Selection of Olduvai Domains during Evolution: A Role for Primate-Specific Splicing Super-Enhancer and RNA Guanine Quadruplex in Bipartite NBPF Exons
Abstract
1. Introduction
2. Hypothesis
2.1. Giant Exonic Splicing Enhancers Created by an Intra-Exon Duplication of a Purine-Rich Motif in the Expanded HLS Exons
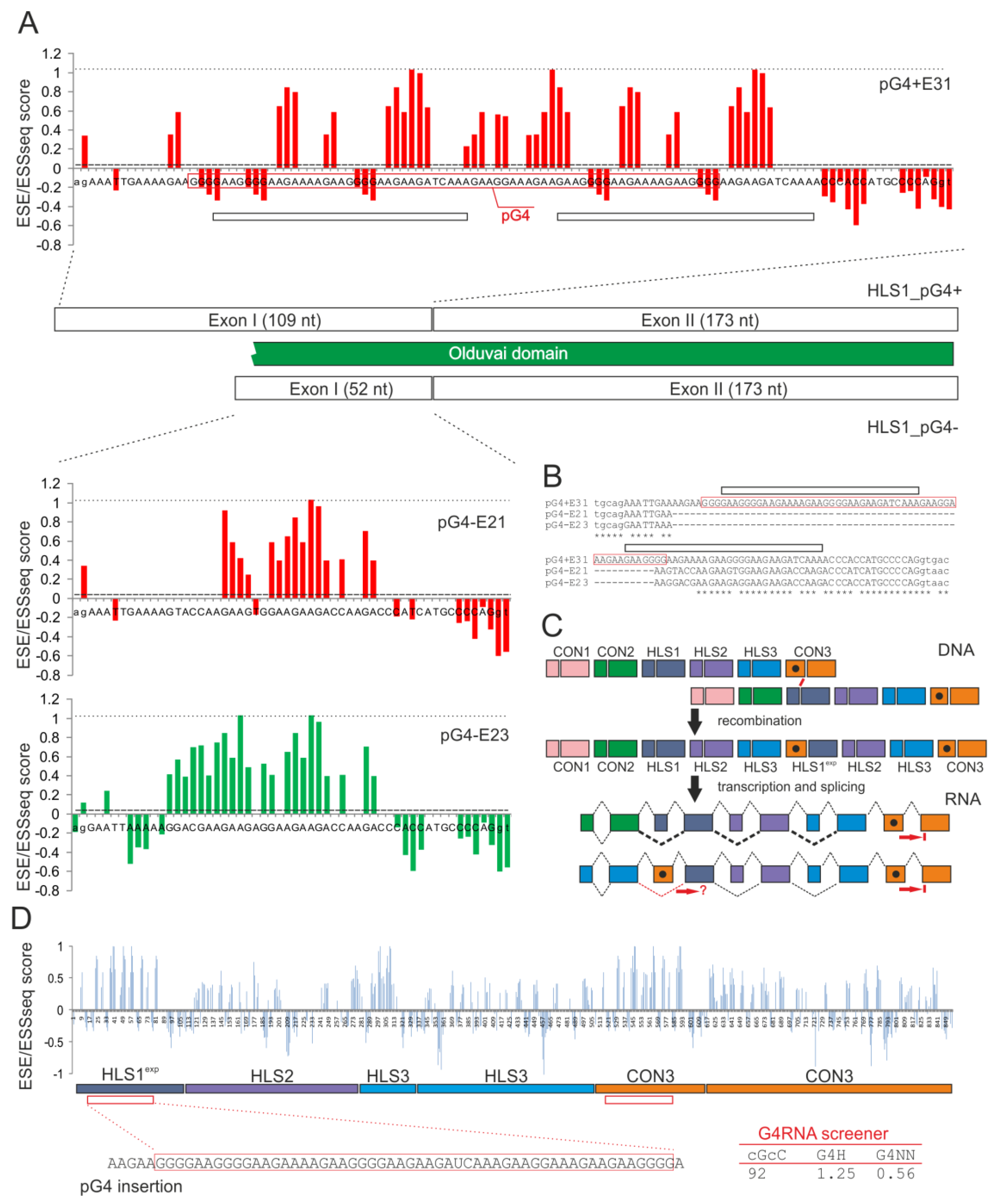
2.2. Intragenic Duplications of Olduvai Exon Doublets and Their Possible Impact on Alternative Splicing
2.3. Is Access to the Super-Enhancer in the First HLS1 Exons Regulated by RNA G4 Formation?
2.4. Conclusions
3. Material and Methods
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Popesco, M.C.; MacLaren, E.J.; Hopkins, J.; Dumas, L.; Cox, M.; Meltesen, L.; McGavran, L.; Wyckoff, G.J.; Sikela, J.M. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science 2006, 313, 1304–1307. [Google Scholar] [CrossRef] [PubMed]
- O’Bleness, M.S.; Dickens, C.M.; Dumas, L.; Kehrer-Sawatzki, H.; Wyckoff, G.J.; Sikela, J.M. Evolutionary history and genome organization of DUF1220 protein domains. G3 2012, 2, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, B.; Montgomery, S.H. Phylogenetic analysis supports a link between DUF1220 domain number and primate brain expansion. Genome Biol. Evol. 2015, 7, 2083–2088. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.M.; Searles, V.B.; Anderson, N.; Keeney, J.; Dumas, L.; Sikela, J.M. DUF1220 dosage is linearly associated with increasing severity of the three primary symptoms of autism. PLoS Genet. 2014, 10, e1004241. [Google Scholar] [CrossRef]
- Davis, J.M.; Searles Quick, V.B.; Sikela, J.M. Replicated linear association between DUF1220 copy number and severity of social impairment in autism. Hum. Genet. 2015, 134, 569–575. [Google Scholar] [CrossRef]
- Heft, I.E.; Mostovoy, Y.; Levy-Sakin, M.; Ma, W.; Stevens, A.J.; Pastor, S.; McCaffrey, J.; Boffelli, D.; Martin, D.I.; Xiao, M.; et al. The driver of extreme human-specific Olduvai repeat expansion remains highly active in the human genome. Genetics 2020, 214, 179–191. [Google Scholar] [CrossRef]
- Vandepoele, K.; Van Roy, N.; Staes, K.; Speleman, F.; van Roy, F. A novel gene family NBPF: Intricate structure generated by gene duplications during primate evolution. Mol. Biol. Evol. 2005, 22, 2265–2274. [Google Scholar] [CrossRef]
- Ke, S.; Shang, S.; Kalachikov, S.M.; Morozova, I.; Yu, L.; Russo, J.J.; Ju, J.; Chasin, L.A. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res. 2011, 21, 1360–1374. [Google Scholar] [CrossRef]
