L-Carnosine Stimulation of Coenzyme Q10 Biosynthesis Promotes Improved Mitochondrial Function and Decreases Hepatic Steatosis in Diabetic Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Animal Experiments
2.2. RNA Purification and Quantitative Real-Time qPCR
2.3. [3H] Mevalonate Incorporation Experiments
2.4. Lipid Extraction and HPLC
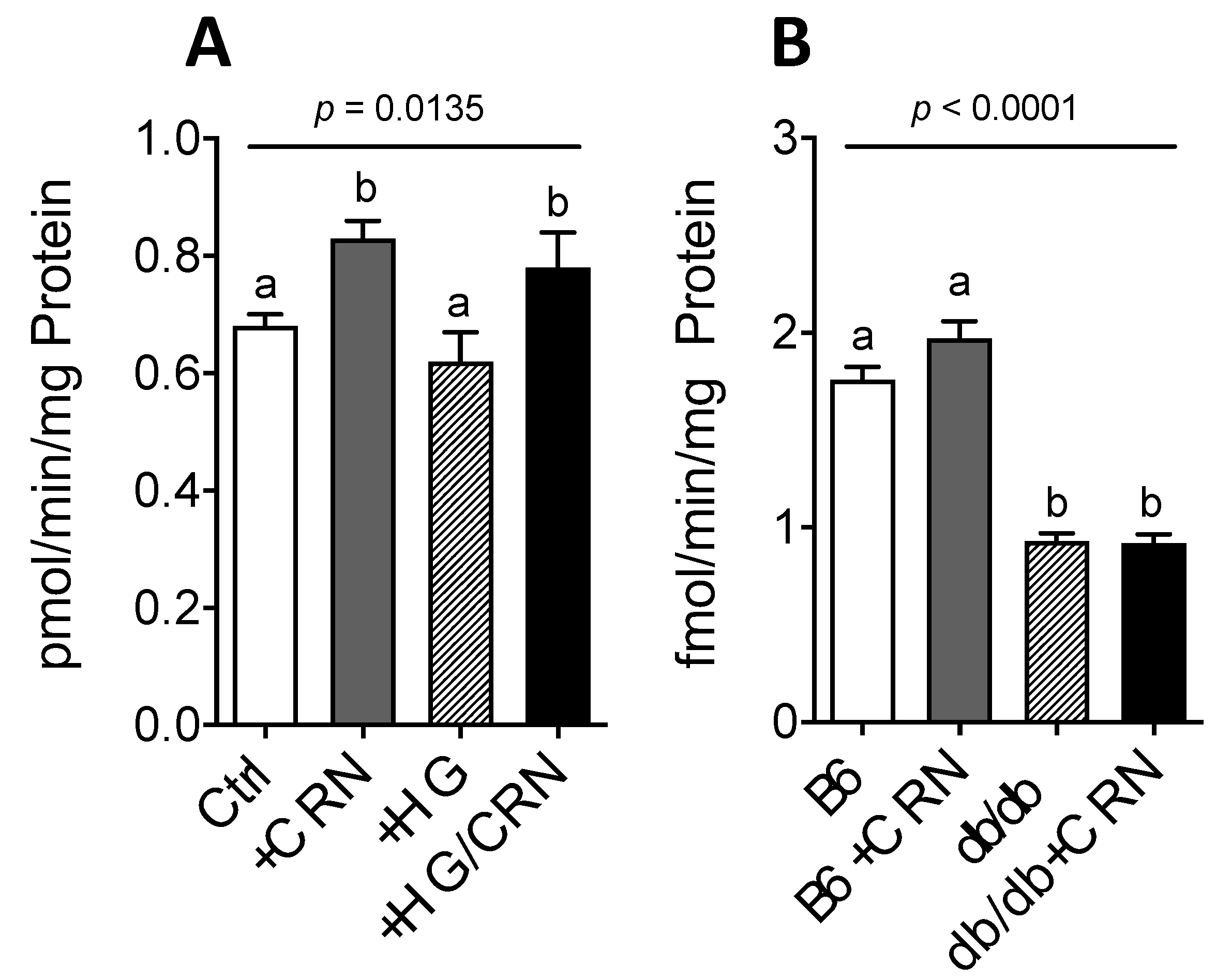
2.5. β-oxidation Activity In Vitro and In Vivo
2.6. Oxygen Consumption Rate, Oxidative Stress and ROS Formation
2.7. Statistical Analysis
3. Results
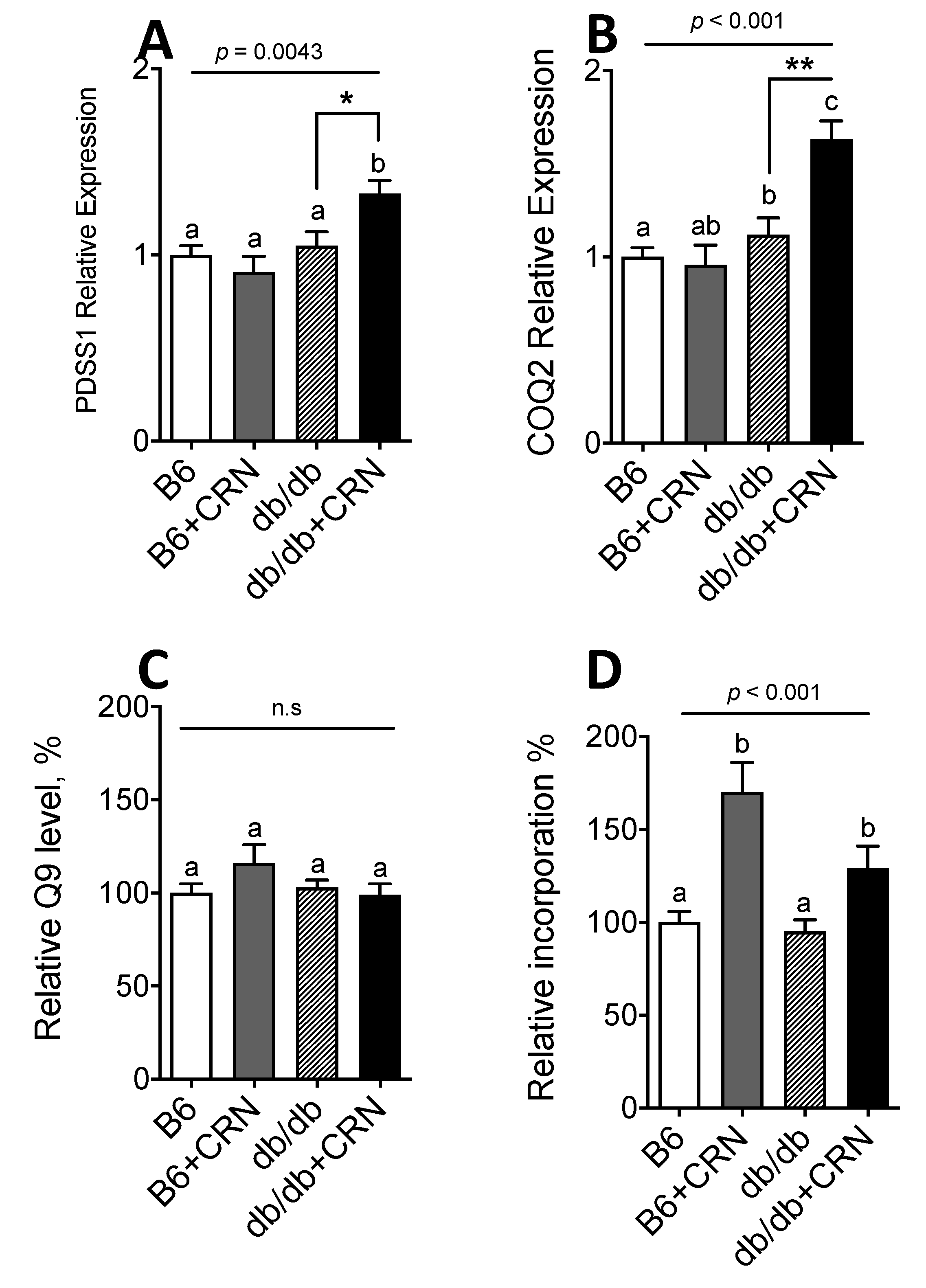
3.1. Carnosine Modulates Genes Regulating CoQ Expression and Stimulates CoQ Biosynthesis
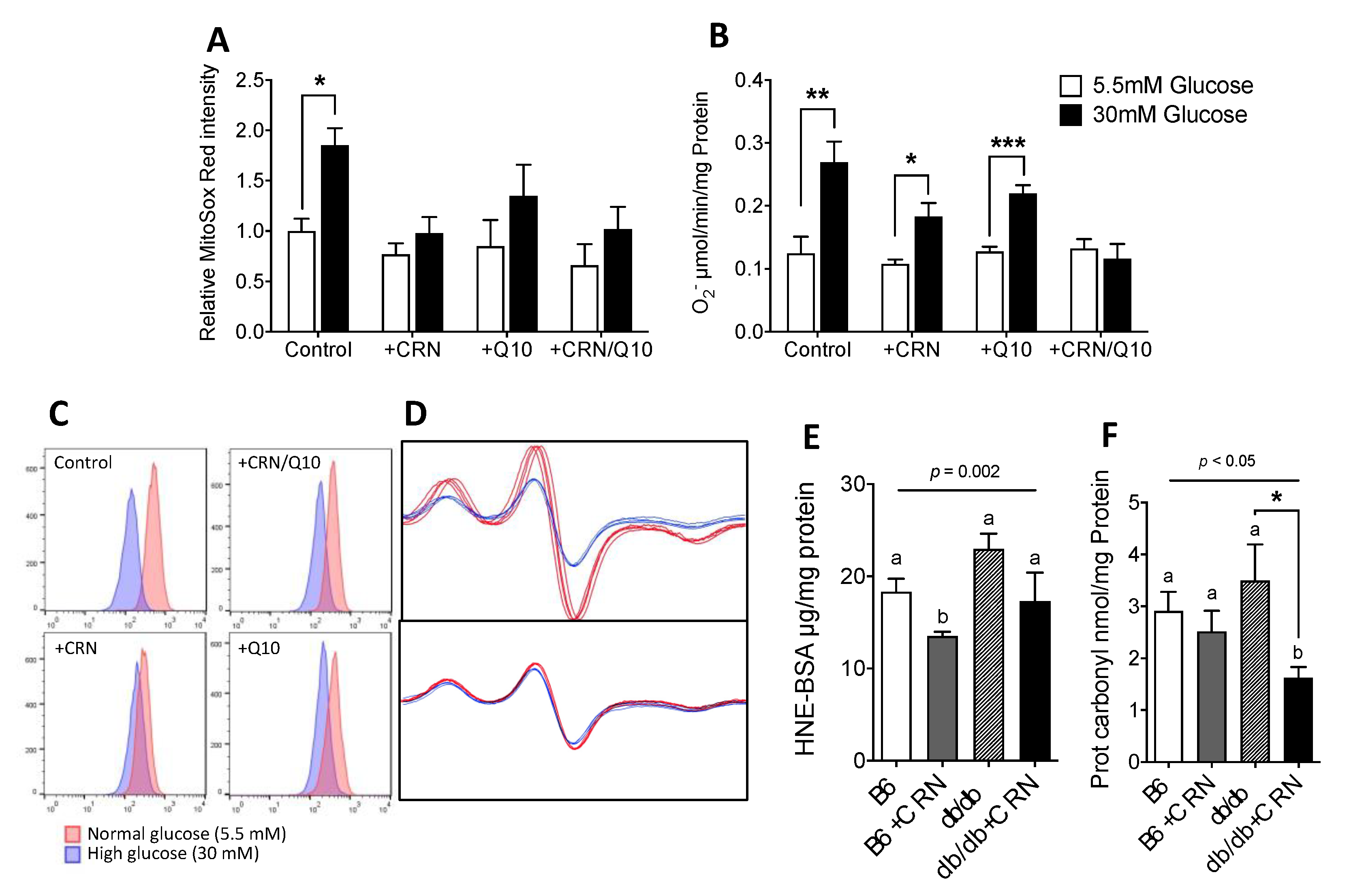
3.2. Carnosine Treatment Reduced In Vitro ROS Formation and Oxidative Stress
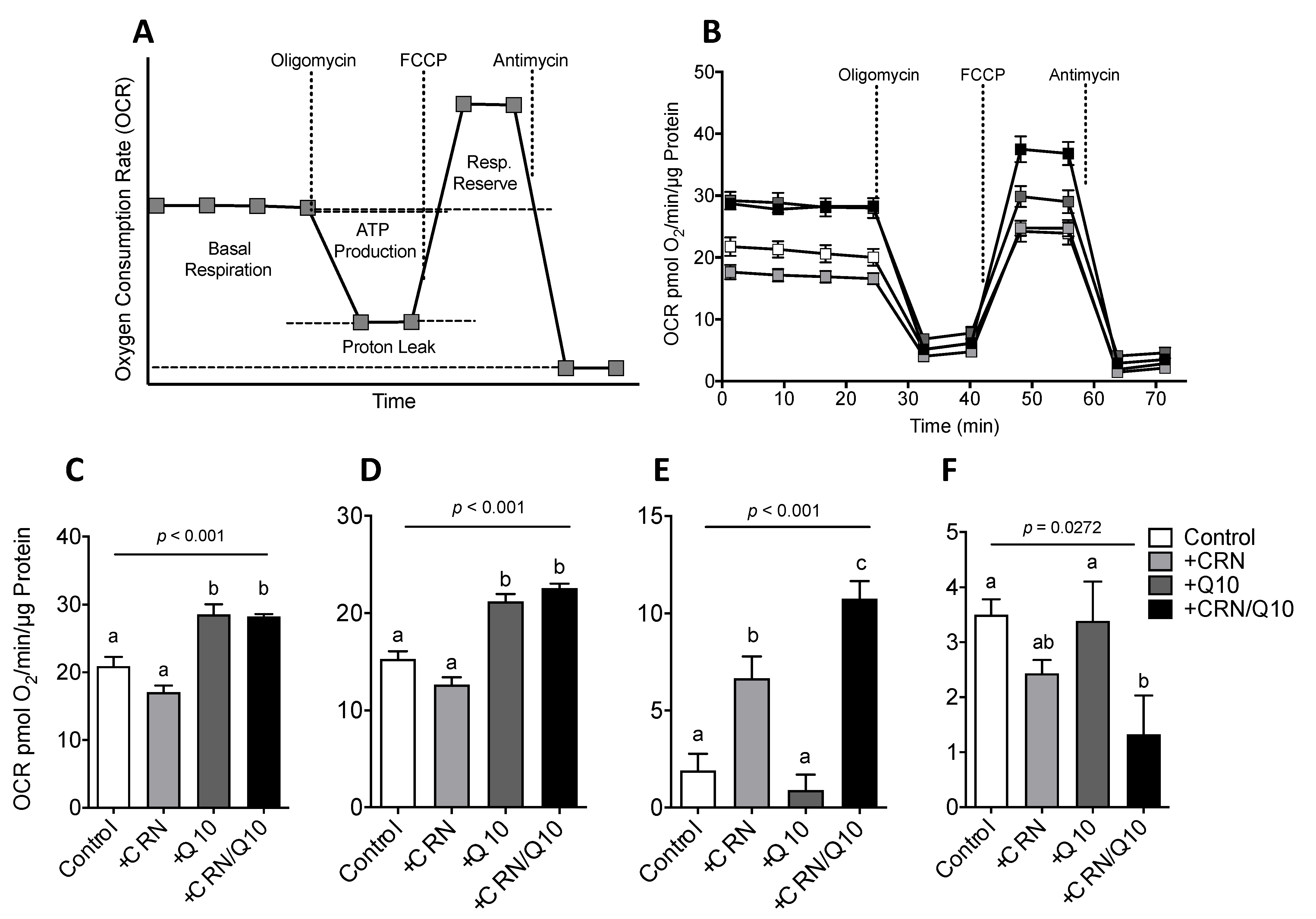
3.3. Carnosine Treatment Enhanced CoQ Effect on Mitochondrial Function
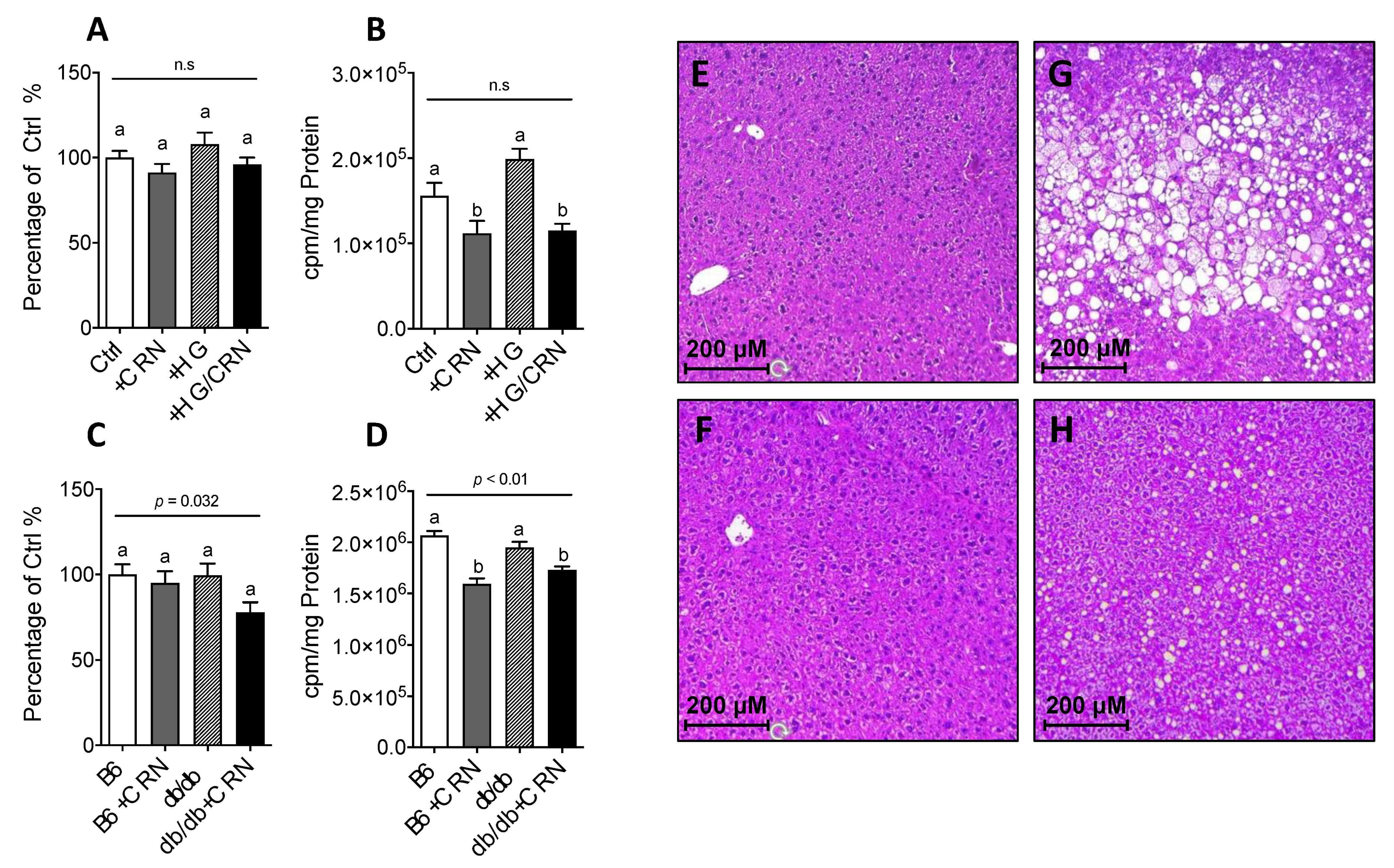
3.4. Carnosine Treatment Decreased Cholesterol Synthesis and Reduced Hepatic Steatosis
3.5. Carnosine Treatment Did Not Affect β-Oxidation in db/db Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Budzen, S.; Rymaszewska, J. The biological role of carnosine and its possible applications in medicine. Adv. Clin. Exp. Med. 2013, 22, 739–744. [Google Scholar] [PubMed]
- Turunen, M.; Olsson, J.; Dallner, G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 2004, 1660, 171–199. [Google Scholar] [CrossRef] [PubMed]
- Peters, V.; Schmitt, C.P.; Zschocke, J.; Gross, M.L.; Brismar, K.; Forsberg, E. Carnosine treatment largely prevents alterations of renal carnosine metabolism in diabetic mice. Amino Acids 2012, 42, 2411–2416. [Google Scholar] [CrossRef]
- Forsberg, E.A.; Botusan, I.R.; Wang, J.; Peters, V.; Ansurudeen, I.; Brismar, K.; Catrina, S.B. Carnosine decreases IGFBP1 production in db/db mice through suppression of HIF-1. J. Endocrinol. 2015, 225, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Ansurudeen, I.; Sunkari, V.G.; Grunler, J.; Peters, V.; Schmitt, C.P.; Catrina, S.B.; Brismar, K.; Forsberg, E.A. Carnosine enhances diabetic wound healing in the db/db mouse model of type 2 diabetes. Amino Acids 2012, 43, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, T.; Schilperoort, M.; Zhang, S.; Braun, J.D.; Qiu, J.; Rodriguez, A.; Pastene, D.O.; Kramer, B.K.; Koppel, H.; Baelde, H.; et al. Carnosine Attenuates the Development of both Type 2 Diabetes and Diabetic Nephropathy in BTBR ob/ob Mice. Sci. Rep. 2017, 7, 44492. [Google Scholar] [CrossRef]
