J.-D.; Xu, Z.-S. Maize WRKY Transcription Factor ZmWRKY106 Confers Drought and Heat Tolerance in Transgenic Plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef] [Green Version]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2017, 16, 354–366. [Google Scholar] [CrossRef] [Green Version]
- Borgohain, P.; Saha, B.; Agrahari, R.; Chowardhara, B.; Sahoo, S.; van der Vyver, C.; Panda, S.K. SlNAC2 overexpression in Arabidopsis results in enhanced abiotic stress tolerance with alteration in glutathione metabolism. Protoplasma 2019, 256, 1065–1077. [Google Scholar] [CrossRef]
- Niu, F.; Wang, C.; Yan, J.; Guo, X.; Wu, F.; Yang, B.; Deyholos, M.K.; Jiang, Y.-Q. Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napus L.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Mol. Biol. 2016, 92, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Fan, Z.; Qiu, L.; Che, Q.; Wang, T.; Li, Y.; Wang, Y. MdbHLH130, an Apple bHLH Transcription Factor, Confers Water Stress Resistance by Regulating Stomatal Closure and ROS Homeostasis in Transgenic Tobacco. Front. Plant Sci. 2020, 11, 543696. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, Z.; Zhang, H.; Wang, X.-C.; Huang, R. Transcriptional Modulation of Ethylene Response Factor Protein JERF3 in the Oxidative Stress Response Enhances Tolerance of Tobacco Seedlings to Salt, Drought, and Freezing. Plant Physiol. 2008, 148, 1953–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Li, W.; Hu, X.; Guo, J.; Liu, A.; Zhang, B. A Cotton MYB Transcription Factor, GbMYB5, is Positively Involved in Plant Adaptive Response to Drought Stress. Plant Cell Physiol. 2015, 56, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Zhang, F.; Sun, F.; Luo, Q.; Wang, R.; Hu, R.; Chen, M.; Chang, J.; Yang, G.; He, G. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. 2017, 265, 112–123. [Google Scholar] [CrossRef]
- Li, K.; Xing, C.; Yao, Z.; Huang, X. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 2017, 15, 1186–1203. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Yan, C.; Sun, Q.; Wang, J.; Yuan, C.; Mou, Y.; Shan, S.; Zhao, X. The bHLH transcription factor AhbHLH112 improves the drought tolerance of peanut. BMC Plant Biol. 2021, 21, 540. [Google Scholar] [CrossRef]
- Cui, X.-Y.; Gao, Y.; Guo, J.; Yu, T.-F.; Zheng, W.-J.; Liu, Y.-W.; Chen, J.; Xu, Z.-S.; Ma, Y.-Z. BES/BZR Transcription Factor TaBZR2 Positively Regulates Drought Responses by Activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef] [Green Version]
- Meng, D.; Li, Y.; Bai, Y.; Li, M.; Cheng, L. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 2016, 103, 71–83. [Google Scholar] [CrossRef]
- Liu, B.; Jiang, Y.; Tang, H.; Tong, S.; Lou, S.; Shao, C.; Zhang, J.; Song, Y.; Chen, N.; Bi, H.; et al. The ubiquitin E3 ligase SR1 modulates the submergence response by degrading phosphorylated WRKY33 in Arabidopsis. Plant Cell 2021, 33, 1771–1789. [Google Scholar] [CrossRef]
- Gonzali, S.; Loreti, E.; Cardarelli, F.; Novi, G.; Parlanti, S.; Pucciariello, C.; Bassolino, L.; Banti, V.; Licausi, F.; Perata, P. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 2015, 1, 15151. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-Y.; Seok, H.-Y.; Woo, D.-H.; Lee, S.-Y.; Tarte, V.N.; Lee, E.-H.; Lee, C.-H.; Moon, Y.-H. AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 414, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The Submergence Tolerance Regulator SUB1A Mediates Crosstalk between Submergence and Drought Tolerance in Rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Jiang, Y.; Guo, Y.; Huang, J.; Zhou, M.; Tang, Y.; Sui, J.; Wang, J.; Qiao, L. A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 2021, 160, 175–183. [Google Scholar] [CrossRef]
- Bao, W.; Wang, X.; Chen, M.; Chai, T.; Wang, H. A WRKY transcription factor, PcWRKY33, from Polygonum cuspidatum reduces salt tolerance in transgenic Arabidopsis thaliana. Plant Cell Rep. 2018, 37, 1033–1048. [Google Scholar] [CrossRef]
- Xiang, X.-Y.; Chen, J.; Xu, W.-X.; Qiu, J.-R.; Song, L.; Wang, J.-T.; Tang, R.; Chen, D.; Jiang, C.-Z.; Huang, Z. Dehydration-Induced WRKY Transcriptional Factor MfWRKY70 of Myrothamnus flabellifolia Enhanced Drought and Salinity Tolerance in Arabidopsis. Biomolecules 2021, 11, 327. [Google Scholar] [CrossRef]
- Kang, G.; Yan, D.; Chen, X.; Yang, L.; Zeng, R. HbWRKY82, a novel IIc WRKY transcription factor from Hevea brasiliensis associated with abiotic stress tolerance and leaf senescence in Arabidopsis. Physiol. Plant. 2020, 171, 151–160. [Google Scholar] [CrossRef]
- Lv, B.; Wu, Q.; Wang, A.; Li, Q.; Dong, Q.; Yang, J.; Zhao, H.; Wang, X.; Chen, H.; Li, C. A WRKY transcription factor, FtWRKY46, from Tartary buckwheat improves salt tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2019, 147, 43–53. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2020, 105, 333–345. [Google Scholar] [CrossRef]
- Ma, B.; Liu, X.; Guo, S.; Xie, X.; Zhang, J.; Wang, J.; Zheng, L.; Wang, Y. RtNAC100 involved in the regulation of ROS, Na+ accumulation and induced salt-related PCD through MeJA signal pathways in recretohalophyte Reaumuria trigyna. Plant Sci. 2021, 310, 110976. [Google Scholar] [CrossRef]
- Ma, J.; Wang, L.-Y.; Dai, J.-X.; Wang, Y.; Lin, D. The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 2021, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Melo, B.P.; Lourenço-Tessutti, I.T.; Fraga, O.T.; Pinheiro, L.B.; Lins, C.B.D.J.; Morgante, C.V.; Engler, J.A.; Reis, P.A.B.; Grossi-De-Sá, M.F.; Fontes, E.P.B. Contrasting roles of GmNAC065 and GmNAC085 in natural senescence, plant development, multiple stresses and cell death responses. Sci. Rep. 2021, 11, 11178. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Zhu, C.; Wang, C.; Liu, L.; Shen, Q.; Xu, D.; Wang, Q. Maize transcription factor ZmEREB20 enhanced salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 159, 257–267. [Google Scholar] [CrossRef]
- Lu, L.; Qanmber, G.; Li, J.; Pu, M.; Chen, G.; Li, S.; Liu, L.; Qin, W.; Ma, S.; Wang, Y.; et al. Identification and Characterization of the ERF Subfamily B3 Group Revealed GhERF13.12 Improves Salt Tolerance in Upland Cotton. Front. Plant Sci. 2021, 12, 705883. [Google Scholar] [CrossRef]