- Pengelly, R.; Bakhtiar, D.; Borovská, I.; Královičová, J.; Vořechovský, I. Exonic splicing code and protein binding sites for calcium. Nucleic Acids Res. 2022, 50, 5493–5512. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell. Biol. 2015, 16, 665–677. [Google Scholar] [CrossRef]
- Garant, J.-M.; Perreault, J.-P.; Scott, M.S. Motif independent identification of potential RNA G-quadruplexes by G4RNA screener. Bioinformatics 2017, 33, 3532–3537. [Google Scholar] [CrossRef] [PubMed]
- Fay, M.M.; Lyons, S.M.; Ivanov, P. RNA G-quadruplexes in biology: Principles and molecular mechanisms. J. Mol. Biol. 2017, 429, 2127–2147. [Google Scholar] [CrossRef] [PubMed]
- Královičová, J.; Vořechovský, I. Global control of aberrant splice site activation by auxiliary splicing sequences: Evidence for a gradient in exon and intron definition. Nucleic Acids Res. 2007, 35, 6399–6413. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.; Buerer, L.; Rhine, C.L.; Wang, J.; Cygan, K.J.; Fairbrother, W.G. Mutational bias and the protein code shape the evolution of splicing enhancers. Nat. Comm. 2020, 11, 2845. [Google Scholar] [CrossRef]
- Královičová, J.; Borovská, I.; Pengelly, R.; Lee, E.; Abaffy, P.; Šindelka, R.; Grutzner, F.; Vořechovský, I. Restriction of an intron size en route to endothermy. Nucleic Acids Res. 2021, 49, 2460–2487. [Google Scholar] [CrossRef] [PubMed]
- Dominski, Z.; Kole, R. Selection of splice sites in pre-mRNAs with short internal exons. Mol. Cell. Biol. 1991, 11, 6075–6083. [Google Scholar] [PubMed]
- Sterner, D.A.; Berget, S.M. In vivo recognition of a vertebrate mini-exon as an exon-intron-exon unit. Mol. Cell. Biol. 1993, 13, 2677–2687. [Google Scholar]
- Sterner, D.A.; Carlo, T.; Berget, S.M. Architectural limits on split genes. Proc. Natl. Acad. Sci. USA 1996, 93, 15081–15085. [Google Scholar] [CrossRef]
- Crabb, T.L.; Lam, B.J.; Hertel, K.J. Retention of spliceosomal components along ligated exons ensures efficient removal of multiple introns. RNA 2010, 16, 1786–1796. [Google Scholar] [CrossRef]
- Vecellio Reane, D.; Vallese, F.; Checchetto, V.; Acquasaliente, L.; Butera, G.; De Filippis, V.; Szabo, I.; Zanotti, G.; Rizzuto, R.; Raffaello, A. A MICU1 splice variant confers high sensitivity to the mitochondrial Ca2+ uptake machinery of skeletal muscle. Mol. Cell 2016, 64, 760–773. [Google Scholar] [CrossRef]
- Knebelmann, B.; Breillat, C.; Forestier, L.; Arrondel, C.; Jacassier, D.; Giatras, I.; Drouot, L.; Deschenes, G.; Grunfeld, J.P.; Broyer, M.; et al. Spectrum of mutations in the COL4A5 collagen gene in X-linked Alport syndrome. Am. J. Hum. Genet. 1996, 59, 1221–1232. [Google Scholar] [PubMed]