- Tran, U.C.; Clarke, C.F. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 2007, 7, S62–S71. [Google Scholar] [CrossRef]
- Bentinger, M.; Tekle, M.; Dallner, G. Coenzyme Q–biosynthesis and functions. Biochem. Biophys. Res. Commun. 2010, 396, 74–79. [Google Scholar] [CrossRef]
- Ernster, L.; Dallner, G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim. Biophys. Acta 1995, 1271, 195–204. [Google Scholar] [CrossRef]
- Persson, M.F.; Franzen, S.; Catrina, S.B.; Dallner, G.; Hansell, P.; Brismar, K.; Palm, F. Coenzyme Q10 prevents GDP-sensitive mitochondrial uncoupling, glomerular hyperfiltration and proteinuria in kidneys from db/db mice as a model of type 2 diabetes. Diabetologia 2012, 55, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.J.; Zhang, M.D.; Zeberg, H.; Nilsson, J.; Grunler, J.; Liu, S.X.; Xiang, Q.; Persson, J.; Fried, K.J.; Catrina, S.B.; et al. Coenzyme Q10 prevents peripheral neuropathy and attenuates neuron loss in the db-/db- mouse, a type 2 diabetes model. Proc. Natl. Acad. Sci. USA 2013, 110, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Montano, S.J.; Grunler, J.; Nair, D.; Tekle, M.; Fernandes, A.P.; Hua, X.; Holmgren, A.; Brismar, K.; Ungerstedt, J.S. Glutaredoxin mediated redox effects of coenzyme Q10 treatment in type 1 and type 2 diabetes patients. BBA Clin. 2015, 4, 14–20. [Google Scholar] [CrossRef]
- Brauner, H.; Luthje, P.; Grunler, J.; Ekberg, N.R.; Dallner, G.; Brismar, K.; Brauner, A. Markers of innate immune activity in patients with type 1 and type 2 diabetes mellitus and the effect of the anti-oxidant coenzyme Q10 on inflammatory activity. Clin. Exp. Immunol. 2014, 177, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Alehagen, U.; Johansson, P.; Björnstedt, M.; Rosén, A.; Dahlström, U. Cardiovascular mortality and N-terminal-proBNP reduced after combined selenium and coenzyme Q10 supplementation: A 5-year prospective randomized double-blind placebo-controlled trial among elderly Swedish citizens. Int. J. Cardiol. 2013, 167, 1860–1866. [Google Scholar] [CrossRef]
- Abe, H.; Okuma, E.; Sekine, H.; Maeda, A.; Yoshiue, S. Human urinary excretion of L-histidine-related compounds after ingestion of several meats and fish muscle. Int. J. Biochem. 1993, 25, 1245–1249. [Google Scholar] [PubMed]
