A Combinatorial Protein Microarray for Probing Materials Interaction with Pancreatic Islet Cell Populations
Abstract
:1. Introduction
2. Materials and Methods
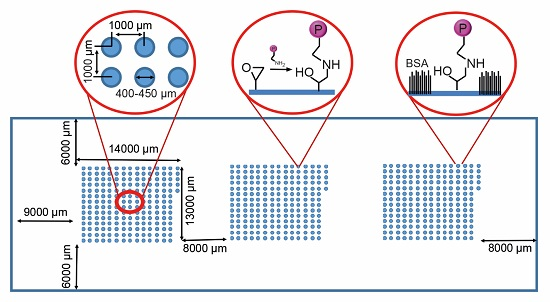
2.1. Protein Microarray Fabrication
2.2. Insulinoma Cell Culture
2.3. Primary Human Islet Culture
2.4. Primary Mouse Islet Isolation and Culture
2.5. Fluorescence Microscopy and Cell Viability
2.6. Immunocytochemistry
3. Results
3.1. Protein Microarray Fabrication
3.2. Profiling Cell Adhesion to Different Protein Combinations
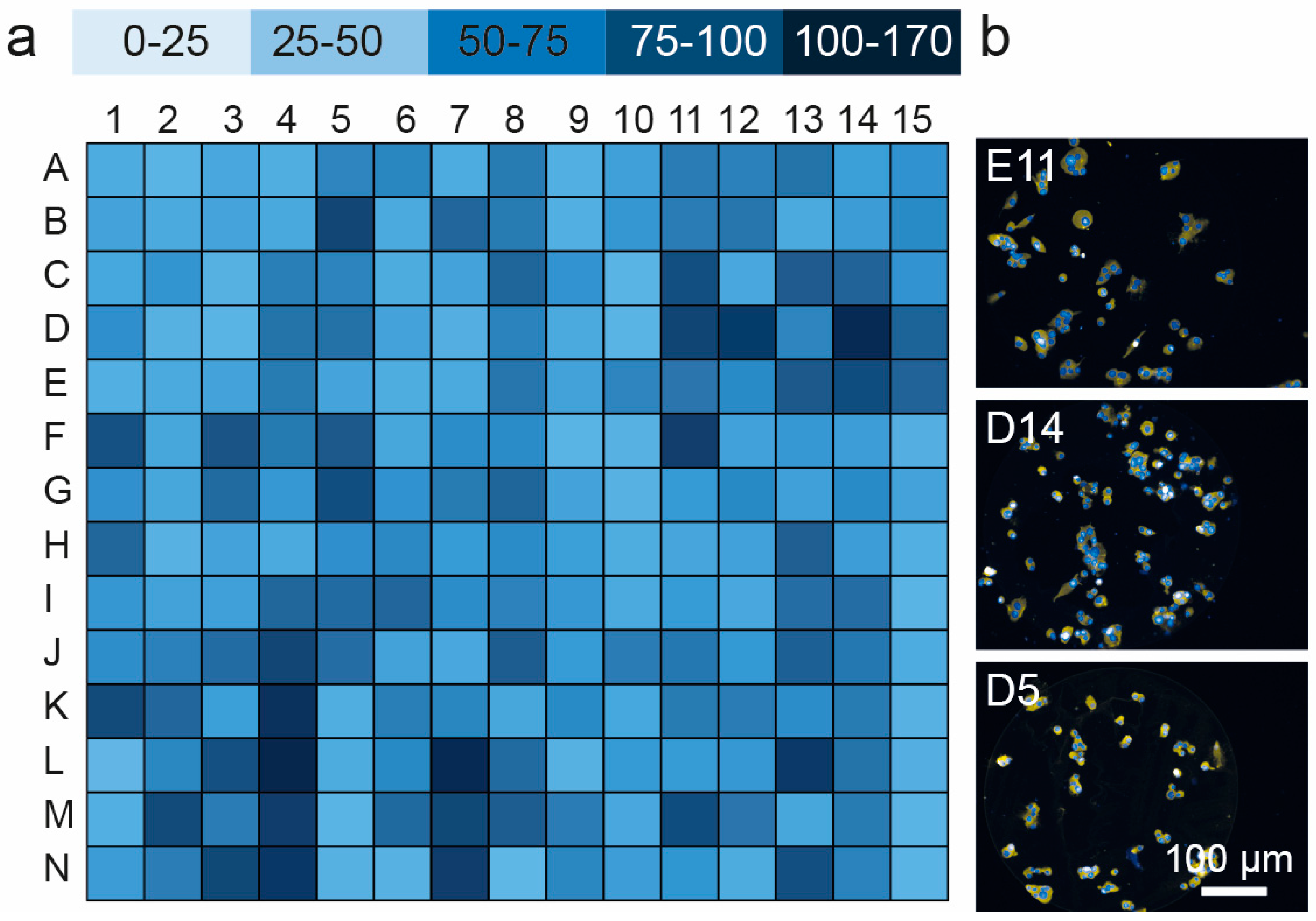
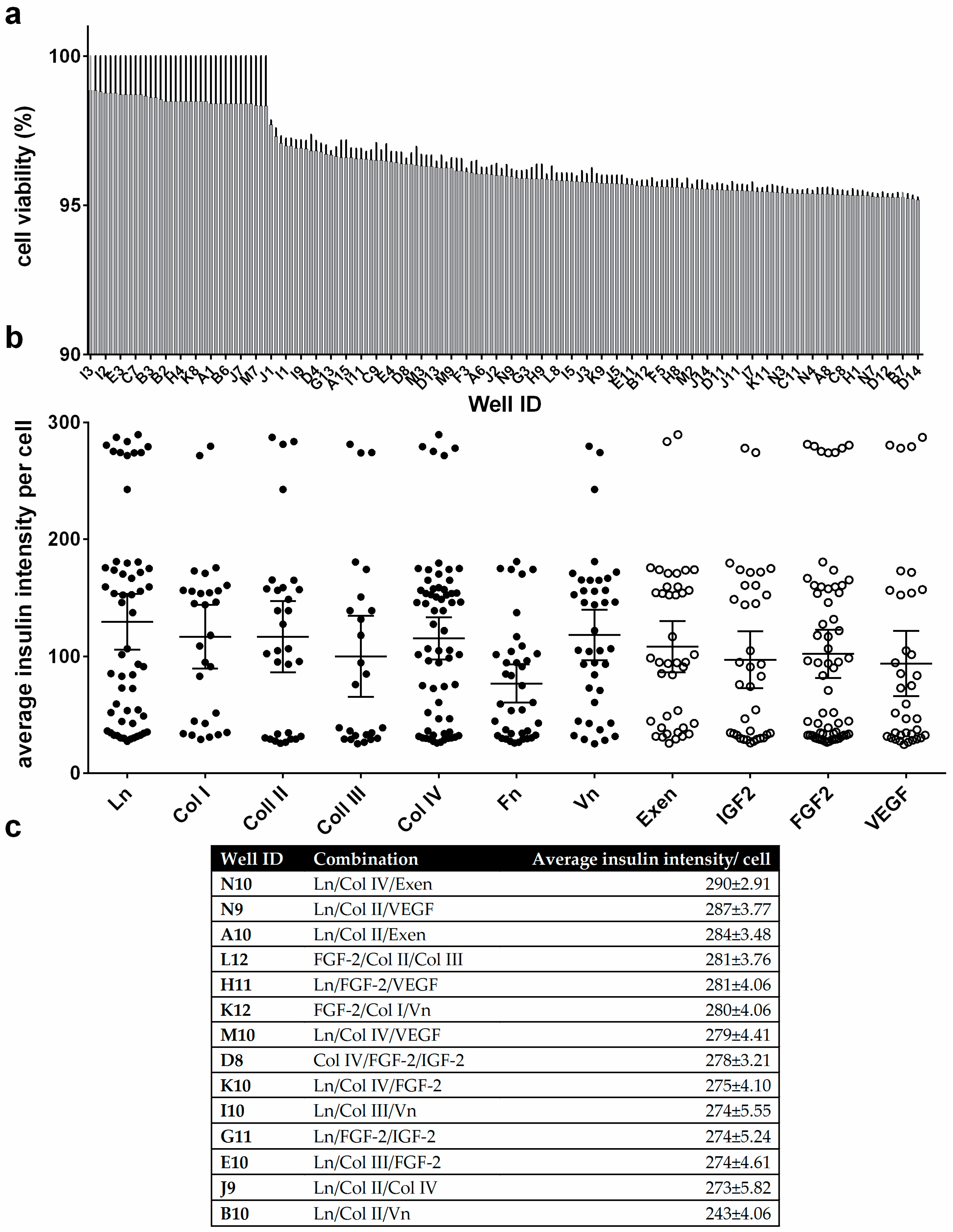
3.3. Cell Viability and Insulin Synthesis by MIN6 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ECM | Extracellular matrix |
| HMDSO | Hexamethyldisiloxane |
| AGE | Allyl glycidyl ether |
| Col I | Type I collagen |
| Col II | Type II collagen |
| Col III | Type III collagen |
| Col IV | Type IV collagen |
| Ln | Laminin 111 |
| Fn | Fibronectin |
| FGF-2 | Fibroblast growth factor 2 |
| IGF-2 | Insulin-like growth factor 2 |
| VEGF | Vascular endothelial growth factor |
| Exen | Exenatide |
| Vn | Vitronectin |
| TRITC | Tetramethylrhodamine |
References
- NCDRF Collaboration. Worldwide trends in diabetes since 1980: A pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 2016, 387, 1513–1530. [Google Scholar]
- Roep, B.O.; Peakman, M. Antigen targets of type 1 diabetes autoimmunity. Cold Spring Harb. Perspect. Med. 2012, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, T.A. Pancreatic β-cell loss and preservation in type 2 diabetes. Clin. Ther. 2003, 25, B32–B46. [Google Scholar] [CrossRef]
- Duran-Salgado, M.B.; Rubio-Guerra, A.F. Diabetic nephropathy and inflammation. World J. Diabetes 2014, 5, 393–398. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37, S81. [Google Scholar] [CrossRef]