- Haire, R.N.; Ohta, Y.; Strong, S.J.; Litman, R.T.; Liu, Y.; Prchal, J.T.; Cooper, M.D.; Litman, G.W. Unusual patterns of exon skipping in Bruton tyrosine kinase are associated with mutations involving the intron 17 3’ splice site. Am. J. Hum. Genet. 1997, 60, 798–807. [Google Scholar] [PubMed]
- Schwarze, U.; Starman, B.J.; Byers, P.H. Redefinition of exon 7 in the COL1A1 gene of type I collagen by an intron 8 splice-donor-site mutation in a form of osteogenesis imperfecta: Influence of intron splice order on outcome of splice-site mutation. Am. J. Hum. Genet. 1999, 65, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Taggart, A.J.; Heintzelman, C.; Cygan, K.J.; Hull, C.G.; Wang, J.; Shrestha, B.; Fairbrother, W.G. Widespread intra-dependencies in the removal of introns from human transcripts. Nucleic Acids Res. 2017, 45, 9503–9513. [Google Scholar] [CrossRef]
- Baralle, M.; Skoko, N.; Knezevich, A.; De Conti, L.; Motti, D.; Bhuvanagiri, M.; Baralle, D.; Buratti, E.; Baralle, F.E. NF1 mRNA biogenesis: Effect of the genomic milieu in splicing regulation of the NF1 exon 37 region. FEBS Lett. 2006, 580, 4449–4456. [Google Scholar] [CrossRef]
- Hershkovitz, D.; Mandel, H.; Ishida-Yamamoto, A.; Chefetz, I.; Hino, B.; Luder, A.; Indelman, M.; Bergman, R.; Sprecher, E. Defective lamellar granule secretion in arthrogryposis, renal dysfunction, and cholestasis syndrome caused by a mutation in VPS33B. Arch. Dermatol. 2008, 144, 334–340. [Google Scholar] [CrossRef]
- Vandepoele, K.; van Roy, F. Insertion of an HERV(K) LTR in the intron of NBPF3 is not required for its transcriptional activity. Virology 2007, 362, 1–5. [Google Scholar] [CrossRef]
- Qin, Z.; Zhang, X. The identification of switch-like alternative splicing exons among mulitple samples with RNA-Seq data. PLoS ONE 2017, 12, e0178320. [Google Scholar] [CrossRef]
- Hale, M.A.; Johnson, N.A.; Berglund, J.A. Repeat-associated RNA structure and aberrant splicing. BBA Gene Regul. Mech. 2019, 1862, 194405. [Google Scholar] [CrossRef]
- Sznajder, L.J.; Thomas, J.D.; Carrell, E.M.; Reid, T.; McFarland, K.N.; Cleary, J.D.; Oliveira, R.; Nutter, C.A.; Bhatt, K.; Sobczak, K.; et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc. Natl. Acad. Sci. USA 2018, 115, 4234–4239. [Google Scholar] [CrossRef]
- Didiot, M.C.; Tian, Z.; Schaeffer, C.; Subramanian, M.; Mandel, J.L.; Moine, H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008, 36, 4902–4912. [Google Scholar] [CrossRef] [PubMed]
- Carlo, T.; Sierra, R.; Berget, S.M. A 5’ splice site-proximal enhancer binds SF1 and activates exon bridging of a microexon. Mol. Cell. Biol. 2000, 20, 3988–3995. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Královičová, J.; Vořechovský, I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J. Immunol. 2006, 176, 2381–2388. [Google Scholar] [CrossRef] [PubMed]