- Nierobisz, L.S.; Hentz, N.G.; Felts, J.V.; Mozdziak, P.E. Fiber phenotype and coenzyme Q10 content in Turkey skeletal muscles. Cells Tissues Organs 2010, 192, 382–394. [Google Scholar] [CrossRef]
- Boldyrev, A.A.; Aldini, G.; Derave, W. Physiology and pathophysiology of carnosine. Physiol. Rev. 2013, 93, 1803–1845. [Google Scholar] [CrossRef]
- Quinn, P.J.; Boldyrev, A.A.; Formazuyk, V.E. Carnosine: Its properties, functions and potential therapeutic applications. Mol. Aspects Med. 1992, 13, 379–444. [Google Scholar] [CrossRef]
- Babizhayev, M.A. Generation of reactive oxygen species in the anterior eye segment. Synergistic codrugs of N-acetylcarnosine lubricant eye drops and mitochondria-targeted antioxidant act as a powerful therapeutic platform for the treatment of cataracts and primary open-angle glaucoma. BBA Clin. 2016, 6, 49–68. [Google Scholar]
- Babizhayev, M.A.; Yegorov, Y.E. Telomere Attrition in Human Lens Epithelial Cells Associated with Oxidative Stress Provide a New Therapeutic Target for the Treatment, Dissolving and Prevention of Cataract with N-Acetylcarnosine Lubricant Eye Drops. Kinetic, Pharmacological and Activity-Dependent Separation of Therapeutic Targeting: Transcorneal Penetration and Delivery of L-Carnosine in the Aqueous Humor and Hormone-Like Hypothalamic Antiaging Effects of the Instilled Ophthalmic Drug Through a Safe Eye Medication Technique. Recent Pat. Drug Deliv. Formul. 2016, 10, 82–129. [Google Scholar]
- Ginsberg, H.N.; MacCallum, P.R. The obesity, metabolic syndrome, and type 2 diabetes mellitus pandemic: Part I. Increased cardiovascular disease risk and the importance of atherogenic dyslipidemia in persons with the metabolic syndrome and type 2 diabetes mellitus. J. Cardiometab. Syndr. 2009, 4, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Bentinger, M.; Savu, O.; Moshfegh, A.; Sunkari, V.; Dallner, G.; Swiezewska, E.; Catrina, S.; Brismar, K.; Tekle, M. Mono-epoxy-tocotrienol-alpha enhances wound healing in diabetic mice and stimulates in vitro angiogenesis and cell migration. J. Diabetes Complicat. 2017, 31, 4–12. [Google Scholar] [CrossRef]