- Goland, R.; Egli, D. Stem cell-derived β cells for treatment of type 1 diabetes? EBioMedicine 2014, 1, 93–94. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.J.; Ryan, E.A.; Lakey, J.R. Islet cell transplantation. Lancet 2001, 358, S21. [Google Scholar] [CrossRef]
- Halban, P.A.; German, M.S.; Kahn, S.E.; Weir, G.C. Current status of islet cell replacement and regeneration therapy. J. Clin. Endocrinol. Metab. 2010, 95, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Barton, F.B.; Rickels, M.R.; Alejandro, R.; Hering, B.J.; Wease, S.; Naziruddin, B.; Oberholzer, J.; Odorico, J.S.; Garfinkel, M.R.; Levy, M. Improvement in outcomes of clinical islet transplantation: 1999–2010. Diabetes Care 2012, 35, 1436–1445. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, G.D.; Fischbach, R.L. Stem cells: Science, policy, and ethics. J. Clin. Investig. 2004, 114, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Vogt, C.; Leong, K.W.; Zhao, F. Highly aligned nanofibrous scaffold derived from decellularized human fibroblasts. Adv. Funct. Mater. 2014, 24, 3027–3035. [Google Scholar] [CrossRef] [PubMed]
- Riopel, M.; Wang, R. Collagen matrix support of pancreatic islet survival and function. Front. Biosci. 2014, 19. [Google Scholar] [CrossRef]
- Mirmalek-Sani, S.-H.; Orlando, G.; McQuilling, J.P.; Pareta, R.; Mack, D.L.; Salvatori, M.; Farney, A.C.; Stratta, R.J.; Atala, A.; Opara, E.C. Porcine pancreas extracellular matrix as a platform for endocrine pancreas bioengineering. Biomaterials 2013, 34, 5488–5495. [Google Scholar] [CrossRef] [PubMed]
- Goh, S.-K.; Bertera, S.; Olsen, P.; Candiello, J.E.; Halfter, W.; Uechi, G.; Balasubramani, M.; Johnson, S.A.; Sicari, B.M.; Kollar, E. Perfusion-decellularized pancreas as a natural 3D scaffold for pancreatic tissue and whole organ engineering. Biomaterials 2013, 34, 6760–6772. [Google Scholar] [CrossRef] [PubMed]