- Li, Z.; Tian, Y.; Xu, J.; Fu, X.; Gao, J.; Wang, B.; Han, H.; Wang, L.; Peng, R.; Yao, Q. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol. Biochem. 2018, 132, 683–695. [Google Scholar] [CrossRef]
- Wang, D.; Sun, Z.; Hu, X.; Xiong, J.; Hu, L.; Xu, Y.; Tang, Y.; Wu, Y. The key regulator LcERF056 enhances salt tolerance by modulating reactive oxygen species-related genes in Lotus corniculatus. BMC Plant Biol. 2021, 21, 605. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Shi, Q.; Ren, Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2019, 291, 110356. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, N.; Zhou, X.; Hou, X.; Yang, G.; Meng, J.; Luan, Y. Tomato MYB49 enhances resistance to Phytophthora infestans and tolerance to water deficit and salt stress. Planta 2018, 248, 1487–1503. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; He, Q.; Guo, D.; Liu, C.; Cao, J.; Wu, Z.; Kang, Z.; Wang, X. TaMYB29: A Novel R2R3-MYB Transcription Factor Involved in Wheat Defense Against Stripe Rust. Front. Plant Sci. 2021, 12, 783388. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Tian, Y.; Wang, Q.; Chen, S.; Li, H.; Ma, C.; Li, H. Functional Characterization of a Sugar Beet BvbHLH93 Transcription Factor in Salt Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 3669. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Niu, M.-X.; Feng, C.-H.; Li, H.-G.; Su, Y.; Su, W.-L.; Pang, H.; Yang, Y.; Yu, X.; Wang, H.-L.; et al. PeSTZ1 confers salt stress tolerance by scavenging the accumulation of ROS through regulating the expression of PeZAT12 and PeAPX2 in Populus. Tree Physiol. 2020, 40, 1292–1311. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Wang, Y.-S.; Cheng, H.; Su, Y.-B.; Zhong, Y.-J.; Zheng, L. The Kandelia obovata transcription factor KoWRKY40 enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 2022, 22, 274. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.-M.; Zhang, H.-F.; Liu, S.-Y.; Wang, X.-K.; Zhang, Y.-M.; Meng, Y.-C.; Luo, D.; Chen, R.-G. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Liu, Y.; Zhao, L.; Zhang, S.; Huang, X. A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ. 2018, 42, 832–845. [Google Scholar] [CrossRef] [PubMed]
- Lattanzio, V.; Di Venere, D.; Linsalata, V.; Bertolini, P.; Ippolito, A.; Salerno, M. Low Temperature Metabolism of Apple Phenolics and Quiescence of Phlyctaena vagabunda. J. Agric. Food Chem. 2001, 49, 5817–5821. [Google Scholar] [CrossRef]
- An, J.-P.; Wang, X.-F.; Zhang, X.-W.; Xu, H.-F.; Bi, S.-Q.; You, C.-X.; Hao, Y.-J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2019, 18, 337–353. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Li, J.; Zhao, S.; Yu, X.; Du, W.; Li, H.; Sun, Y.; Sun, H.; Ruan, C. Role of Xanthoceras sorbifolium MYB44 in tolerance to combined drought and heat stress via modulation of stomatal closure and ROS homeostasis. Plant Physiol. Biochem. 2021, 162, 410–420. [Google Scholar] [CrossRef]
- Singh, S.; Chopperla, R.; Shingote, P.; Chhapekar, S.S.; Deshmukh, R.; Khan, S.; Padaria, J.C.; Sharma, T.R.; Solanke, A.U. Overexpression of EcDREB2A transcription factor from finger millet in tobacco enhances tolerance to heat stress through ROS scavenging. J. Biotechnol. 2021, 336, 10–24. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, Y.; Yang, X.; Zhang, B.; Xu, F.; Wang, Y.; Song, C.; Yi, M.; Ma, N.; Zhou, X.; et al. The Heat Stress Transcription Factor LlHsfA4 Enhanced Basic Thermotolerance through Regulating ROS Metabolism in Lilies (Lilium Longiflorum). Int. J. Mol. Sci. 2022, 23, 572. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, L.; Long, Y.; Si, W.; Cheng, B.; Jiang, H. A Novel Heat Shock Transcription Factor (ZmHsf08) Negatively Regulates Salt and Drought Stress Responses in Maize. Int. J. Mol. Sci. 2021, 22, 11922. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 Transcription Factor Regulates Heat Stress Tolerance Through FERONIA Receptor-Like Kinase-Mediated Reactive Oxygen Species Signaling in Tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.V.; Vo, K.T.X.; Rahman, M.; Choi, S.-H.; Jeon, J.-S. Heat stress transcription factor OsSPL7 plays a critical role in reactive oxygen species balance and stress responses in rice. Plant Sci. 2019, 289, 110273. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.-R.; Huang, Z.; Xiang, X.-Y.; Xu, W.-X.; Wang, J.-T.; Chen, J.; Song, L.; Xiao, Y.; Li, X.; Ma, J.; et al. MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol. 2020, 20, 542. [Google Scholar] [CrossRef]
- Qiu, J.-R.; Xiang, X.-Y.; Wang, J.-T.; Xu, W.-X.; Chen, J.; Xiao, Y.; Jiang, C.-Z.; Huang, Z. MfPIF1 of Resurrection Plant Myrothamnus flabellifolia Plays a Positive Regulatory Role in Responding to Drought and Salinity Stresses in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 3011. [Google Scholar] [CrossRef]
- Wang, D.; Chen, Q.; Chen, W.; Liu, X.; Xia, Y.; Guo, Q.; Jing, D.; Liang, G. A WRKY Transcription Factor, EjWRKY17, from Eriobotrya japonica Enhances Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 5593. [Google Scholar] [CrossRef]
- Tang, H.; Bi, H.; Liu, B.; Lou, S.; Song, Y.; Tong, S.; Chen, N.; Jiang, Y.; Liu, J.; Liu, H.; et al. WRKY33 interacts with WRKY12 protein to up-regulate RAP2. 2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 2020, 229, 106–125. [Google Scholar] [CrossRef]
- Gulzar, F.; Fu, J.; Zhu, C.; Yan, J.; Li, X.; Meraj, T.A.; Shen, Q.; Hassan, B.; Wang, Q. Maize WRKY Transcription Factor ZmWRKY79 Positively Regulates Drought Tolerance through Elevating ABA Biosynthesis. Int. J. Mol. Sci. 2021, 22, 10080. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Chuong, N.N.; Hoa, T.T.K.; Doan, H.; Van, P.H.P.; Trang, L.D.M.; Huyen, P.N.T.; Le, D.T.; Tran, L.-S.P.; Thao, N.P. The Drought-Mediated Soybean GmNAC085 Functions as a Positive Regulator of Plant Response to Salinity. Int. J. Mol. Sci. 2021, 22, 8986. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef]