- Johansen, J.S.; Harris, A.K.; Rychly, D.J.; Ergul, A. Oxidative stress and the use of antioxidants in diabetes: Linking basic science to clinical practice. Cardiovasc. Diabetol. 2005, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Schrauwen, P.; Hesselink, M.K. Oxidative capacity, lipotoxicity, and mitochondrial damage in type 2 diabetes. Diabetes 2004, 53, 1412–1417. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, M.K.; Turner, N. Mitochondrial dysfunction and insulin resistance: An update. Endocr. Connect. 2015, 4, R1–R15. [Google Scholar] [CrossRef]
- Kim, J.A.; Wei, Y.; Sowers, J.R. Role of mitochondrial dysfunction in insulin resistance. Circ. Res. 2008, 102, 401–414. [Google Scholar] [CrossRef]
- Hartman, M.L.; Shirihai, O.S.; Holbrook, M.; Xu, G.; Kocherla, M.; Shah, A.; Fetterman, J.L.; Kluge, M.A.; Frame, A.A.; Hamburg, N.M.; et al. Relation of mitochondrial oxygen consumption in peripheral blood mononuclear cells to vascular function in type 2 diabetes mellitus. Vasc. Med. 2014, 19, 67–74. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; Chandel, N.S. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 2010, 35, 505–513. [Google Scholar] [CrossRef]
- Mohammadi-Bardbori, A.; Najibi, A.; Amirzadegan, N.; Gharibi, R.; Dashti, A.; Omidi, M.; Saeedi, A.; Ghafarian-Bahreman, A.; Niknahad, H. Coenzyme Q10 remarkably improves the bio-energetic function of rat liver mitochondria treated with statins. Eur. J. Pharmacol. 2015, 762, 270–274. [Google Scholar] [CrossRef]
- Keller, R.K. The mechanism and regulation of dolichyl phosphate biosynthesis in rat liver. J. Biol. Chem. 1986, 261, 12053–12059. [Google Scholar] [CrossRef]
- Bentinger, M.; Turunen, M.; Zhang, X.X.; Wan, Y.J.; Dallner, G. Involvement of retinoid X receptor alpha in coenzyme Q metabolism. J. Mol. Biol. 2003, 326, 795–803. [Google Scholar] [CrossRef]
- Tekle, M.; Turunen, M.; Dallner, G.; Chojnacki, T.; Swiezewska, E. Investigation of coenzyme Q biosynthesis in human fibroblast and HepG2 cells. J. Biochem. Biophys. Methods 2008, 70, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Kvannes, J.; Flatmark, T. A fluorometric assay of acyl-CoA oxidase activity by a coupled peroxidatic reaction: Elimination of interfering side reactions. J. Biochem. Biophys. Methods 1991, 23, 135–149. [Google Scholar] [CrossRef]
- Bleier, L.; Wittig, I.; Heide, H.; Steger, M.; Brandt, U.; Drose, S. Generator-specific targets of mitochondrial reactive oxygen species. Free Radic. Biol. Med. 2015, 78, 1–10. [Google Scholar] [CrossRef]
