5-Hydroxymaltol Derived from Beetroot Juice through Lactobacillus Fermentation Suppresses Inflammatory Effect and Oxidant Stress via Regulating NF-kB, MAPKs Pathway and NRF2/HO-1 Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Plant Materials
2.3. Fermentation of Beet Using Lactic Acid Bacteria
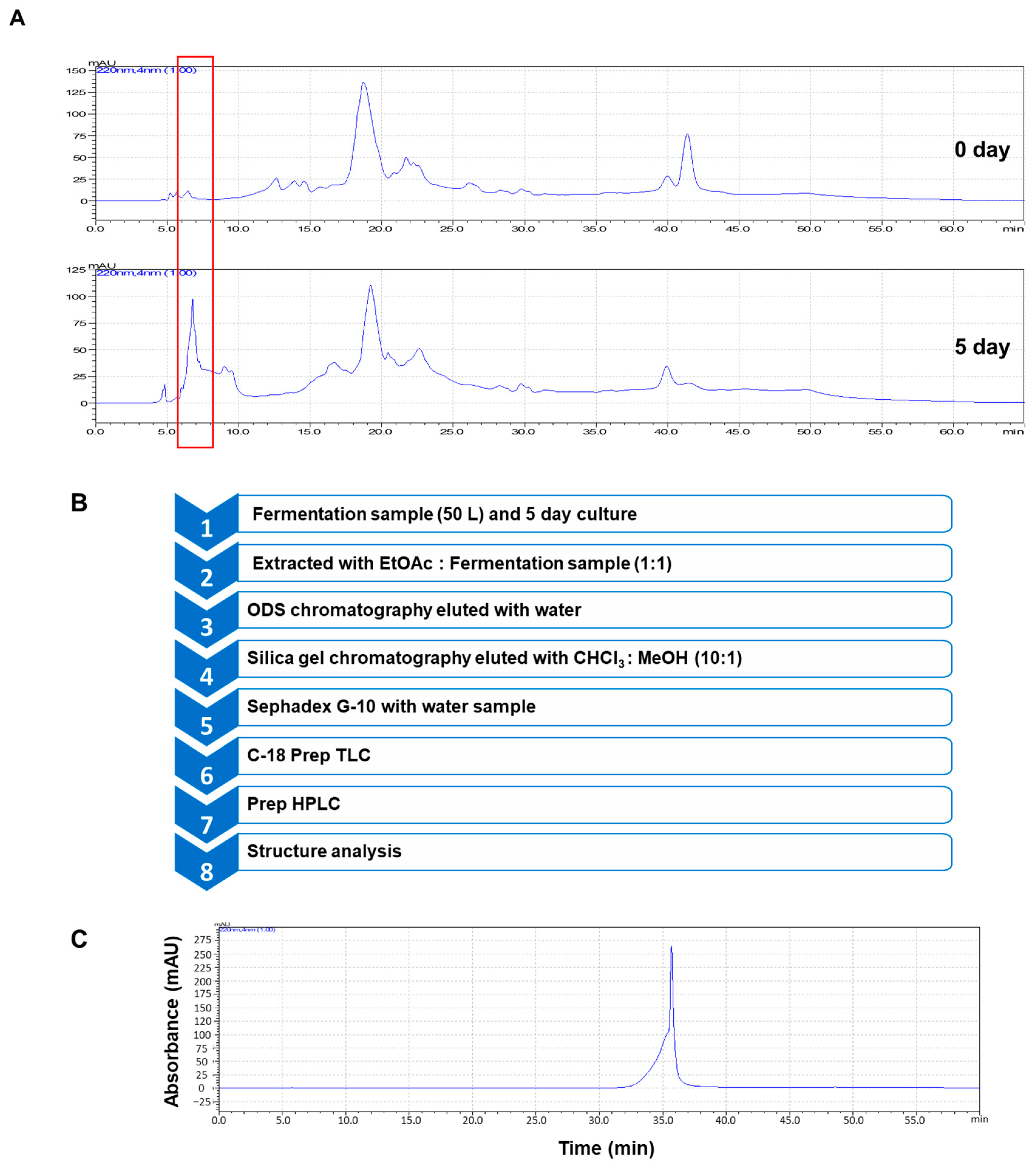
2.4. Extraction and Purification
2.5. Structure Analysis of the Isolated Sample
2.6. Antioxidant Assay
2.7. Cell Line and Culture Conditions
2.8. Cell Proliferation
2.9. NO Assay
2.10. RT-qPCR
2.11. ELISA
2.12. Immuno Blot Analysis
2.13. Measurement of ROS Activity Using CellROX Green Dye
2.14. Statistical Analysis
3. Results
3.1. Isolation of an Antioxidant Compound from Beet Fermentation Using L. rhamnosus GG
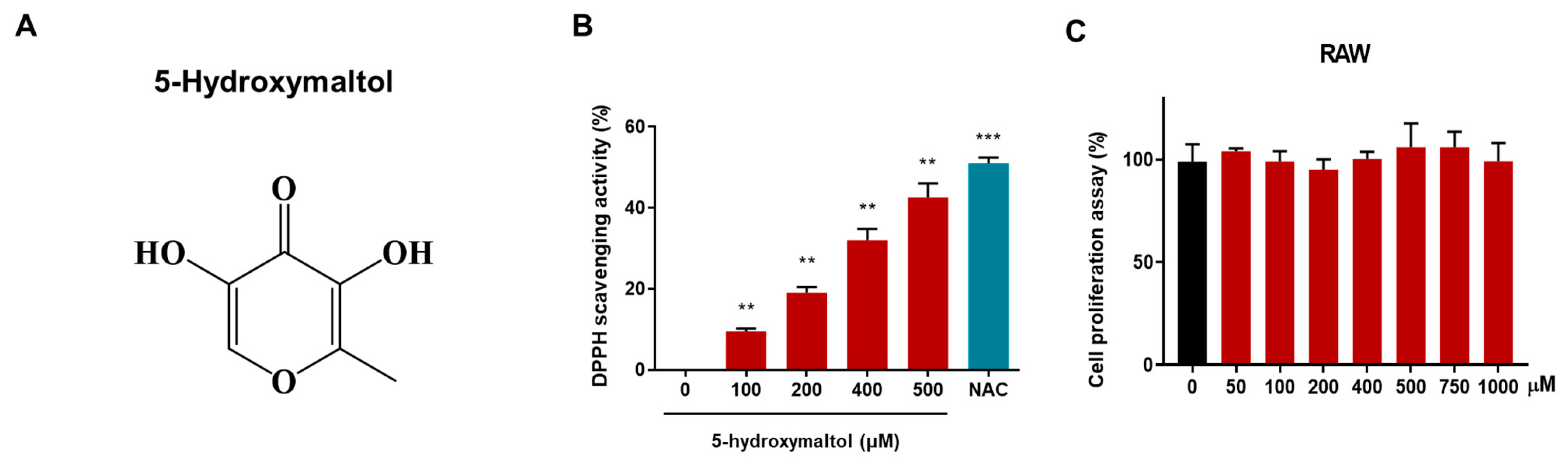
3.2. Structural Analysis of Purified Compound and Effect of the Isolated Compound on Antioxidant Activity and Cell Proliferation
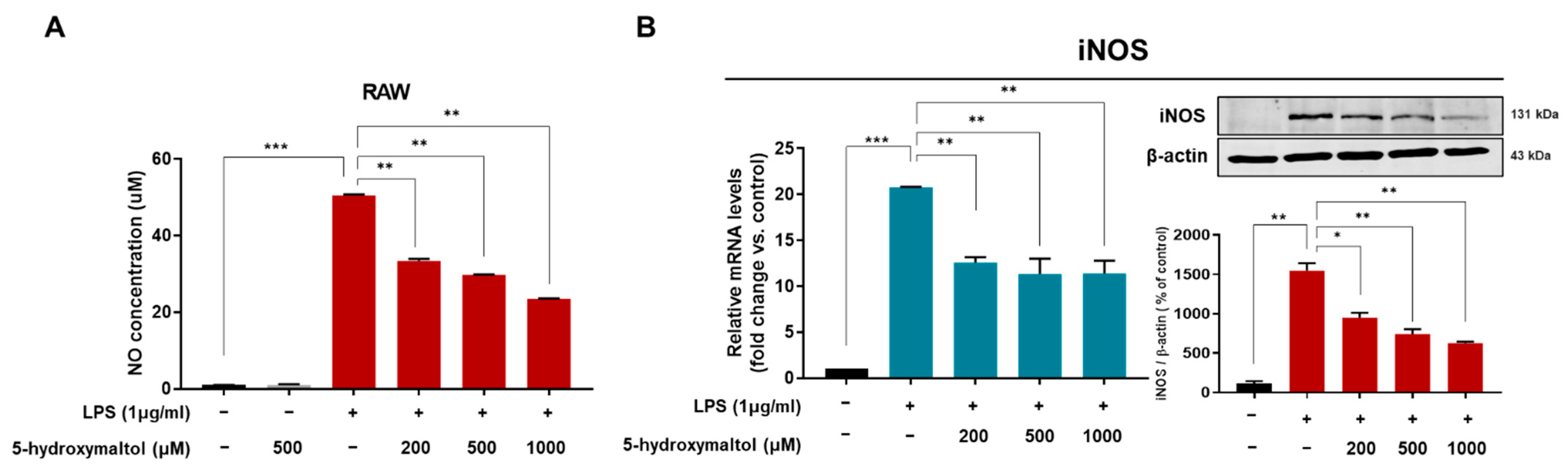
3.3. Effect of 5-Hydoxylmaltol on NO Production and Inducible NO Synthase Expression in RAW 264.7 Macrophage Cells
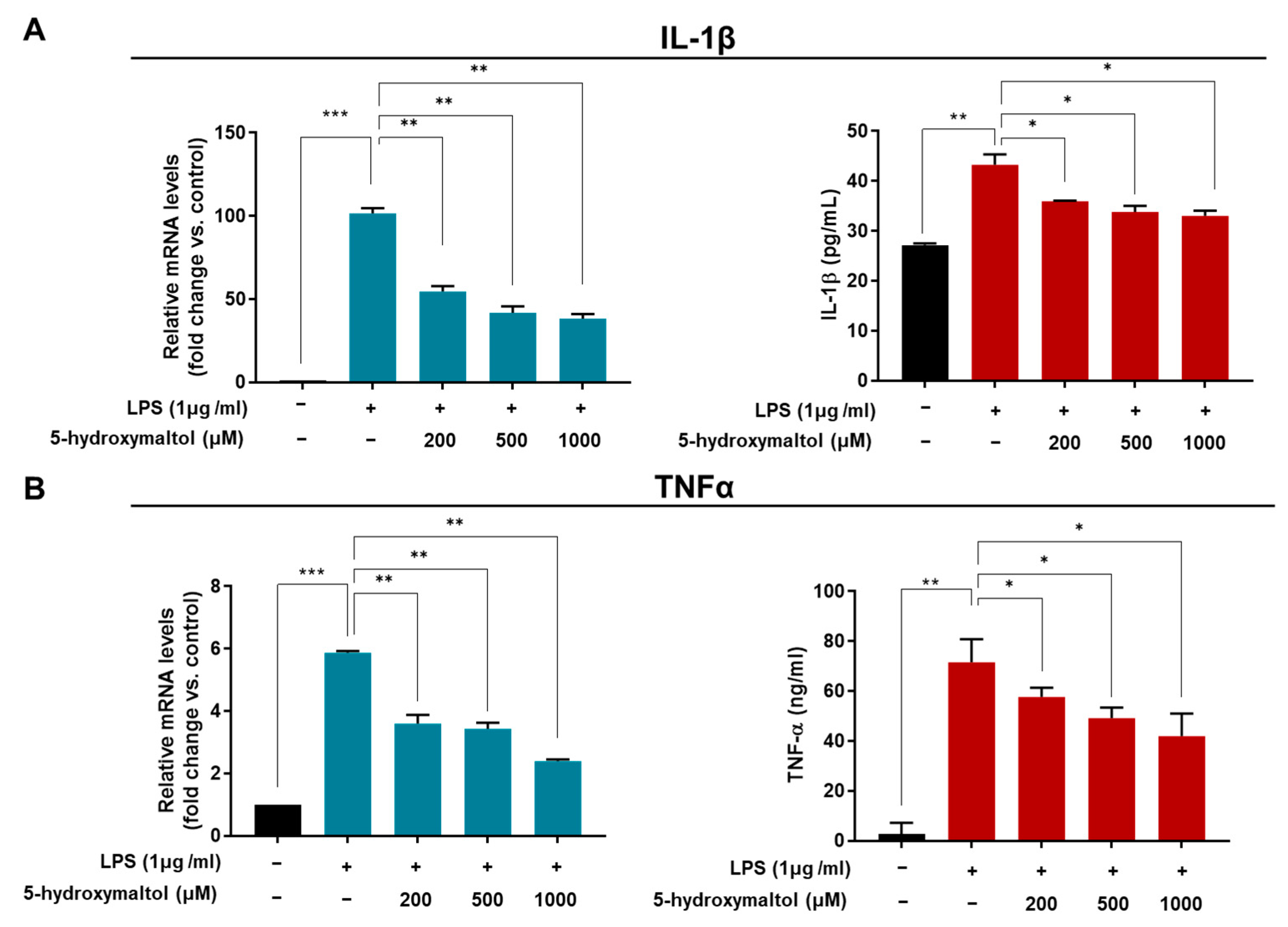
3.4. 5-Hydroxymaltol Suppressed the LPS-Stimulated TNF-α and IL-1β Production and mRNA Level in RAW 264.7 Cells
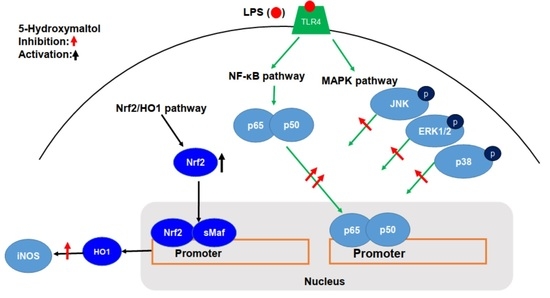
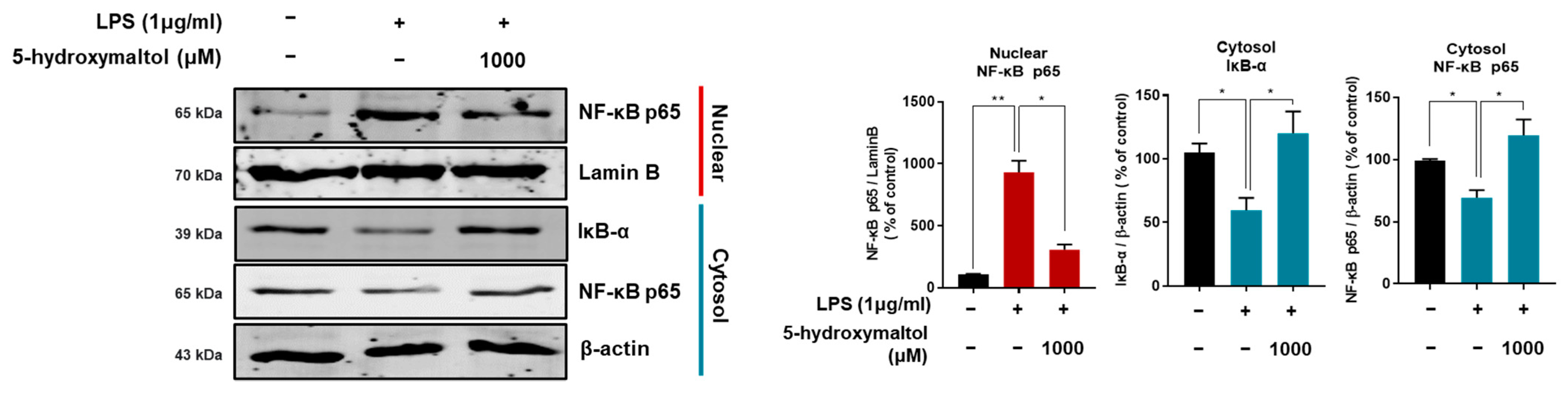
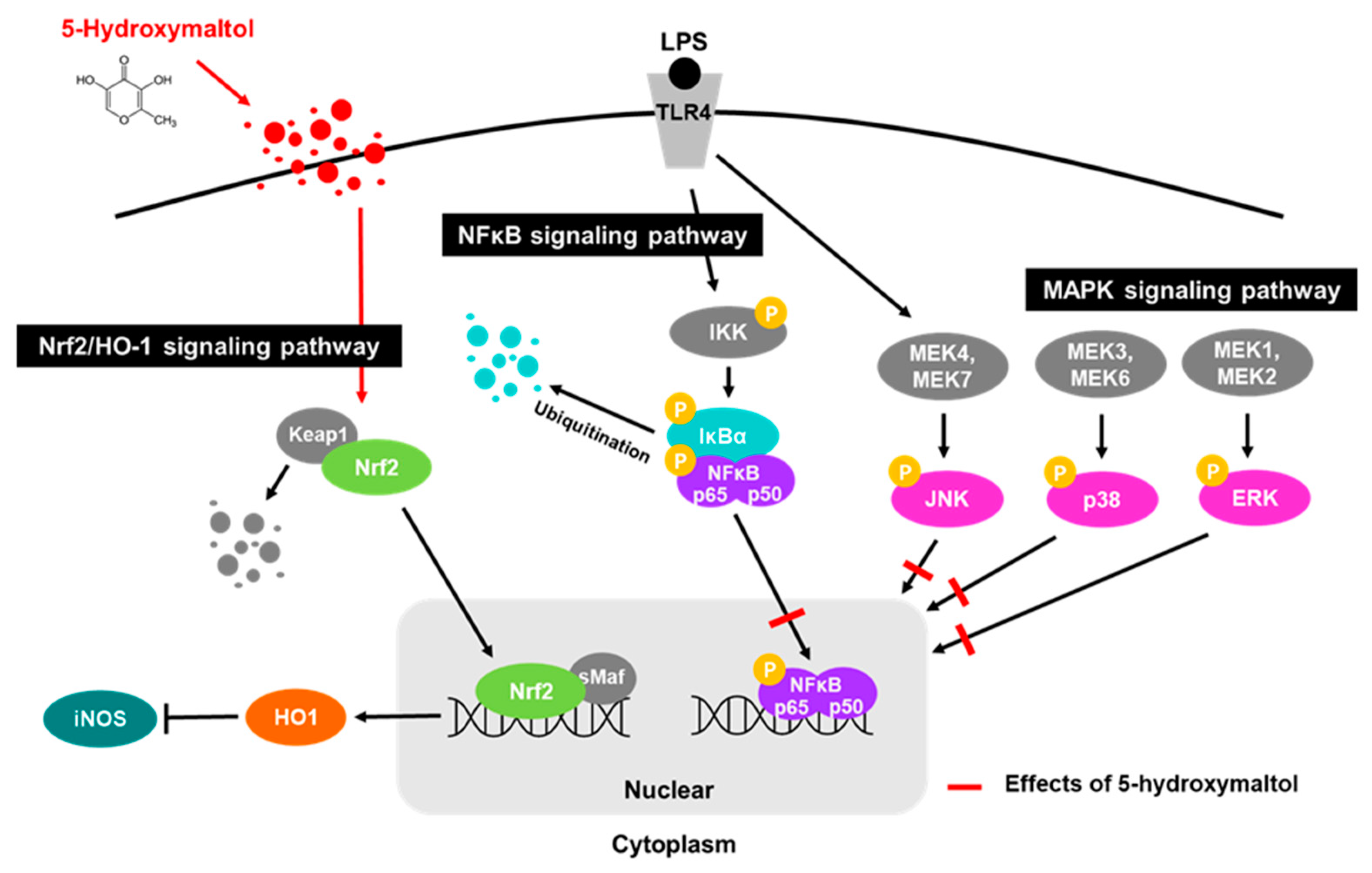
3.5. 5-Hydroxymaltol Suppressed LPS-Stimulated NF-κB Activation in RAW 264.7 Cells
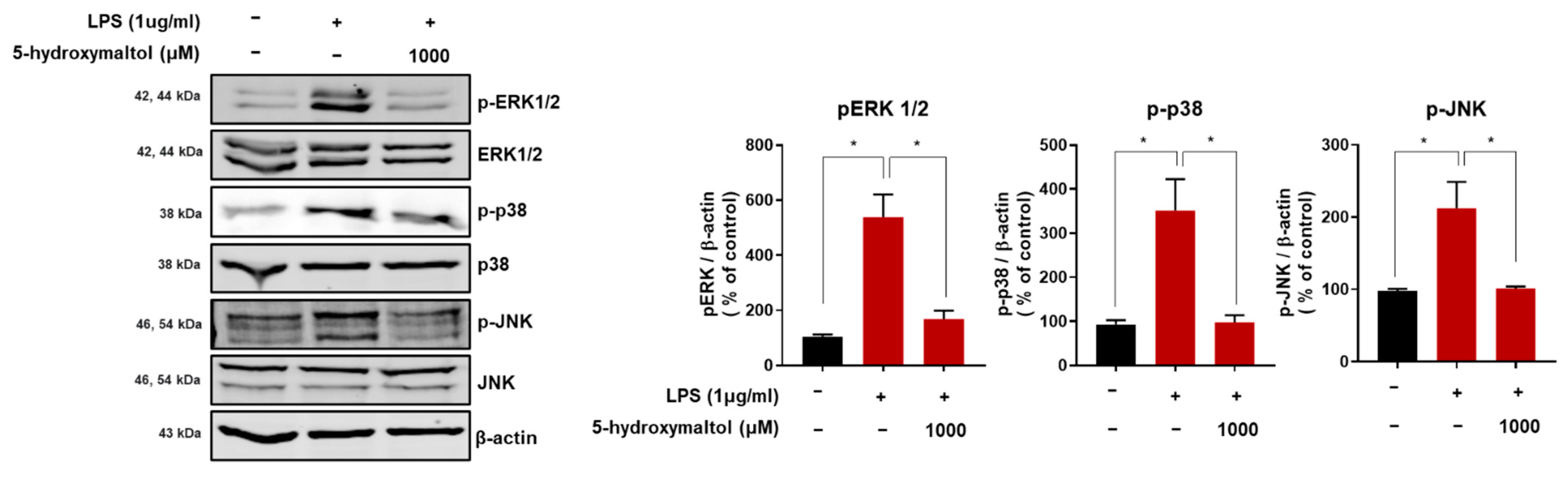
3.6. 5-Hydroxymaltol Inhibited LPS-Stimulated MAPK Activation in RAW 264.7 Cells