- Roca-Cusachs, P.; Sunyer, R.; Trepat, X. Mechanical guidance of cell migration: Lessons from chemotaxis. Curr. Opin. Cell Biol. 2013, 25, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Llacua, A.; de Haan, B.J.; Smink, S.A.; Smink, A.M.; de Vos, P. Extracellular matrix components supporting human islet function in alginate based immunoprotective microcapsules for treatment of diabetes. J. Biomed. Mater. Res. A 2016, 104, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Stendahl, J.C.; Kaufman, D.B.; Stupp, S.I. Extracellular matrix in pancreatic islets: Relevance to scaffold design and transplantation. Cell Transplant. 2009, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bogdani, M.; Korpos, E.; Simeonovic, C.J.; Parish, C.R.; Sorokin, L.; Wight, T.N. Extracellular matrix components in the pathogenesis of type 1 diabetes. Curr. Diabetes Rep. 2014, 14, 552–552. [Google Scholar] [CrossRef] [PubMed]
- Weber, L.M.; Hayda, K.N.; Anseth, K.S. Cell-matrix interactions improve β-cell survival and insulin secretion in three-dimensional culture. Tissue Eng. Part. A 2008, 14, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Saha, K.; Bogatyrev, S.R.; Yang, J.; Hook, A.L.; Kalcioglu, Z.I.; Cho, S.-W.; Mitalipova, M.; Pyzocha, N.; Rojas, F.; et al. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat. Mater. 2010, 9, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Hook, A.L.; Thissen, H.; Voelcker, N.H. Surface manipulation of biomolecules for cell microarray applications. Trends Biotechnol. 2006, 24, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Hook, A.L.; Anderson, D.G.; Langer, R.; Williams, P.; Davies, M.C.; Alexander, M.R. High throughput methods applied in biomaterial development and discovery. Biomaterials 2010, 31, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Hook, A.L.; Thissen, H.; Voelcker, N.H. Advanced substrate fabrication for cell