- Traverso, N.; Menini, S.; Odetti, P.; Pronzato, M.A.; Cottalasso, D.; Marinari, U.M. Diabetes impairs the enzymatic disposal of 4-hydroxynonenal in rat liver. Free Radic. Biol. Med. 2002, 32, 350–359. [Google Scholar] [CrossRef]
- Sarkar, P.; Kar, K.; Mondal, M.C.; Chakraborty, I.; Kar, M. Elevated level of carbonyl compounds correlates with insulin resistance in type 2 diabetes. Ann. Acad. Med. Singap. 2010, 39, 909. [Google Scholar]
- Dikalov, S.I.; Kirilyuk, I.A.; Voinov, M.; Grigor’ev, I.A. EPR detection of cellular and mitochondrial superoxide using cyclic hydroxylamines. Free Radic. Res. 2011, 45, 417–430. [Google Scholar] [CrossRef]
- Evans, J.L.; Maddux, B.A.; Goldfine, I.D. The molecular basis for oxidative stress-induced insulin resistance. Antioxid. Redox Signal. 2005, 7, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Rosen, P.; Nawroth, P.P.; King, G.; Moller, W.; Tritschler, H.J.; Packer, L. The role of oxidative stress in the onset and progression of diabetes and its complications: A summary of a Congress Series sponsored by UNESCO-MCBN, the American Diabetes Association and the German Diabetes Society. Diabetes Metab. Res. Rev. 2001, 17, 189–212. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zhu, X.; Hu, Y.; Li, T.; Gao, Y.; Shi, Y.; Tang, S. Mitochondrial DNA oxidative damage triggering mitochondrial dysfunction and apoptosis in high glucose-induced HRECs. Investig. Ophthalmol. Vis. Sci. 2008, 49, 4203–4209. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, H.; He, Y.L.; Xie, B.; Xu, Y.F.; Tang, H.Y.; Li, M.; Hao, W.; Wang, X.; Zhang, M.; et al. Mental health system in China: Histoty, recent 210 service reform and future challenges. World Psychiatry World Psychiatry 2011, 10, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Fresta, C.G.; Fidilio, A.; O’Donnell, F.; Musso, N.; Lazzarino, G.; Grasso, M.; Amorini, A.M.; Tascedda, F.; Bucolo, C.; et al. Carnosine Decreases PMA-Induced Oxidative Stress and Inflammation in Murine Macrophages. Antioxidants 2019, 8, 281. [Google Scholar] [CrossRef]
- Aydin, A.F.; Bingul, I.; Kucukgergin, C.; Dogan-Ekici, I.; Dogru Abbasoglu, S.; Uysal, M. Carnosine decreased oxidation and glycation products in serum and liver of high-fat diet and low-dose streptozotocin-induced diabetic rats. Int. J. Exp. Pathol. 2017, 98, 278–288. [Google Scholar] [CrossRef]