- Sewelam, N.; Kazan, K.; Thomas-Hall, S.R.; Kidd, B.N.; Manners, J.M.; Schenk, P.M. Ethylene Response Factor 6 Is a Regulator of Reactive Oxygen Species Signaling in Arabidopsis. PLoS ONE 2013, 8, e70289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Yang, J.; Liu, C.; Chen, Z.; Yao, Z.; Cao, S. Overexpression of ethylene response factor ERF96 gene enhances selenium tolerance in Arabidopsis. Plant Physiol. Biochem. 2020, 149, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.-J.; Geng, Z.; Wang, Y.; Wang, Y.; Liu, S.; Chen, C.; Song, A.; Jiang, J.; Chen, S.; Chen, F. A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in chrysanthemum. Hortic. Res. 2021, 8, 248. [Google Scholar] [CrossRef]
- An, J.-P.; Li, R.; Qu, F.-J.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Li, T.; Cheng, Z.; Zhao, D.; Tao, J. R2R3-MYB Transcription Factor PlMYB108 Confers Drought Tolerance in Herbaceous Peony (Paeonia lactiflora Pall.). Int. J. Mol. Sci. 2021, 22, 11884. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yang, W.; Fan, H.; Zhang, X.; Sui, N. TaMYB86B encodes a R2R3-type MYB transcription factor and enhances salt tolerance in wheat. Plant Sci. 2020, 300, 110624. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Gao, S.; Li, J.; Song, P.; Zhang, Q.; Guo, J.; Wang, X.; Han, X.; Wang, X.; Zhu, Y.; et al. The bHLH transcription factor regulated gene OsWIH2 is a positive regulator of drought tolerance in rice. Plant Physiol. Biochem. 2021, 169, 269–279. [Google Scholar] [CrossRef]
- Akmakjian, G.Z.; Riaz, N.; Guerinot, M.L. Photoprotection during iron deficiency is mediated by the bHLH transcription factors PYE and ILR3. Proc. Natl. Acad. Sci. USA 2021, 118, e2024918118. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Zhu, W.-C.; Feng, X.-H.; Jin, J.-H.; Wei, A.-M.; Gong, Z.-H. Transcription Factor CaSBP12 Negatively Regulates Salt Stress Tolerance in Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2020, 21, 444. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Sun, Q.; Lu, S.; Chai, L.; Ye, J.; Deng, X. Citrus transcription factor CsHB5 regulates abscisic acid biosynthetic genes and promotes senescence. Plant J. 2021, 108, 151–168. [Google Scholar] [CrossRef] [PubMed]
- Bellegarde, F.; Maghiaoui, A.; Boucherez, J.; Krouk, G.; Lejay, L.; Bach, L.; Gojon, A.; Martin, A. The Chromatin Factor HNI9 and ELONGATED HYPOCOTYL5 Maintain ROS Homeostasis under High Nitrogen Provision. Plant Physiol. 2019, 180, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, C.-Y.; An, J.-P.; Wang, D.-R.; Wang, X.; Wang, C.-K.; You, C.-X. The C2H2-type zinc finger transcription factor MdZAT10 negatively regulates drought tolerance in apple. Plant Physiol. Biochem. 2021, 167, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, H.; Jia, X.; Gao, H.; Mao, K.; Ma, F. The HD-Zip I transcription factor MdHB7 -like confers tolerance to salinity in transgenic apple (Malus domestica). Physiol. Plant. 2021, 172, 1452–1464. [Google Scholar] [CrossRef]
- Wu, J.; Yu, C.; Huang, L.; Gan, Y. A rice transcription factor, OsMADS57, positively regulates high salinity tolerance in transgenic Arabidopsis thaliana and Oryza sativa plants. Physiol. Plant. 2021, 173, 1120–1135. [Google Scholar] [CrossRef]
- Yin, M.; Wang, Y.; Zhang, L.; Li, J.; Quan, W.; Yang, L.; Wang, Q.; Chan, Z. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 2017, 68, 2991–3005. [Google Scholar] [CrossRef]
- Fan, F.; Zhou, Z.; Qin, H.; Tan, J.; Ding, G. Exogenous Brassinosteroid Facilitates Xylem Development in Pinus massoniana Seedlings. Int. J. Mol. Sci. 2021, 22, 7615. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Khan, R.; Wu, X.; Zhou, L.; Xu, N.; Du, S.; Ma, X. Exogenous application of brassinosteroids regulates tobacco leaf size and expansion via modulation of endogenous hormones content and gene expression. Physiol. Mol. Biol. Plants 2021, 27, 847–860. [Google Scholar] [CrossRef]
- Zeng, G.; Gao, F.; Li, C.; Li, D.; Xi, Z. Characterization of 24-epibrassinolide-mediated modulation of the drought stress responses: Morphophysiology, antioxidant metabolism and hormones in grapevine (Vitis vinifera L.). Plant Physiol. Biochem. 2022, 184, 98–111. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, H. Epibrassinolide improves the growth performance of Sedum lineare upon Zn stress through boosting antioxidative capacities. PLoS ONE 2021, 16, e0257172. [Google Scholar] [CrossRef]
- Alam, P.; Kohli, S.K.; Al Balawi, T.; Altalayan, F.; Alam, P.; Ashraf, M.; Bhardwaj, R.; Ahmad, P. Foliar Application of 24-Epibrassinolide Improves Growth, Ascorbate-Glutathione Cycle, and Glyoxalase System in Brown Mustard (Brassica juncea (L.) Czern.) Under Cadmium Toxicity. Plants 2020, 9, 1487. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Bai, L.; Miao, L.; Liu, Y.; Li, S.; Yu, X.; Li, Y. 24-Epibrassinolide Ameliorates Endogenous Hormone Levels to Enhance Low-Temperature Stress Tolerance in Cucumber Seedlings. Int. J. Mol. Sci. 2018, 19, 2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, S.; Huang, S.; Wang, S.; Mao, Y.; Liu, J.; Ma, R.; Wang, X. Enhanced brassinosteroid signaling intensity via SlBRI1 overexpression negatively regulates drought resistance in a manner opposite of that via exogenous BR application in tomato. Plant Physiol. Biochem. 2019, 138, 36–47. [Google Scholar] [CrossRef]
- Ferrero, M.; Pagliarani, C.; Novák, O.; Ferrandino, A.; Cardinale, F.; Visentin, I.; Schubert, A. Exogenous strigolactone interacts with abscisic acid-mediated accumulation of anthocyanins in grapevine berries. J. Exp. Bot. 2018, 69, 2391–2401. [Google Scholar] [CrossRef] [Green Version]
- Mayzlish-Gati, E.; LekKala, S.P.; Resnick, N.; Wininger, S.; Bhattacharya, C.; Lemcoff, J.H.; Kapulnik, Y.; Koltai, H. Strigolactones are positive regulators of light-harvesting genes in tomato. J. Exp. Bot. 2010, 61, 3129–3136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, C.-W.; Zhang, C.; Wang, N.-H.; Mao, W.; Wu, F. Strigolactone GR24 improves cadmium tolerance by regulating cadmium uptake, nitric oxide signaling and antioxidant metabolism in barley (Hordeum vulgare L.). Environ. Pollut. 2021, 273, 116486. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, Y.; Xi, X.; Ma, C.; Sun, Z.; Yang, X.; Li, X.; Tian, Y.; Wang, C. Exogenous Strigolactones alleviate KCl stress by regulating photosynthesis, ROS migration and ion transport in Malus hupehensis Rehd. Plant Physiol. Biochem. 2020, 159, 113–122. [Google Scholar] [CrossRef]