microarrays. Biomacromolecules 2009, 10, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Anglin, E.; Davey, R.; Herrid, M.; Hope, S.; Kurkuri, M.; Pasic, P.; Hor, M.; Fenech, M.; Thissen, H.; Voelcker, N.H. Cell microarrays for the screening of factors that allow the enrichment of bovine testicular cells. Cytom. A 2010, 77, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Kurkuri, M.D.; Driever, C.; Johnson, G.; McFarland, G.; Thissen, H.; Voelcker, N.H. Multifunctional polymer coatings for cell microarray applications. Biomacromolecules 2009, 10, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Coad, B.R.; Jasieniak, M.; Griesser, S.S.; Griesser, H.J. Controlled covalent surface immobilisation of proteins and peptides using plasma methods. Surf. Coat. Tech. 2013, 233, 169–177. [Google Scholar] [CrossRef]
- Rasi Ghaemi, S.; Harding, F.; Delalat, B.; Vasani, R.; Voelcker, N.H. Surface engineering for long-term culturing of mesenchymal stem cell microarrays. Biomacromolecules 2013, 14, 2675–2683. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, G.; Jabs, N.; Konstantinova, I.; Domogatskaya, A.; Tryggvason, K.; Sorokin, L.; Fässler, R.; Gu, G.; Gerber, H.-P.; Ferrara, N.; et al. The vascular basement membrane: A niche for insulin gene expression and β cell proliferation. Dev. Cell 2006, 10, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Daoud, J.T.; Petropavlovskaia, M.S.; Patapas, J.M.; Degrandpre, C.E.; Diraddo, R.W.; Rosenberg, L.; Tabrizian, M. Long-term in vitro human pancreatic islet culture using three-dimensional microfabricated scaffolds. Biomaterials 2011, 32, 1536–1542. [Google Scholar] [CrossRef] [PubMed]
- Daoud, J.; Petropavlovskaia, M.; Rosenberg, L.; Tabrizian, M. The effect of extracellular matrix components on the preservation of human islet function in vitro. Biomaterials 2010, 31, 1676–1682. [Google Scholar] [CrossRef] [PubMed]