- Palin, M.F.; Lapointe, J.; Gariepy, C.; Beaudry, D.; Kalbe, C. Characterisation of intracellular molecular mechanisms modulated by carnosine in porcine myoblasts under basal and oxidative stress conditions. PLoS ONE 2020, 15, e0239496. [Google Scholar] [CrossRef]
- Bentinger, M.; Brismar, K.; Dallner, G. The antioxidant role of coenzyme Q. Mitochondrion 2007, 7, S41–S50. [Google Scholar] [CrossRef]
- Sohet, F.M.; Neyrinck, A.M.; Pachikian, B.D.; de Backer, F.C.; Bindels, L.B.; Niklowitz, P.; Menke, T.; Cani, P.D.; Delzenne, N.M. Coenzyme Q10 supplementation lowers hepatic oxidative stress and inflammation associated with diet-induced obesity in mice. Biochem. Pharmacol. 2009, 78, 1391–1400. [Google Scholar] [CrossRef]
- Mezawa, M.; Takemoto, M.; Onishi, S.; Ishibashi, R.; Ishikawa, T.; Yamaga, M.; Fujimoto, M.; Okabe, E.; He, P.; Kobayashi, K.; et al. The reduced form of coenzyme Q10 improves glycemic control in patients with type 2 diabetes: An open label pilot study. BioFactors 2012, 38, 416–421. [Google Scholar] [CrossRef]
- Forsberg, E.; Xu, C.; Grunler, J.; Frostegard, J.; Tekle, M.; Brismar, K.; Karvestedt, L. Coenzyme Q10 and oxidative stress, the association with peripheral sensory neuropathy and cardiovascular disease in type 2 diabetes mellitus. J. Diabetes Complicat. 2015, 29, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Kwolek-Mirek, M.; Molon, M.; Kaszycki, P.; Zadrag-Tecza, R. L-carnosine enhanced reproductive potential of the Saccharomyces cerevisiae yeast growing on medium containing glucose as a source of carbon. Biogerontology 2016, 17, 737–747. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Macarini, J.R.; Maravai, S.G.; Cararo, J.H.; Dimer, N.W.; Goncalves, C.L.; Kist, L.W.; Bogo, M.R.; Schuck, P.F.; Streck, E.L.; Ferreira, G.C. Impairment of electron transfer chain induced by acute carnosine administration in skeletal muscle of young rats. Biomed. Res. Int. 2014, 2014, 632986. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, M. Biochemistry and molecular cell biology of diabetic complications. Nature 2001, 414, 813–820. [Google Scholar] [CrossRef]
- Soto-Urquieta, M.G.; Lopez-Briones, S.; Perez-Vazquez, V.; Saavedra-Molina, A.; Gonzalez-Hernandez, G.A.; Ramirez-Emiliano, J. Curcumin restores mitochondrial functions and decreases lipid peroxidation in liver and kidneys of diabetic db/db mice. Biol. Res. 2014, 47, 74. [Google Scholar] [CrossRef]