- Song, X.; Zhao, Y.; Han, B.; Li, T.; Zhao, P.; Xu, J.-W.; Yu, X. Strigolactone mediates jasmonic acid-induced lipid production in microalga Monoraphidium sp. QLY-1 under nitrogen deficiency conditions. Bioresour. Technol. 2020, 306, 123107. [Google Scholar] [CrossRef]
- Lu, T.; Yu, H.; Li, Q.; Chai, L.; Jiang, W. Improving plant growth and alleviating photosynthetic inhibition and oxidative stress from low-light stress with exogenous GR24 in tomato (Solanum lycopersicum L.) seedlings. Front Plant. Sci. 2019, 10, 490. [Google Scholar] [CrossRef]
- Yu, L.; Gao, B.; Li, Y.; Tan, W.; Li, M.; Zhou, L.; Peng, C.; Xiao, L.; Liu, Y. The synthesis of strigolactone is affected by endogenous ascorbic acid in transgenic rice for l-galactono-1, 4-lactone dehydrogenase suppressed or overexpressing. J. Plant Physiol. 2020, 246–247, 153139. [Google Scholar] [CrossRef]
- Min, Z.; Li, R.; Chen, L.; Zhang, Y.; Li, Z.; Liu, M.; Ju, Y.; Fang, Y. Alleviation of drought stress in grapevine by foliar-applied strigolactones. Plant Physiol. Biochem. 2018, 135, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Pérez, S.G.; Gutiérrez, J.; Garcia-Garcia, F.; Osuna, D.; Dopazo, J.; Lorenzo, O.; Revuelta, J.L.; Arellano, J.B. Early Transcriptional Defense Responses in Arabidopsis Cell Suspension Culture under High-Light Conditions. Plant Physiol. 2011, 156, 1439–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, C.; Zhang, F.; Sun, X.; Shang, D.; He, F.; Li, X.; Jiang, X. Effects of S-Abscisic Acid (S-ABA) on Seed Germination, Seedling Growth, and Asr1 Gene Expression Under Drought Stress in Maize. J. Plant Growth Regul. 2019, 38, 1300–1313. [Google Scholar] [CrossRef]
- Wu, S.; Hu, C.; Tan, Q.; Zhao, X.; Xu, S.; Xia, Y.; Sun, X. Nitric oxide acts downstream of abscisic acid in molybdenum-induced oxidative tolerance in wheat. Plant Cell Rep. 2018, 37, 599–610. [Google Scholar] [CrossRef]
- Dong, N.; Qi, J.; Li, Y.; Chen, Y.; Hao, Y. Effects of Abscisic Acid and Nitric Oxide on Chilling Resistance and Activation of the Antioxidant System in Walnut Shoots In Vitro. J. Am. Soc. Hortic. Sci. Am. Soc. Hortic. Sci. 2017, 142, 376–384. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Wang, J.; Li, S.; Kakan, X.; Zhou, Y.; Miao, Y.; Wang, F.; Qin, H.; Huang, R. Ascorbic Acid Integrates the Antagonistic Modulation of Ethylene and Abscisic Acid in the Accumulation of Reactive Oxygen Species. Plant Physiol. 2019, 179, 1861–1875. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Brouard, E.; Prodhomme, D.; Hilbert, G.; Renaud, C.; Petit, J.-P.; Edwards, E.; Betts, A.; Delrot, S.; Ollat, N.; et al. Regulation of anthocyanin and sugar accumulation in grape berry through carbon limitation and exogenous ABA application. Food Res. Int. 2022, 160, 111478. [Google Scholar] [CrossRef]
- Schussler, J.R.; Brenner, M.L.; Brun, W.A. Relationship of Endogenous Abscisic Acid to Sucrose Level and Seed Growth Rate of Soybeans. Plant Physiol. 1991, 96, 1308–1313. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Cao, P.; Ren, A.; Wang, S.; Yang, T.; Zhu, T.; Shi, L.; Zhu, J.; Jiang, A.-L.; Zhao, M.-W. SA inhibits complex III activity to generate reactive oxygen species and thereby induces GA overproduction in Ganoderma lucidum. Redox Biol. 2018, 16, 388–400. [Google Scholar] [CrossRef]
- Yang, X.; Chen, Z.; Yin, X.; Wang, Y.; Yang, Y.; Yang, Y. Genome-Wide Survey Indicates Diverse Physiological Roles of Dendrobium officinale Calcium-Dependent Protein Kinase Genes. Int. J. Mol. Sci. 2022, 23, 1298. [Google Scholar] [CrossRef]
- Norman, C.; Howell, K.A.; Millar, A.H.; Whelan, J.M.; Day, D.A. Salicylic Acid Is an Uncoupler and Inhibitor of Mitochondrial Electron Transport. Plant Physiol. 2004, 134, 492–501. [Google Scholar] [CrossRef] [Green Version]
- Durner, J.; Klessig, D.F. Salicylic Acid Is a Modulator of Tobacco and Mammalian Catalases. J. Biol. Chem. 1996, 271, 28492–28501. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.; Liang, Y.; Zhu, Y.; Zhao, F. Role of salicylic acid in alleviating oxidative damage in rice roots (Oryza sativa) subjected to cadmium stress. Environ. Pollut. 2007, 147, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Liang, Y.; Zhu, Y. Does salicylic acid regulate antioxidant defense system, cell death, cadmium uptake and partitioning to acquire cadmium tolerance in rice? J. Plant Physiol. 2009, 166, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Kaya, C. Salicylic acid-induced hydrogen sulphide improves lead stress tolerance in pepper plants by upraising the ascorbate-glutathione cycle. Physiol. Plant. 2020, 173, 8–19. [Google Scholar] [CrossRef]
- Kaya, C. Nitrate reductase is required for salicylic acid-induced water stress tolerance of pepper by upraising the AsA-GSH pathway and glyoxalase system. Physiol. Plant. 2020, 172, 351–370. [Google Scholar] [CrossRef] [PubMed]
- Kaya, C.; Ashraf, M.; Alyemeni, M.N.; Ahmad, P. The role of endogenous nitric oxide in salicylic acid-induced up-regulation of ascorbate-glutathione cycle involved in salinity tolerance of pepper (Capsicum annuum L.) plants. Plant Physiol. Biochem. 2019, 147, 10–20. [Google Scholar] [CrossRef]
- Chen, M.; Gu, H.; Wang, L.; Shao, Y.; Li, R.; Li, W. Exogenous Ethylene Promotes Peel Color Transformation by Regulating the Degradation of Chlorophyll and Synthesis of Anthocyanin in Postharvest Mango Fruit. Front. Nutr. 2022, 9, 911542. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, F.; Naghavi, M.R.; Peighambari, S.A.; Dehaghi, N.K.; Khaldari, I.; Bravi, E.; Marconi, O.; Perretti, G. Abscisic acid crosstalk with auxin and ethylene in biosynthesis and degradation of inulin-type fructans in chicory. Plant Biol. 2021, 23, 636–642. [Google Scholar] [CrossRef]