3.7. 5-Hydroxymaltol Decreased LPS-Induced ROS Production in RAW 264.7 Cells
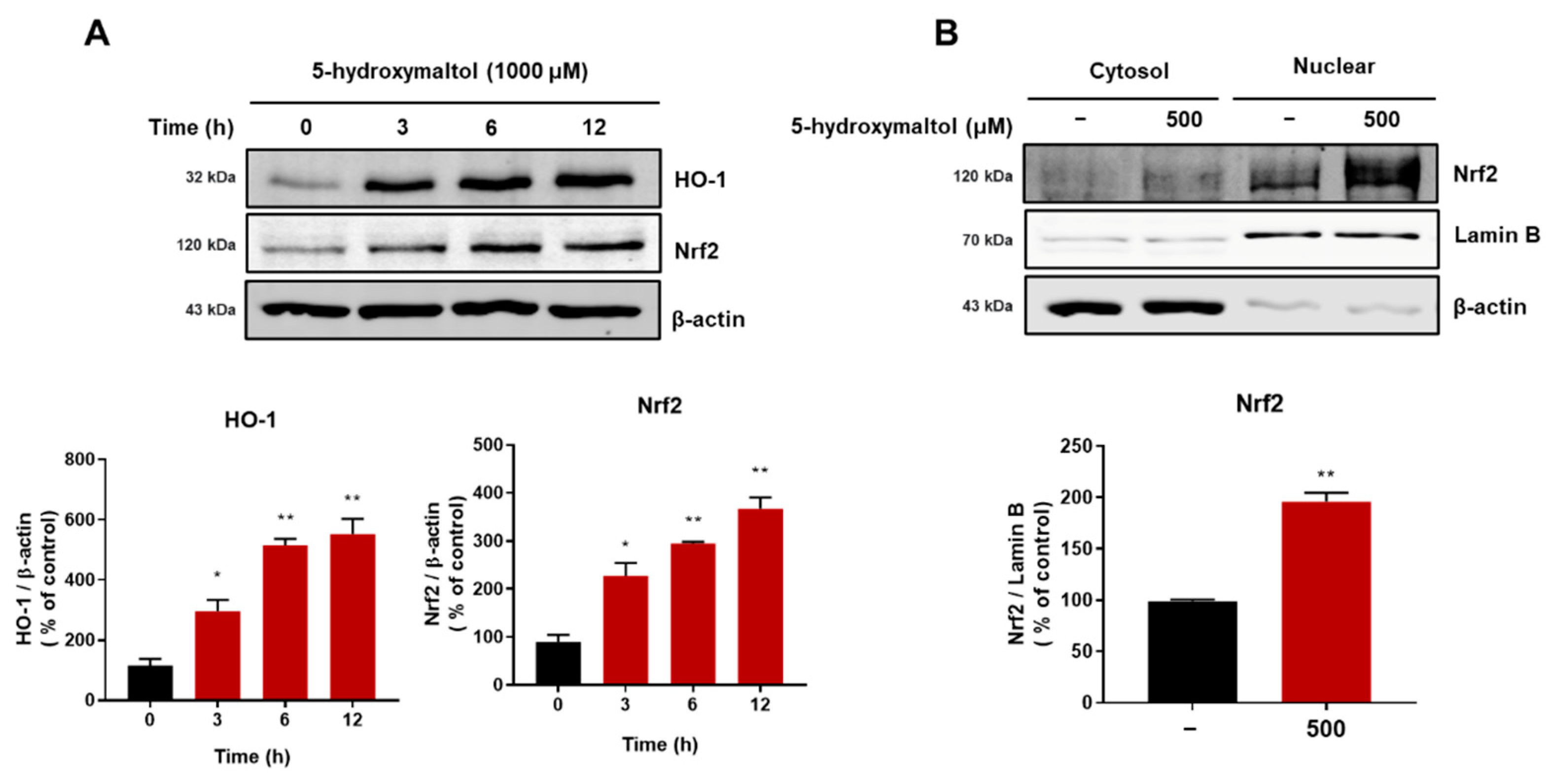
3.8. 5-Hydroxymaltol Increased Protein Exprerssion of HO-1 and Nrf2 in RAW 264.7 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, H.M.; Fu, L.; Cheng, C.C.; Gao, R.; Lin, M.Y.; Su, H.L.; Belinda, N.E.; Nguyen, T.H.; Lin, W.H.; Lee, P.C.; et al. Inhibition of LPS-Induced Oxidative Damages and Potential Anti-Inflammatory Effects of Phyllanthus emblica Extract via Down-Regulating NF-kappaB, COX-2, and iNOS in RAW 264.7 Cells. Antioxidants 2019, 8, 270. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.Y.; Feng, C.W.; Hung, H.C.; Chakraborty, C.; Chen, C.H.; Chen, W.F.; Jean, Y.H.; Wang, H.M.; Sung, C.S.; Sun, Y.M.; et al. A novel zebrafish model to provide mechanistic insights into the inflammatory events in carrageenan-induced abdominal edema. PLoS ONE 2014, 9, e104414. [Google Scholar]
- Du, C.; Bhatia, M.; Tang, S.C.; Zhang, M.; Steiner, T. Mediators of Inflammation: Inflammation in Cancer, Chronic Diseases, and Wound Healing. Mediat. Inflamm. 2015, 2015, 570653. [Google Scholar]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Lee, W.S.; Shin, J.S.; Jang, D.S.; Lee, K.T. Xanthotoxin suppresses LPS-induced expression of iNOS, COX-2, TNF-alpha, and IL-6 via AP-1, NF-kappaB, and JAK-STAT inactivation in RAW 264.7 macrophages. Int. Immunopharmacol. 2017, 49, 21–29. [Google Scholar] [CrossRef]
- Guha, M.; Mackman, N. LPS induction of gene expression in human monocytes. Cell Signal. 2001, 13, 85–94. [Google Scholar] [CrossRef]