- Durr, J.; Goodman, S.; Potocnik, A.; von der Mark, H.; von der Mark, K. Localization of β 1-integrins in human cartilage and their role in chondrocyte adhesion to collagen and fibronectin. Exp. Cell Res. 1993, 207, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Hardikar, A.A.; Marcus-Samuels, B.; Geras-Raaka, E.; Raaka, B.M.; Gershengorn, M.C. Human pancreatic precursor cells secrete FGF2 to stimulate clustering into hormone-expressing islet-like cell aggregates. Proc. Natl. Acad. Sci. USA 2003, 100, 7117–7122. [Google Scholar] [CrossRef] [PubMed]
- Inchovska, M.; Ogneva, V.; Martinova, Y. Role of FGF1, FGF2 and FGF7 in the development of the pancreas from control and streptozotocin-treated hamsters. Cell Prolif. 2006, 39, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Irving-Rodgers, H.F.; Choong, F.J.; Hummitzsch, K.; Parish, C.R.; Rodgers, R.J.; Simeonovic, C.J. Pancreatic islet basement membrane loss and remodeling after mouse islet isolation and transplantation: Impact for allograft rejection. Cell Transplant. 2014, 23, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Rosenberg, L. Maintenance of β-cell function and survival following islet isolation requires re-establishment of the islet-matrix relationship. J. Endocrinol. 1999, 163, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Flaim, C.J.; Teng, D.; Chien, S.; Bhatia, S.N. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev. 2008, 17, 29–40. [Google Scholar] [CrossRef] [PubMed]
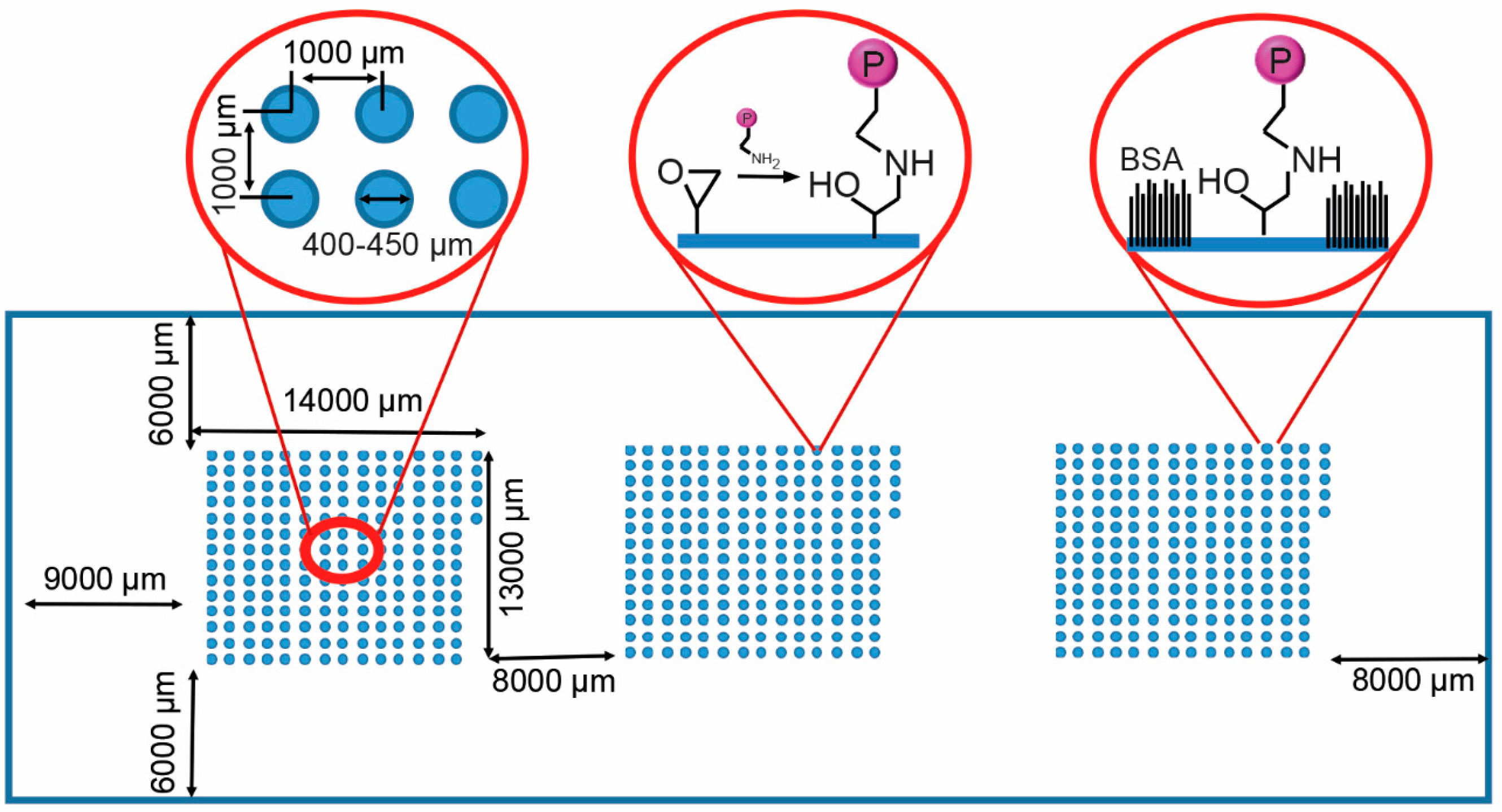
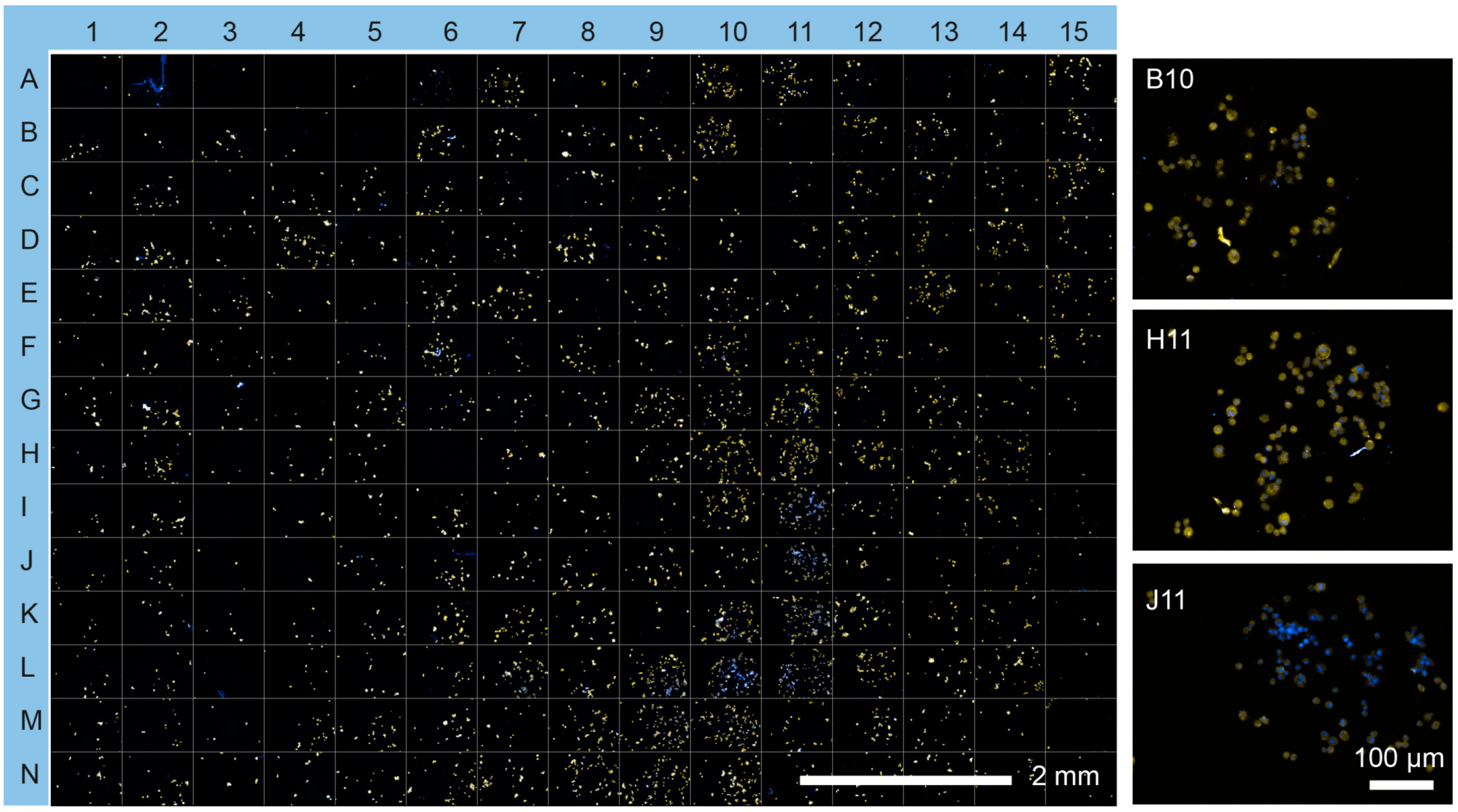
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Col I | Col I/ Ln | Col II/ Exen | Col IV/ VEGF | FGF-2/ GF-2 | Col IV/ Col I/ FGF-2 | Col IV/ Col III/ Fn | Col IV/ Fn/ VEGF | Ln/ Col I/ Col III | Ln/ Col II/ Exen | Ln/ Col IV/ Vn | Ln/ VEGF/ Vn | FGF-2/ Col II/ Fn | FGF-2/ Col IV/ IGF-2 | Col IV/ IGF-2/ Exen |