- Jung, J.Y.; Shin, R.; Schachtman, D.P. Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 2009, 21, 607–621. [Google Scholar] [CrossRef]
- Schachtman, D.P. The role of ethylene in plant responses to K+ deficiency. Front Plant Sci. 2015, 6, 1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, S.; Davies, W.J. Ozone suppresses soil drying- and abscisic acid (ABA)-induced stomatal closure via an ethylene-dependent mechanism. Plant Cell Environ. 2009, 32, 949–959. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yue, X.; Wang, R.; Zhang, Y. Ethylene mediates UV-B-induced stomatal closure via peroxidase-dependent hydrogen peroxide synthesis in Vicia faba L. J. Exp. Bot. 2011, 62, 2657–2666. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yuan, M.; Li, Z.; Niu, Y.; Jin, Q.; Zhu, B.; Xu, Y. Effects of ethylene biosynthesis and signaling on oxidative stress and antioxidant defense system in Nelumbo nucifera G. under cadmium exposure. Environ. Sci. Pollut. Res. 2020, 27, 40156–40170. [Google Scholar] [CrossRef]
- Hou, J.; Riaz, M.; Yan, L.; Lu, K.; Jiang, C. Effect of exogenous l-aspartate nano-calcium on root growth, calcium forms and cell wall metabolism of Brassica napus L. NanoImpact 2022, 27, 100415. [Google Scholar] [CrossRef]
- Ban, Q.; Liu, T.; Ning, K.; Fan, J.; Cui, Q.; Guo, Y.; Zai, X. Effect of calcium treatment on the browning of harvested eggplant fruits and its relation to the metabolisms of reactive oxygen species (ROS) and phenolics. Food Sci. Nutr. 2021, 9, 5567–5574. [Google Scholar] [CrossRef]
- He, L.; Li, B.; Lu, X.; Yuan, L.; Yang, Y.; Yuan, Y.; Du, J.; Guo, S. The effect of exogenous calcium on mitochondria, respiratory metabolism enzymes and ion transport in cucumber roots under hypoxia. Sci. Rep. 2015, 5, 11391. [Google Scholar] [CrossRef] [Green Version]
- Erinle, K.O.; Jiang, Z.; Ma, B.; Li, J.; Chen, Y.; Ur-Rehman, K.; Shahla, A.; Zhang, Y. Exogenous calcium induces tolerance to atrazine stress in Pennisetum seedlings and promotes photosynthetic activity, antioxidant enzymes and psbA gene transcripts. Ecotoxicol. Environ. Saf. 2016, 132, 403–412. [Google Scholar] [CrossRef]
- Rui, Z.; Huali, X.; Min, S.; Yang, B.; Mina, N.; Yuanyuan, Z.; Haitao, L.; Prusky, D.; Xiaoyan, C. Mechanism of Ca2+-mediated NOX modulated in ROS metabolism induced by T-2 toxin in potato tuber. Food Chem. 2020, 317, 126416. [Google Scholar] [CrossRef]
- Snedden, W.A.; Arazi, T.; Fromm, H.; Shelp, B.J. Calcium/Calmodulin Activation of Soybean Glutamate Decarboxylase. Plant Physiol. 1995, 108, 543–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Liu, Y.; Ni, Y.; Meng, Z.; Lu, T.; Li, T. Exogenous Calcium Alleviates Low Night Temperature Stress on the Photosynthetic Apparatus of Tomato Leaves. PLoS ONE 2014, 9, e97322. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; He, Y.; Yin, X.; Zhong, X.; Yan, B.; Wu, Y.; Chen, J.; Li, X.; Zhai, K.; Huang, Y.; et al. Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 2021, 184, 5391–5404.e17. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.H.; Alamri, S.; Alsubaie, Q.D.; Ali, H.M.; Khan, M.N.; Al-Ghamdi, A.; Ibrahim, A.A.; Alsadon, A. Exogenous nitric oxide alleviates sulfur deficiency-induced oxidative damage in tomato seedlings. Nitric Oxide 2019, 94, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Silveira, N.M.; Frungillo, L.; Marcos, F.C.C.; Pelegrino, M.T.; Miranda, M.T.; Seabra, A.B.; Salgado, I.; Machado, E.C.; Ribeiro, R.V. Exogenous nitric oxide improves sugarcane growth and photosynthesis under water deficit. Planta 2016, 244, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Mahjoubi, Y.; Rzigui, T.; Kharbech, O.; Mohamed, S.N.; Abaza, L.; Chaoui, A.; Nouairi, I.; Djebali, W. Exogenous nitric oxide alleviates manganese toxicity in bean plants by modulating photosynthesis in relation to leaf lipid composition. Protoplasma 2021, 259, 949–964. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.; Zheng, H.; Jia, S.; Jiang, Y.; Yang, Q.; Kang, G. Nitric Oxide Enhancing Resistance to PEG-Induced Water Deficiency is Associated with the Primary Photosynthesis Reaction in Triticum aestivum L. Int. J. Mol. Sci. 2018, 19, 2819. [Google Scholar] [CrossRef] [Green Version]
- Soliman, M.; Alhaithloul, H.A.; Hakeem, K.R.; Alharbi, B.M.; El-Esawi, M.; Elkelish, A. Exogenous Nitric Oxide Mitigates Nickel-Induced Oxidative Damage in Eggplant by Upregulating Antioxidants, Osmolyte Metabolism, and Glyoxalase Systems. Plants 2019, 8, 562. [Google Scholar] [CrossRef] [Green Version]
- Gautam, H.; Sehar, Z.; Rehman, T.; Hussain, A.; AlAjmi, M.; Khan, N. Nitric Oxide Enhances Photosynthetic Nitrogen and Sulfur-Use Efficiency and Activity of Ascorbate-Glutathione Cycle to Reduce High Temperature Stress-Induced Oxidative Stress in Rice (Oryza sativa L.) Plants. Biomolecules 2021, 11, 305. [Google Scholar] [CrossRef]
- Savvides, A.; Ali, S.; Tester, M.; Fotopoulos, V. Chemical Priming of Plants Against Multiple Abiotic Stresses: Mission Possible? Trends Plant Sci. 2016, 21, 329–340. [Google Scholar] [CrossRef]
- Molassiotis, A.; Tanou, G.; Diamantidis, G. NO says more than ‘YES’ to salt tolerance: Salt priming and systemic nitric oxide signaling in plants. Plant Signal. Behav. 2010, 5, 209–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valderrama, R.; Corpas, F.J.; Carreras, A.; Fernández-Ocaña, A.; Chaki, M.; Luque, F.; Gómez-Rodríguez, M.V.; Colmenero-Varea, P.; Del Río, L.A.; Barroso, J.B. Nitrosative stress in plants. FEBS Lett. 2007, 581, 453–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Bel, A.J.E.; Gaupels, F. Pathogen-induced resistance and alarm signals in the phloem. Mol. Plant Pathol. 2004, 5, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wei, X.; Ji, X.; Ma, W. Endogenous NO-mediated transcripts involved in photosynthesis and carbohydrate metabolism in alfalfa (Medicago sativa L.) seedlings under drought stress. Plant Physiol. Biochem. 2019, 141, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ye, T.; Zhu, J.-K.; Chan, Z. Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J. Exp. Bot. 2014, 65, 4119–4131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.; Hou, D.; O’Connor, D.; Pan, S.; Zhu, J.; Bolan, N.S.; Mulder, J. Exogenous phosphorus treatment facilitates chelation-mediated cadmium detoxification in perennial ryegrass (Lolium perenne L.). J. Hazard. Mater. 2019, 389, 121849. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Huo, J.; Liao, W. Hydrogen sulfide: Roles in plant abiotic stress response and crosstalk with other signals. Plant Sci. 2020, 302, 110733. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Ji, J.; Chen, W.; Yue, J.; Du, C.; Sun, J.; Chen, L.; Jiang, Z.; Shi, S. GABA negatively regulates adventitious root development in poplar. J. Exp. Bot. 2019, 71, 1459–1474. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Ashraf, U.; Li, G.; Li, Y.; Lu, W.; Gao, L.; Han, F.; Hu, J. Exogenous γ-aminobutyric Acid (GABA) Application Improved Early Growth, Net Photosynthesis, and Associated Physio-Biochemical Events in Maize. Front. Plant Sci. 2016, 7, 919. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zhu, J.; Sun, L.; Cheng, Y.; Hou, J.; Fan, Y.; Ge, Y. Exogenous γ-aminobutyric acid maintains fruit quality of apples through regulation of ethylene anabolism and polyamine metabolism. Plant Physiol. Biochem. 2021, 169, 92–101. [Google Scholar] [CrossRef]
- Chen, X.; Li, N.; Liu, C.; Wang, H.; Li, Y.; Xie, Y.; Ma, F.; Liang, J.; Li, C. Exogenous GABA improves the resistance of apple seedlings to long-term drought stress by enhancing GABA shunt and secondary cell wall biosynthesis. Tree Physiol. 2022, tpac096. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, U.; Mahmood, S.; Anjum, S.A.; Abbas, R.N.; Rasul, F.; Iqbal, J.; Mo, Z.; Tang, X. Exogenous Gamma-Aminobutyric Acid Application Induced Modulations in the Performance of Aromatic Rice Under Lead Toxicity. Front. Plant Sci. 2022, 13, 933694. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Wu, X.; Tian, Z.; Jia, K.; Pan, Y.; Li, J.; Gao, H. Comparative proteomic analysis of gamma-aminobutyric acid responses in hypoxia-treated and untreated melon roots. Phytochemistry 2015, 116, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Fan, L.; Gao, H.; Wu, X.; Li, J.; Lv, G.; Gong, B. Polyamine biosynthesis and degradation are modulated by exogenous gamma-aminobutyric acid in root-zone hypoxia-stressed melon roots. Plant Physiol. Biochem. 2014, 82, 17–26. [Google Scholar] [CrossRef]
- Zhu, X.; Liao, J.; Xia, X.; Xiong, F.; Li, Y.; Shen, J.; Wen, B.; Ma, Y.; Wang, Y.; Fang, W. Physiological and iTRAQ-based proteomic analyses reveal the function of exogenous γ-aminobutyric acid (GABA) in improving tea plant (Camellia sinensis L.) tolerance at cold temperature. BMC Plant Biol. 2019, 19, 1–20. [Google Scholar] [CrossRef]
- Bouché, N.; Fait, A.; Bouchez, D.; Møller, S.G.; Fromm, H. Mitochondrial succinic-semialdehyde dehydrogenase of the γ-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. USA 2003, 100, 6843–6848. [Google Scholar] [CrossRef] [Green Version]
- Qu, Y.; Jiang, L.; Wuyun, T.; Mu, S.; Xie, F.; Chen, Y.; Zhang, L. Effects of Exogenous Putrescine on Delaying Senescence of Cut Foliage of Nephrolepis cordifolia. Front. Plant Sci. 2020, 11, 933694. [Google Scholar] [CrossRef]
- Tavallali, V.; Alhavi, N.; Gholami, H.; Abarghuei, F.M. Developmental and phytochemical changes in pot marigold (Calendula officinalis L.) using exogenous application of polyamines. Plant Physiol. Biochem. 2022, 183, 128–137. [Google Scholar] [CrossRef]
- Aziz, A.; Larher, F. Changes in polyamine titers associated with the proline response and osmotic adjustment of rape leaf discs submitted to osmotic stresses. Plant Sci. 1995, 112, 175–186. [Google Scholar] [CrossRef]
- Drolet, G.; Dumbroff, E.B.; Legge, R.L.; Thompson, J.E. Radical scavenging properties of polyamines. Phytochemistry 1986, 25, 367–371. [Google Scholar] [CrossRef]
- Dondini, L.; Del Duca, S.; Dall’Agata, L.; Bassi, R.; Gastaldelli, M.; Della Mea, M.; Serafini-Fracassini, D. Suborganellar localisation and effect of light on Helianthus tuberosus chloroplast transglutaminases and their substrates. Planta 2003, 217, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Tachibana, N.S. Involvement of Polyamines in the Chilling Tolerance of Cucumber Cultivars. Plant Physiol. 2000, 124, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, S.; Liu, L.; Wang, H.; Li, D.; Bai, Z.; Zhang, Y.; Sun, H.; Zhang, K.; Li, C. Exogenous melatonin accelerates seed germination in cotton (Gossypium hirsutum L.). PLoS ONE 2019, 14, e0216575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Zhang, H.J.; Sun, Q.Q.; Cao, Y.Y.; Li, X.; Zhao, B.; Wu, P.; Guo, Y.D. Proteomic analysis reveals a role of melatonin in promoting cucumber seed germination under high salinity by regulating energy production. Sci. Rep. 2017, 7, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; He, Q.; Zhang, F.; Yu, J.; Li, C.; Zhao, T.; Zhang, Y.; Xie, Q.; Su, B.; Mei, L.; et al. Melatonin enhances cotton immunity to Verticillium wilt via manipulating lignin and gossypol biosynthesis. Plant J. 2019, 100, 784–800. [Google Scholar] [CrossRef] [Green Version]
- Weeda, S.; Na Zhang, N.; Zhao, X.; Ndip, G.; Guo, Y.; Buck, G.A.; Fu, C.; Ren, S. Arabidopsis Transcriptome Analysis Reveals Key Roles of Melatonin in Plant Defense Systems. PLoS ONE 2014, 9, e93462. [Google Scholar] [CrossRef] [Green Version]
- Ramel, F.; Sulmon, C.; Bogard, M.; Couée, I.; Gouesbet, G. Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and sucrose-induced tolerance in Arabidopsis thaliana plantlets. BMC Plant Biol. 2009, 9, 28. [Google Scholar] [CrossRef] [Green Version]
- Van Den Ende, W.; Valluru, R. Sucrose, sucrosyl oligosaccharides, and oxidative stress: Scavenging and salvaging? J. Exp. Bot. 2009, 60, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Tian, Y.; Zhang, B.; Hassan, M.J.; Li, Z.; Zhu, Y. Chitosan (CTS) Alleviates Heat-Induced Leaf Senescence in Creeping Bentgrass by Regulating Chlorophyll Metabolism, Antioxidant Defense, and the Heat Shock Pathway. Molecules 2021, 26, 5337. [Google Scholar] [CrossRef]