- Shi, Q.; Cao, J.; Fang, L.; Zhao, H.; Liu, Z.; Ran, J.; Zheng, X.; Li, X.; Zhou, Y.; Ge, D.; et al. Geniposide suppresses LPS-induced nitric oxide, PGE2 and inflammatory cytokine by downregulating NF-kappaB, MAPK and AP-1 signaling pathways in macrophages. Int. Immunopharmacol. 2014, 20, 298–306. [Google Scholar] [PubMed]
- Kim, J.B.; Han, A.R.; Park, E.Y.; Kim, J.Y.; Cho, W.; Lee, J.; Seo, E.K.; Lee, K.T. Inhibition of LPS-induced iNOS, COX-2 and cytokines expression by poncirin through the NF-kappaB inactivation in RAW 264.7 macrophage cells. Biol. Pharm. Bull. 2007, 30, 2345–2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leiro, J.; Alvarez, E.; Arranz, J.A.; Laguna, R.; Uriarte, E.; Orallo, F. Effects of cis-resveratrol on inflammatory murine macrophages: Antioxidant activity and down-regulation of inflammatory genes. J. Leukoc. Biol. 2004, 75, 1156–1165. [Google Scholar] [PubMed] [Green Version]
- Su, Y.W.; Chiou, W.F.; Chao, S.H.; Lee, M.H.; Chen, C.C.; Tsai, Y.C. Ligustilide prevents LPS-induced iNOS expression in RAW 264.7 macrophages by preventing ROS production and down-regulating the MAPK, NF-kappaB and AP-1 signaling pathways. Int. Immunopharmacol. 2011, 11, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Cha, H.J.; Choi, E.O.; Leem, S.H.; Kim, G.Y.; Moon, S.K.; Chang, Y.C.; Yun, S.J.; Hwang, H.J.; Kim, B.W.; et al. Schisandrin A suppresses lipopolysaccharide-induced inflammation and oxidative stress in RAW 264.7 macrophages by suppressing the NF-kappaB, MAPKs and PI3K/Akt pathways and activating Nrf2/HO-1 signaling. Int. J. Mol. Med. 2018, 41, 264–274. [Google Scholar]