| B | Col II | Col I/ Fn | Col II/ /Vn | Col IV/ Exen | FGF-2 / VEGF | Col IV/ Col I/ IGF-2 | Col IV/ Col III/ FGF-2 | Col IV/ Fn/ Exen | Ln/ Col I/ Col IV | Ln/ Col II/ Vn | Ln/ Fn/ FGF-2 | Ln/ Exen/ Vn | FGF-2/ Col II/ IGF-2 | FGF-2/ Col IV/ VEGF | Col IV/ IGF-2/ Vn |
| C | Col III | Col I/ FGF-2 | Col III/ Col IV | Col IV/ Vn | FGF-2/ Exen | Col IV/ Col I/ VEGF | Col IV/ Col III/ IGF-2 | Col IV/ Fn/ Vn | Ln/ Col I / Fn | Ln/ Col III/ Col IV | Ln/ Fn / IGF-2 | FGF-2/ Col I/ Col II | FGF-2/ Col II/ VEGF | FGF-2/ Col IV/ Exen | Col IV/ VEGF/ Exen |
| D | Col IV | Col I/ IGF-2 | Col III/ Ln | Ln/ Fn | FGF-2/ Vn | Col IV/ Col I/ Exen | Col IV/ Col III/ VEGF | Col IV/ FGF-2/ IGF-2 | Ln/ Col I/ FGF-2 | Ln/ Col III/ Fn | Ln/ Fn VEGF | FGF-2/ Col I/ Col III | FGF-2/ Col II/ Exen | FGF-2/ Col IV/ Vn | Col IV/ VEGF/ Vn |
| E | Ln | Col I/ VEGF | Col III/ Fn | Ln/ FGF-2 | IGF-2/ VEGF | Col IV/ Col I/ Vn | Col IV/ Col III/ Exen | Col IV/ FGF-2/ VEGF | Ln/ Col I / IGF-2 | Ln/ Col III/ FGF-2 | Ln/ Fn/ Exen | FGF-2/ Col I/ Col IV | FGF-2/ Col II/ Vn | FGF-2/ L / Fn | Col IV/ Exen/ Vn |
| F | Fn | Col I/ Exen | Col III/ FGF-2 | Ln/ IGF-2 | IGF-2/ Exen | Col IV/ Col II/ Col III | Col IV/ Col III/ Vn | Col IV/ FGF-2/ Exen | Ln/ Col I/ VEGF | Ln/ Col III/ IGF-2 | Ln/ Fn/ Vn | FGF-2/ Col I/ Ln | FGF-2/ Col III/ Col IV | FGF-2/ Ln/ IGF-2 | |
| G | FGF-2 | Col I/ Vn | Col III/ IGF-2 | Ln/ VEGF | IGF-2/ Vn | Col IV/ Col II/ Ln | Col IV/ Ln/ Fn | Col IV/ FGF-2/ Vn | Ln/ Col I/ Exen | Ln/ Col III/ VEGF | Ln/ FGF-2/ IGF-2 | FGF-2/ Col I/ Fn | FGF-2/ Col III/ Ln | FGF-2/ Ln/ VEGF | |
| H | IGF-2 | Col II/ Col III | Col III/ VEGF | Ln/ Exen | VEGF/ Exen | Col IV/ Col II/ Fn | Col IV/ Ln/ FGF-2 | Col I/ IGF-2/ VEGF | Ln/ Col I/ Vn | Ln/ Col III/ Exen | Ln/ FGF-2/ VEGF | FGF-2/ Col I/ IGF-2 | FGF-2/ Col III/ Fn | FGF-2/ Ln/ Exen | |