- Forbes, J.M.; Cooper, M.E. Mechanisms of diabetic complications. Physiol. Rev. 2013, 93, 137–188. [Google Scholar] [CrossRef]
- Tyagi, S.; Gupta, P.; Saini, A.S.; Kaushal, C.; Sharma, S. The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J. Adv. Pharm. Technol. Res. 2011, 2, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Giris, M.; Dogru-Abbasoglu, S.; Kumral, A.; Olgac, V.; Kocak-Toker, N.; Uysal, M. Effect of carnosine alone or combined with alpha-tocopherol on hepatic steatosis and oxidative stress in fructose-induced insulin-resistant rats. J. Physiol. Biochem. 2014, 70, 385–395. [Google Scholar] [CrossRef]
- Mong, M.C.; Chao, C.Y.; Yin, M.C. Histidine and carnosine alleviated hepatic steatosis in mice consumed high saturated fat diet. Eur. J. Pharmacol. 2011, 653, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Oppermann, H.; Schnabel, L.; Meixensberger, J.; Gaunitz, F. Pyruvate attenuates the anti-neoplastic effect of carnosine independently from oxidative phosphorylation. Oncotarget 2016, 7, 85848–85860. [Google Scholar] [CrossRef]
- Myers, M.G.; Cowley, M.A.; Munzberg, H. Mechanisms of leptin action and leptin resistance. Annu Rev. Physiol. 2008, 70, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Hsu, C.C.; Lin, M.H.; Liu, K.S.; Yin, M.C. Histidine and carnosine delay diabetic deterioration in mice and protect human low density lipoprotein against oxidation and glycation. Eur. J. Pharmacol. 2005, 513, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Aberg, F.; Zhang, Y.; Teclebrhan, H.; Appelkvist, E.L.; Dallner, G. Increases in tissue levels of ubiquinone in association with peroxisome proliferation. Chem. Biol. Interact. 1996, 99, 205–218. [Google Scholar] [CrossRef]
- Farsi, F.; Mohammadshahi, M.; Alavinejad, P.; Rezazadeh, A.; Zarei, M.; Engali, K.A. Functions of Coenzyme Q10 Supplementation on Liver Enzymes, Markers of Systemic Inflammation, and Adipokines in Patients Affected by Nonalcoholic Fatty Liver Disease: A Double-Blind, Placebo-Controlled, Randomized Clinical Trial. J. Am. Coll Nutr. 2016, 35, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Santos, M.A.; Juarez-Rojop, I.E.; Tovilla-Zarate, C.A.; Espinosa-Garcia, M.T.; Juarez-Oropeza, M.A.; Ramon-Frias, T.; Bermúdez-Ocaña, D.Y.; Díaz-Zagoya, J.C. Coenzyme Q10 supplementation improves metabolic parameters, liver function and mitochondrial respiration in rats with high doses of atorvastatin and a cholesterol-rich diet. Lipids Health Dis. 2014, 13, 22. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| PDSS1 (Human) | 5′-TCA TAG GCG GAA GGG ACT TGA-3′ | 5′-GGT TGT GTG ATG AAA CCG TGA T-3′ |
| PDSS1 (Mouse) | 5′-CGG TTC AGT TTG CCA GGA GAT-3′ | 5′-GCG TCC CTT TCT GTA GAT GGT-3′ |
| COQ2 (Human) | 5′CGG TTG GCA AAG CCC ATT G-3′ | 5′-GGA CGA TTG GCT GTT CTT GTA-3′ |
| COQ2 (Mouse) | 5′-ACA AGC CCA TAG GAA CCT GG-3′ | 5′-CTC CAC GCA TCA GAA TAG CTC-3′ |
| PBGD (Human) | 5′-AGG ATG GGC AAC TGT ACC-3′ | 5′-GTT TTG GCT CCT TTG CTC AG-3′ |
| PBGD (Mouse) | 5′-ACT CTG CTT CGC TGC ATT G-3′ | 5′-AGT TGC CCA TCT TTC ATC ACT G-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwank-Xu, C.; Forsberg, E.; Bentinger, M.; Zhao, A.; Ansurudeen, I.; Dallner, G.; Catrina, S.-B.; Brismar, K.; Tekle, M. L-Carnosine Stimulation of Coenzyme Q10 Biosynthesis Promotes Improved Mitochondrial Function and Decreases Hepatic Steatosis in Diabetic Conditions. Antioxidants 2021, 10, 793. https://doi.org/10.3390/antiox10050793
Schwank-Xu C, Forsberg E, Bentinger M, Zhao A, Ansurudeen I, Dallner G, Catrina S-B, Brismar K, Tekle M. L-Carnosine Stimulation of Coenzyme Q10 Biosynthesis Promotes Improved Mitochondrial Function and Decreases Hepatic Steatosis in Diabetic Conditions. Antioxidants. 2021; 10(5):793. https://doi.org/10.3390/antiox10050793
Chicago/Turabian StyleSchwank-Xu, Cheng, Elisabete Forsberg, Magnus Bentinger, Allan Zhao, Ishrath Ansurudeen, Gustav Dallner, Sergiu-Bogdan Catrina, Kerstin Brismar, and Michael Tekle. 2021. "L-Carnosine Stimulation of Coenzyme Q10 Biosynthesis Promotes Improved Mitochondrial Function and Decreases Hepatic Steatosis in Diabetic Conditions" Antioxidants 10, no. 5: 793. https://doi.org/10.3390/antiox10050793
APA StyleSchwank-Xu, C., Forsberg, E., Bentinger, M., Zhao, A., Ansurudeen, I., Dallner, G., Catrina, S.-B., Brismar, K., & Tekle, M. (2021). L-Carnosine Stimulation of Coenzyme Q10 Biosynthesis Promotes Improved Mitochondrial Function and Decreases Hepatic Steatosis in Diabetic Conditions. Antioxidants, 10(5), 793. https://doi.org/10.3390/antiox10050793