- Zhao, D.-Q.; Li, T.-T.; Hao, Z.-J.; Cheng, M.-L.; Tao, J. Exogenous trehalose confers high temperature stress tolerance to herbaceous peony by enhancing antioxidant systems, activating photosynthesis, and protecting cell structure. Cell Stress Chaperon 2019, 24, 247–257. [Google Scholar] [CrossRef]
- Rohman, M.; Islam, R.; Monsur, M.B.; Amiruzzaman, M.; Fujita, M.; Hasanuzzaman, M. Trehalose Protects Maize Plants from Salt Stress and Phosphorus Deficiency. Plants 2019, 8, 568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostofa, M.G.; Hossain, M.A.; Fujita, M.; Tran, L.-S.P. Physiological and biochemical mechanisms associated with trehalose-induced copper-stress tolerance in rice. Sci. Rep. 2015, 5, 11433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.; Li, J.; Al-Huqail, A.A.; Al-Harbi, M.S.; Ali, E.F.; Wang, J.; Ding, Z.; Rekaby, S.A.; Ghoneim, A.M.; Eissa, M.A. Mechanisms of Chitosan Nanoparticles in the Regulation of Cold Stress Resistance in Banana Plants. Nanomaterials 2021, 11, 2670. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Zhang, L.; Duan, X.; Chai, X.; Huang, R.; Kang, Y.; Yang, X. Effects of exogenous sucrose and selenium on plant growth, quality, and sugar metabolism of pea sprouts. J. Sci. Food Agric. 2021, 102, 2855–2863. [Google Scholar] [CrossRef]
- Nkomo, M.; Gokul, A.; Keyster, M.; Klein, A. Exogenous p-Coumaric Acid Improves Salvia hispanica L. Seedling Shoot Growth. Plants 2019, 8, 546. [Google Scholar] [CrossRef] [Green Version]
- Xiang, C.; Werner, B.L.; Christensen, E.M.; Oliver, D.J. The Biological Functions of Glutathione Revisited in Arabidopsis Transgenic Plants with Altered Glutathione Levels. Plant Physiol. 2001, 126, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [Green Version]
- Oney-Birol, S. Exogenous L-Carnitine Promotes Plant Growth and Cell Division by Mitigating Genotoxic Damage of Salt Stress. Sci. Rep. 2019, 9, 17229. [Google Scholar] [CrossRef]
| Transcription Factor Family | Genes Name | Description |
|---|---|---|
| WRKY | AhWRKY75 [85] | Peanut AhWRKY75 gene conferred salt tolerance in transgenic peanut lines by improving the efficiency of the ROS scavenging system and photosynthesis. |
| EjWRKY17 [118] | Overexpression of Eriobotrya japonica EjWRKY17 led to enhanced drought tolerance in transgenic Arabidopsis, which was lower levels of ROS. | |
| FtWRKY46 [89] | Overexpression of Tartary buckwheat FtWRKY46 enhanced the stress tolerance of transgenic Tartary buckwheat by modulating ROS clearance and stress-related gene expression. | |
| HbWRKY82 [88] | Hevea brasiliensis HbWRKY82 regulated the transcriptional expression of ROS-related genes (RbohD, CSD1, CSD2, FSD3) against salt and drought stress in Hevea brasiliensis. | |
| KoWRKY40 [104] | Mangrove plant K. obovate KoWRKY40 transgenic Arabidopsis exhibited higher proline content, SOD, POD and CAT activities, and lower H2O2 content under cold stress conditions. | |
| MfWRKY70 [87] | Myrothamnus flabellifolia MfWRKY70 could significantly increase tolerance to drought, osmotic and salinity stresses by enhancing the antioxidant enzyme system and maintaining ROS homeostasis in Myrothamnus flabellifolia. | |
| PcWRKY33 [86] | Polygonum cuspidatum PcWRKY33 negatively regulates the salt tolerance by increasing the level of cellular ROS in Arabidopsis thaliana. | |
| WRKY33 [119] | Arabidopsis thaliana WRKY33 can bind to and activate RAP2.2 and activate towards RAP2.2 to increase hypoxia tolerance of Arabidopsis thaliana. | |
| ZmWRKY40 [68] | Overexpression of maize ZmWRKY40 improved drought tolerance in transgenic Arabidopsis by enhancing the activities of POD and CAT under drought stress. | |
| ZmWRKY79 [120] | Maize ZmWRKY79 boost ROS scavenging to result in less H2O2 and MDA accumulation and increased antioxidant enzyme activities under drought stress in Arabidopsis. | |
| ZmWRKY106 [69] | Overexpression of maize ZmWRKY106 improved the tolerance to drought and heat in transgenic Arabidopsis by reducing ROS content in transgenic lines by enhancing the activities of SOD, POD and CAT under drought stress. | |
| NAC | CaNAC46 [92] | Overexpression of Capsicum annuum CaNAC46 improved the tolerance of transgenic Arabidopsis thaliana plants to drought and salt stresses by promoting the expression of SOD and POD. |
| CaNAC064 [105] | The Capsicum annuum CaNAC064-overexpressing Arabidopsis plants exhibited lower MDA content, chilling injury index under cold stress. | |
| GmNAC06 [90] | Soybean GmNAC06 could cause the accumulation of proline and glycine betaine to alleviate or avoid the negative effects of ROS in soybean. | |
| GmNAC065 [93] | Soybean GmNAC065 expression shows a phenotype associated with enhanced oxidative performance and higher carotenoid contents under salt stress in Arabidopsis. | |
| GmNAC085 [121] | Soybean GmNAC085 mediated drought resistance in transgenic Arabidopsis plants, with higher activities of antioxidant enzymes responsible for scavenging hydrogen peroxide or superoxide radicals. | |
| MbNAC25 [94] | Overexpressing Malus baccata (L.) Borkh MbNAC25 Arabidopsis plants showed enhanced tolerance against cold and drought salinity by increasing proline content, the activities of antioxidant enzymes SOD, POD and CAT. | |
| ONAC066 [122] | Overexpression of rice ONAC066 in transgenic rice improved drought and oxidative stress tolerance, accompanied with increased contents of proline, decreased accumulation of ROS in rice. | |
| RtNAC100 [91] | R. trigyna RtNAC100 overexpression aggravated salt-induced PCD in transgenic R. trigyna lines by promoting ROS. | |
| AP2/ERF | ERF6 [123] | Arabidopsis ERF6 functions as a transcriptional activator and suppressor of genes in response to drought stress and decreased ROS content in Arabidopsis. |
| ERF96 [124] | Arabidopsis ERF96-overexpressing Arabidopsis lines exhibited the significant increases in CAT and GPX activities as well as the glutathione (GSH) content, while having a decrease in ROS accumulation compared to WT. | |
| ERF74 [63] | Arabidopsis ERF74-overexpressing Arabidopsis lines showed enhanced tolerance to drought, high light, heat and aluminum stresses, and induction of stress marker genes and ROS-scavenging enzyme genes is dependent on the ERF74-RbohD-ROS signal pathway. | |