- Murakami, A.; Ohigashi, H. Targeting NOX, INOS and COX-2 in inflammatory cells: Chemoprevention using food phytochemicals. Int. J. Cancer 2007, 121, 2357–2363. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Takagi, T.; Higashimura, Y. Heme oxygenase-1 and anti-inflammatory M2 macrophages. Arch. Biochem. Biophys. 2014, 564, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Esatbeyoglu, T.; Wagner, A.E.; Schini-Kerth, V.B.; Rimbach, G. Betanin—A food colorant with biological activity. Mol. Nutr. Food Res. 2015, 59, 36–47. [Google Scholar] [CrossRef]
- Rahimi, P.; Abedimanesh, S.; Mesbah-Namin, S.A.; Ostadrahimi, A. Betalains, the nature-inspired pigments, in health and diseases. Crit. Rev. Food Sci. Nutr. 2019, 59, 2949–2978. [Google Scholar] [CrossRef] [PubMed]
- Aratani, Y. Myeloperoxidase: Its role for host defense, inflammation, and neutrophil function. Arch. Biochem. Biophys. 2018, 640, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Choi, H.S.; Zhen, X.; Kim, S.L.; Kim, J.H.; Ko, Y.C.; Yun, B.S.; Lee, D.S. Betavulgarin Isolated from Sugar Beet (Beta vulgaris) Suppresses Breast Cancer Stem Cells through Stat3 Signaling. Molecules 2020, 25, 2999. [Google Scholar] [CrossRef]
- Elliger, C.A.; Halloin, J.M. Phenolics induced in Beta vulgaris by Rhizoctonia solani infection. Phytochemistry 1994, 37, 691–693. [Google Scholar] [CrossRef]