| I | VEGF | Col II/ Col IV | Col III/ Exen | Ln/ Vn | VEGF/ Vn | Col IV/ Col II/ FGF-2 | Col IV/ Ln/ IGF-2 | Col I/ IGF-2/ Exen | Ln/ Col II/ Col III | Ln/ Col III/ Vn | Ln/ FGF-2/ Exen | FGF-2/ Col I/ VEGF | FGF-2/ Col III/ IGF-2 | FGF-2/ Ln / Vn | |
| J | Exen | Col II/ Ln | Col III/ Vn | Fn/ FGF-2 | Exen/ Vn | Col IV/ Col II/ IGF-2 | Col IV/ Ln/ VEGF | Col I/ IGF-2/ Vn | Ln/ Col II/ Col IV | Ln/ Col IV/ Fn | Ln/ FGF-2/ Vn | FGF-2/ Col I/ Exen | FGF-2/ Col III/ VEGF | FGF-2/ Fn/ IGF-2 | |
| K | Vn | Col II/ Fn | Col IV/ Ln | Fn/ IGF-2 | Col IV/ Col I Col II | Col IV/ Col II/ VEGF | Col IV/ Ln/ Exen | Col IV/ EGF/ Exen | Ln/ Col II/ Fn | Ln/ Col IV/ FGF-2 | Ln/ IGF-2/ VEGF | FGF-2/ Col I/ Vn | FGF-2/ Col III/ Exen | FGF-2/ Fn VEGF | |
| L | Col I/ Col II | Col II/ FGF-2 | Col IV/ Fn | Fn/ VEGF | Col IV/ Col I/ Col III | Col IV/ Col II/ Exen | Col IV/ Ln/ Vn | Col I/ VEGF/ Vn | Ln/ Col II/ FGF-2 | Ln// Col IV/ IGF-2 | Ln/ IGF-2/ Exen | FGF-2/ Col II/ Col III | FGF-2/ Col III/ Vn | FGF-2/ Fn/ Exen | |
| M | Col I/ Col III | Col II/ IGF-2 | Col IV/ FGF-2 | Fn/ Exen | Col IV/ Col I/ Ln | Col IV/ Col II/ Vn | Col IV/ Fn/ FGF-2 | Col I/ Exen/ Vn | Ln/ Col II / IGF-2 | Ln/ Col IV/ VEGF | Ln/ IGF-2/ Vn | FGF-2/ Col II/ Col IV | FGF-2/ Col IV/ Ln | FGF-2/ Fn/ Vn | |
| N | Col I/ Col IV | Col II/ VEGF | Col I/ IGF-2 | Fn/ Vn | Col IV/ Col I/ Fn | Col IV/ Col III/ Ln | Col IV/ Fn/ IGF-2 | Ln/ Col I/ Col II | Ln/ Col II/ VEGF | Ln/ Col IV/ Exen | Ln/ VEGF/ Exen | FGF-2/ Col II/ Ln | FGF-2/ Col IV Fn | Col IV/ IGF-2/ VEGF |
| Well ID | Formulation | Cells Attached Per Spot |
|---|---|---|
| D14 | FGF-2/Col IV/Vn | 168 ± 11.7 |
| D11 | Ln/Fn/VEGF | 154 ± 6.39 |
| N7 | Col IV/Fn/IGF-2 | 153 ± 12.4 |
| D12 | FGF-2/Col I/Col III | 152 ± 10.0 |
| L7 | Col IV/Ln/Vn | 152 ± 10.5 |
| L13 | FGF-2/Col III/Vn | 151 ± 9.82 |
| F11 | Ln/Fn/Vn | 151 ± 11.0 |
| L4 | Fn/VEGF | 145 ± 21.5 |