| LcERF056 [98] | Lotus corniculatus LcERF056 plays important roles in salt tolerance in Lotus corniculatus by modulating ROS-related genes | |
| GhERF13.12 [96] | GhERF13.12 from Gossypium hirsutum transgenic Arabidopsis showed enhanced salt stress tolerance and enhanced expression of genes participating in proline biosynthesis, and ROS scavenging. | |
| ZmEREB20 [95] | Maize ZmEREB20 positively regulated salt tolerance through the molecular mechanism associated with ROS scavenging in maize. | |
| SlERF84 [97] | Overexpression of tomato SlERF84 in Arabidopsis endows transgenic plants enhanced tolerance to drought and salt stress. | |
| MYB | CmMYB012 [125] | CmMYB012 from Chrysanthemum morifolium was also found to inhibit anthocyanin biosynthesis by suppressing the expression of CmCHS, CmDFR, CmANS and CmUFGT against heat stress on Chrysanthemum morifolium. |
| MdMYB23 [126] | Transgenic apple calli and Arabidopsis with overexpression of MdMYB23 from apple exhibited increased cold tolerance through active MdANR to promote proanthocyanidin accumulation and ROS scavenging. | |
| MYB44 [110] | Suppression of XsMYB44 expression via virus-induced gene silencing weakened yellowhorn tolerance to both individual and combined drought and heat stress and increased ROS levels and decreased antioxidant enzyme activities and proline content. | |
| MYB49 [100] | Overexpression of SlMYB49 in tomato significantly enhanced the resistance of tomato to salt and drought stress and decreased accumulation of ROS. | |
| PlMYB108 [127] | Overexpression of PlMYB108 from Herbaceous Peony in tobacco plants, showed that the flavonoid content, antioxidant enzyme activities, and photosynthesis were markedly elevated to confer drought stress. | |
| SlMYB102 [99] | The overexpression of SlMYB102 in tomato maintained lower ROS generation and increased the activity of ROS scavenging enzymes, the accumulation of antioxidants and proline was higher under salt stress. | |
| TaMYB86B [128] | Wheat TaMYB86B influences the salt tolerance of wheat by regulating the ion homeostasis to maintain an appropriate osmotic balance and decrease ROS levels. | |
| bHLH | AhbHLH112 [78] | The overexpression of AhbHLH112 from peanut improves the drought tolerance of transgenic Arabidopsis plants both in seedling and adult stages through directly activating the POD gene. |
| bHLH123 [62] | Overexpression of NtbHLH123 from tobacco resulted in greater resistance to salt stress on tobacco through the NtbHLH123-NtRbohE signaling pathway. | |
| BvbHLH93 [102] | Overexpression of sugar beet BvBHLH93 in Arabidopsis enhanced the activities of antioxidant enzymes by positively regulating the expression of antioxidant genes SOD and POD to against to salt stress. | |
| MdbHLH130 [73] | Overexpression of apple MdbHLH130 in tobacco led to lower ROS accumulation and upregulation of the expression of some ROS-scavenging under drought stress. | |
| MfbHLH38 [116] | Heterologous expression of M. flabellifolia MfbHLH38 in Arabidopsis improved the tolerance to drought and salinity stresses, decreased proline and ROS accumulation and increased antioxidant enzyme activities | |
| MfPIF1 [117] | Overexpression of MfPIF1 from M. flabellifolia in Arabidopsis thaliana led to enhanced drought and salinity tolerance, which was attributed to higher contents of proline and activities of antioxidant enzymes, as well as lower ROS accumulation in transgenic lines. | |
| OsWIH2 [129] | Heterologous expression of OsWIH2 in rice resulted in significantly higher drought tolerance, probably due to the decreased ROS accumulation under drought stress. | |
| PYE, ILR 3 [130] | Arabidopsis ILR3 and PYE confer photoprotection during Fe deficiency to prevent the accumulation of singlet oxygen and repair of the photosynthetic machinery. | |
| Other families | CaSBP12 [131] | Silencing the CaSBP12 gene enhanced pepper plant tolerance to salt stress and decreased accumulation of ROS. |
| CsHB5 [132] | Heterologous expression of CsHB5 in citrus calli upregulated the expression of ROS-related genes and increased the content of H2O2 to against to senescence. | |
| HY5 [133] | Arabidopsis ELONGATED HYPOCOTYL5 as a major transcription factor required for activation of the detoxification program under high N. | |
| MdZAT10 [134] | Heterologous expression of MdZAT10 in apple calli decreased the expression level of MdAPX2 and increased sensitivity to drought stress. | |
| MdHB7-like [135] | Heterologous expression of MdHB7-like reduced ROS under salt stress. | |
| OsMADS57 [136] | Overexpression of rice OsMADS57 in both Arabidopsis thaliana and rice could improve their salt tolerance by increasing the activities of antioxidative enzymes. | |
| ZAT18 [137] | Heterologous expression of ZAT18 in Arabidopsis improved drought tolerance and exhibited a lower content of ROS and higher antioxidant enzyme activities. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Wu, X.; Gong, B.; Lü, G.; Li, J.; Gao, H. Review of the Mechanisms by Which Transcription Factors and Exogenous Substances Regulate ROS Metabolism under Abiotic Stress. Antioxidants 2022, 11, 2106. https://doi.org/10.3390/antiox11112106
Liu P, Wu X, Gong B, Lü G, Li J, Gao H. Review of the Mechanisms by Which Transcription Factors and Exogenous Substances Regulate ROS Metabolism under Abiotic Stress. Antioxidants. 2022; 11(11):2106. https://doi.org/10.3390/antiox11112106
Chicago/Turabian StyleLiu, Peng, Xiaolei Wu, Binbin Gong, Guiyun Lü, Jingrui Li, and Hongbo Gao. 2022. "Review of the Mechanisms by Which Transcription Factors and Exogenous Substances Regulate ROS Metabolism under Abiotic Stress" Antioxidants 11, no. 11: 2106. https://doi.org/10.3390/antiox11112106
APA StyleLiu, P., Wu, X., Gong, B., Lü, G., Li, J., & Gao, H. (2022). Review of the Mechanisms by Which Transcription Factors and Exogenous Substances Regulate ROS Metabolism under Abiotic Stress. Antioxidants, 11(11), 2106. https://doi.org/10.3390/antiox11112106