- Nguyen, L.; Hwang, E.S. Quality Characteristics and Antioxidant Activity of Yogurt Supplemented with Aronia (Aronia melanocarpa) Juice. Prev. Nutr. Food Sci. 2016, 21, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Demirci, M.A.; Ipek, Y.; Gul, F.; Ozen, T.; Demirtas, I. Extraction, isolation of heat-resistance phenolic compounds, antioxidant properties, characterization and purification of 5-hydroxymaltol from Turkish apple pulps. Food Chem. 2018, 269, 111–117. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free-Radical Method to Evaluate Antioxidant Activity. Food Sci. Technol. Lebensm. Wiss. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Huang, B.P.; Lin, C.H.; Chen, H.M.; Lin, J.T.; Cheng, Y.F.; Kao, S.H. AMPK activation inhibits expression of proinflammatory mediators through downregulation of PI3K/p38 MAPK and NF-kappaB signaling in murine macrophages. DNA Cell Biol. 2015, 34, 133–141. [Google Scholar] [CrossRef]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar]
- Kwon, D.H.; Jeong, J.W.; Choi, E.O.; Lee, H.W.; Lee, K.W.; Kim, K.Y.; Kim, S.G.; Hong, S.H.; Kim, G.Y.; Park, C.; et al. Inhibitory effects on the production of inflammatory mediators and reactive oxygen species by Mori folium in lipopolysaccharide-stimulated macrophages and zebrafish. Acad. Bras. Cienc. 2017, 89, 661–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Li, W.; Su, Z.Y.; Kong, A.N. The complexity of the Nrf2 pathway: Beyond the antioxidant response. J. Nutr. Biochem. 2015, 26, 1401–1413. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Hyun, J.W. Oxidative Stress, Nrf2, and Epigenetic Modification Contribute to Anticancer Drug Resistance. Toxicol. Res. 2017, 33, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Clifford, T.; Howatson, G.; West, D.J.; Stevenson, E.J. The potential benefits of red beetroot supplementation in health and disease. Nutrients 2015, 7, 2801–2822. [Google Scholar] [CrossRef]
- Yoon, K.Y.; Woodams, E.E.; Hang, Y.D. Fermentation of beet juice by beneficial lactic acid bacteria. Lwt-Food Sci. Technol. 2005, 38, 73–75. [Google Scholar] [CrossRef]
- Kazimierczak, R.; Hallmann, E.; Lipowski, J.; Drela, N.; Kowalik, A.; Pussa, T.; Matt, D.; Luik, A.; Gozdowski, D.; Rembialkowska, E. Beetroot (Beta vulgaris L.) and naturally fermented beetroot juices from organic and conventional production: Metabolomics, antioxidant levels and anticancer activity. J. Sci. Food Agric. 2014, 94, 2618–2629. [Google Scholar] [CrossRef] [PubMed]
- Shobharani, P.; Nanishankar, V.H.; Halami, P.M.; Sachindra, N.M. Antioxidant and anticoagulant activity of polyphenol and polysaccharides from fermented Sargassum sp. Int. J. Biol. Macromol. 2014, 65, 542–548. [Google Scholar] [CrossRef]
- Cuschieri, J.; Maier, R.V. Oxidative stress, lipid rafts, and macrophage reprogramming. Antioxid. Redox Signal. 2007, 9, 1485–1497. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target. 2017, 2, e17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.S.; Choi, M.J.; Lee, Y.Y.; Moon, B.I.; Park, J.S.; Kim, H.S. Suppression of Lipopolysaccharide-Induced Neuroinflammation by Morin via MAPK, PI3K/Akt, and PKA/HO-1 Signaling Pathway Modulation. J. Agric. Food Chem. 