- Královičová, J.; Lages, A.; Patel, A.; Dhir, A.; Buratti, E.; Searle, M.S.; Vořechovský, I. Optimal antisense target reducing INS intron 1 retention is adjacent to a parallel G quadruplex. Nucleic Acids Res. 2014, 42, 8161–8173. [Google Scholar] [CrossRef]
- Pinnavaia, T.J.; Miles, H.T.; Becker, E.D. Self-assembled 5’-guanosine monophosphate, nuclear magnetic resonance evidence for a regular, ordered structure and slow chemical exchange. J. Am. Chem. Soc. 1975, 97, 7198–7200. [Google Scholar] [CrossRef]
- Guiset Miserachs, H.; Donghi, D.; Borner, R.; Johannsen, S.; Sigel, R.K. Distinct differences in metal ion specificity of RNA and DNA G-quadruplexes. J. Biol. Inorg. Chem. 2016, 21, 975–986. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef]
- Miyoshi, D.; Nakao, A.; Sugimoto, N. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+. Nucleic Acids Res. 2003, 31, 1156–1163. [Google Scholar] [CrossRef]
- Jiang, H.-X.; Cui, Y.; Zhao, T.; Fu, H.-W.; Koirala, D.; Punnoose, J.A.; Kong, D.-M.; Mao, H. Divalent cations and molecular crowding buffers stabilize G-triplex at physiologically relevant temperatures. Sci. Rep. 2015, 5, 9255. [Google Scholar] [CrossRef]
- Pacheco, A.; Issaian, A.; Davis, J.; Anderson, N.; Nemkov, T.; Vogeli, B.; Hansen, K.; Sikela, J.M. Proteolytic activation of human-specific Olduvai domains by the furin protease. bioRxiv 2021. [Google Scholar] [CrossRef]
- Yeo, G.; Holste, D.; Kreiman, G.; Burge, C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004, 5, R74. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vořechovský, I. Selection of Olduvai Domains during Evolution: A Role for Primate-Specific Splicing Super-Enhancer and RNA Guanine Quadruplex in Bipartite NBPF Exons. Brain Sci. 2022, 12, 874. https://doi.org/10.3390/brainsci12070874
Vořechovský I. Selection of Olduvai Domains during Evolution: A Role for Primate-Specific Splicing Super-Enhancer and RNA Guanine Quadruplex in Bipartite NBPF Exons. Brain Sciences. 2022; 12(7):874. https://doi.org/10.3390/brainsci12070874
Chicago/Turabian StyleVořechovský, Igor. 2022. "Selection of Olduvai Domains during Evolution: A Role for Primate-Specific Splicing Super-Enhancer and RNA Guanine Quadruplex in Bipartite NBPF Exons" Brain Sciences 12, no. 7: 874. https://doi.org/10.3390/brainsci12070874
APA StyleVořechovský, I. (2022). Selection of Olduvai Domains during Evolution: A Role for Primate-Specific Splicing Super-Enhancer and RNA Guanine Quadruplex in Bipartite NBPF Exons. Brain Sciences, 12(7), 874. https://doi.org/10.3390/brainsci12070874