| N4 | Fn/Vn | 138 ± 11.3 |
| K4 | Fn/IGF-2 | 131 ± 16.6 |
| M11 | Ln/IGF-2/Vn | 126 ± 7.63 |
| E14 | FGF-2/Ln/Fn | 125 ± 22.1 |
| F5 | IGF-2/Exen | 125 ± 23.7 |
| B5 | FGF-2/VEGF | 122 ± 19.1 |
| J4 | Fn/FGF-2 | 120 ± 9.91 |
| N13 | FGF-2/Col IV/Fn | 117 ± 7.94 |
| K1 | Vn | 115 ± 7.94 |
| M2 | Col II/IGF-2 | 113 ± 11.3 |
| E13 | FGF-2/Col II/Vn | 113 ± 22.9 |
| C13 | FGF-2/Col II/VEGF | 112 ± 11.72 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delalat, B.; Rojas-Canales, D.M.; Rasi Ghaemi, S.; Waibel, M.; Harding, F.J.; Penko, D.; Drogemuller, C.J.; Loudovaris, T.; Coates, P.T.H.; Voelcker, N.H. A Combinatorial Protein Microarray for Probing Materials Interaction with Pancreatic Islet Cell Populations. Microarrays 2016, 5, 21. https://doi.org/10.3390/microarrays5030021
Delalat B, Rojas-Canales DM, Rasi Ghaemi S, Waibel M, Harding FJ, Penko D, Drogemuller CJ, Loudovaris T, Coates PTH, Voelcker NH. A Combinatorial Protein Microarray for Probing Materials Interaction with Pancreatic Islet Cell Populations. Microarrays. 2016; 5(3):21. https://doi.org/10.3390/microarrays5030021
Chicago/Turabian StyleDelalat, Bahman, Darling M. Rojas-Canales, Soraya Rasi Ghaemi, Michaela Waibel, Frances J. Harding, Daniella Penko, Christopher J. Drogemuller, Thomas Loudovaris, Patrick T. H. Coates, and Nicolas H. Voelcker. 2016. "A Combinatorial Protein Microarray for Probing Materials Interaction with Pancreatic Islet Cell Populations" Microarrays 5, no. 3: 21. https://doi.org/10.3390/microarrays5030021
APA StyleDelalat, B., Rojas-Canales, D. M., Rasi Ghaemi, S., Waibel, M., Harding, F. J., Penko, D., Drogemuller, C. J., Loudovaris, T., Coates, P. T. H., & Voelcker, N. H. (2016). A Combinatorial Protein Microarray for Probing Materials Interaction with Pancreatic Islet Cell Populations. Microarrays, 5(3), 21. https://doi.org/10.3390/microarrays5030021