2017, 65, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, E.; Sekiyama, A.; Hori, M.; Hara, K.; Takahashi, N.; Konishi, M.; Sato, E.F.; Matsumoto, S.; Okamura, H.; Inoue, M. Mitochondrial density contributes to the immune response of macrophages to lipopolysaccharide via the MAPK pathway. FEBS Lett. 2011, 585, 2263–2268. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Kim, T.B.; Moon, K.A.; Kim, T.J.; Shin, D.; Cho, Y.S.; Moon, H.B.; Lee, K.Y. Regulation of pro-inflammatory responses by lipoxygenases via intracellular reactive oxygen species in vitro and in vivo. Exp. Mol. Med. 2008, 40, 461–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Jia, Z.; Li, Y.R. Nrf2 Signaling in Macrophages. React. Oxyg. Species 2016, 2, 417–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habtemariam, S. The Nrf2/HO-1 Axis as Targets for Flavanones: Neuroprotection by Pinocembrin, Naringenin, and Eriodictyol. Oxid. Med. Cell Longev. 2019, 2019, 4724920. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hao, W.; Hu, J.; Mi, X.; Han, Y.; Ren, S.; Jiang, S.; Wang, Y.; Li, X.; Li, W. Maltol Improves APAP-Induced Hepatotoxicity by Inhibiting Oxidative Stress and Inflammation Response via NF-κB and PI3K/Akt Signal Pathways. Antioxidants 2019, 8, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, H.A.; Bracewell, J.M.; Fraser, A.R.; Jones, D.; Robertson, G.W.; Russell, J.D. 5-Hydroxymaltol and Mycophenolic-Acid, Secondary Metabolites from Penicillium-Echinulatum. Trans. Br. Mycol. Soc. 1988, 91, 649–651. [Google Scholar] [CrossRef]
- Cutzach, I.; Chatonnet, P.; Henry, R.; Dubourdieu, D. Identification of volatile compounds with a “toasty” aroma in heated oak used in barrelmaking. J. Agric. Food Chem. 1997, 45, 2217–2224. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-L.; Choi, H.S.; Ko, Y.-C.; Yun, B.-S.; Lee, D.-S. 5-Hydroxymaltol Derived from Beetroot Juice through Lactobacillus Fermentation Suppresses Inflammatory Effect and Oxidant Stress via Regulating NF-kB, MAPKs Pathway and NRF2/HO-1 Expression. Antioxidants 2021, 10, 1324. https://doi.org/10.3390/antiox10081324
Kim S-L, Choi HS, Ko Y-C, Yun B-S, Lee D-S. 5-Hydroxymaltol Derived from Beetroot Juice through Lactobacillus Fermentation Suppresses Inflammatory Effect and Oxidant Stress via Regulating NF-kB, MAPKs Pathway and NRF2/HO-1 Expression. Antioxidants. 2021; 10(8):1324. https://doi.org/10.3390/antiox10081324
Chicago/Turabian StyleKim, Su-Lim, Hack Sun Choi, Yu-Chan Ko, Bong-Sik Yun, and Dong-Sun Lee. 2021. "5-Hydroxymaltol Derived from Beetroot Juice through Lactobacillus Fermentation Suppresses Inflammatory Effect and Oxidant Stress via Regulating NF-kB, MAPKs Pathway and NRF2/HO-1 Expression" Antioxidants 10, no. 8: 1324. https://doi.org/10.3390/antiox10081324
APA StyleKim, S.-L., Choi, H. S., Ko, Y.-C., Yun, B.-S., & Lee, D.-S. (2021). 5-Hydroxymaltol Derived from Beetroot Juice through Lactobacillus Fermentation Suppresses Inflammatory Effect and Oxidant Stress via Regulating NF-kB, MAPKs Pathway and NRF2/HO-1 Expression. Antioxidants, 10(8), 1324. https://doi.org/10.3390/antiox10081324