Functional Connectivity-Derived Optimal Gestational-Age Cut Points for Fetal Brain Network Maturity
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Acquisition of Resting-State Data
2.3. Preprocessing of Resting-State Data
2.4. Graph Construction
2.5. Graph Analysis
2.6. Statistical Analysis
3. Results
3.1. GEE Modeling
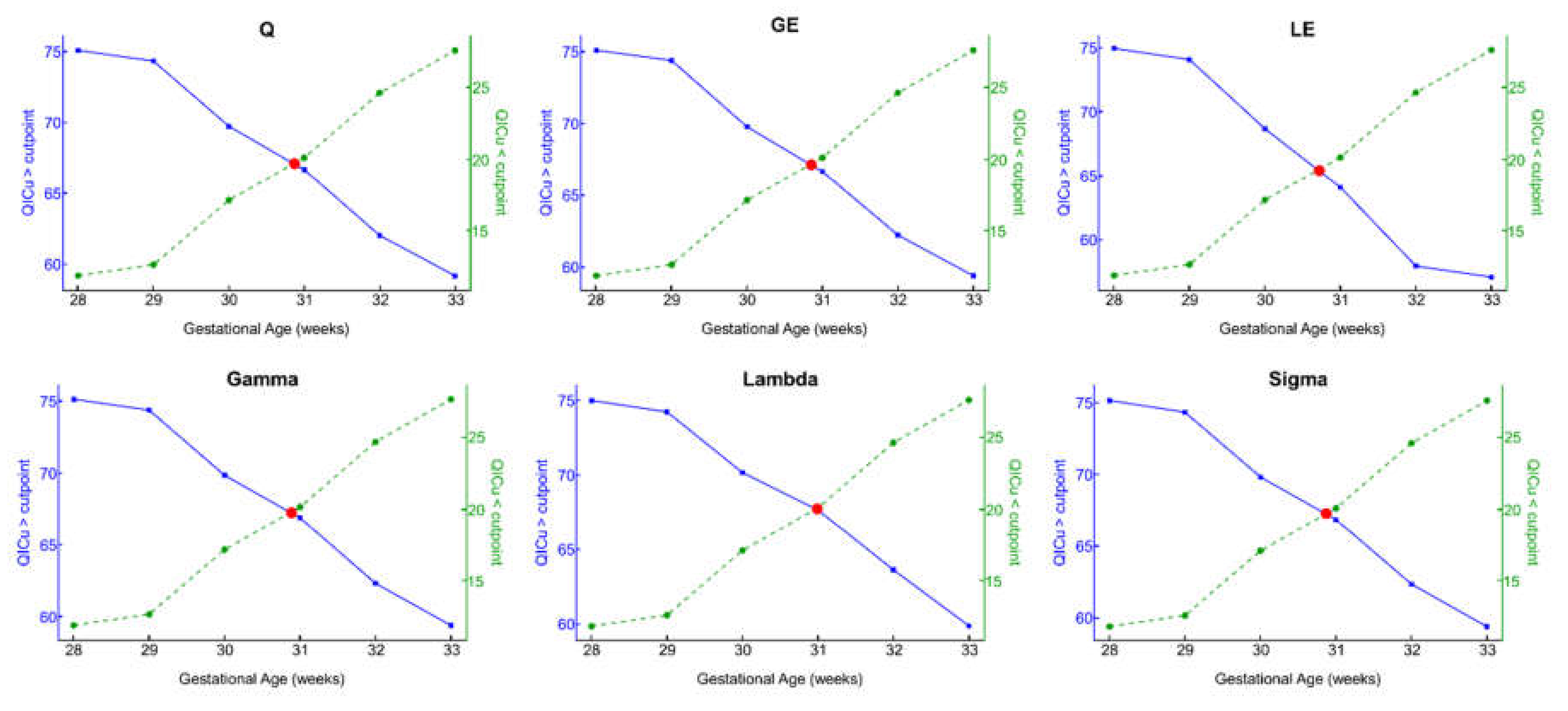
3.2. Optimal GA Cut Point
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biswal, B.; Yetkin, F.Z.; Haughton, V.M.; Hyde, J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 1995, 34, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Raichle, M.E. The restless brain: How intrinsic activity organizes brain function. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140172. [Google Scholar] [CrossRef] [PubMed]
- Schöpf, V.; Kasprian, G.; Brugger, P.C.; Prayer, D. Watching the fetal brain at “rest”. Int. J. Dev. Neurosci. 2012, 30, 11–17. [Google Scholar] [CrossRef]
- Jakab, A.; Schwartz, E.; Kasprian, G.; Gruber, G.M.; Prayer, D.; Schöpf, V.; Langs, G. Fetal functional imaging portrays heterogeneous development of emerging human brain networks. Front. Hum. Neurosci. 2014, 8, 852. [Google Scholar] [CrossRef] [PubMed]
- De Asis-Cruz, J.; Andersen, N.; Kapse, K.; Khrisnamurthy, D.; Quistorff, J.; Lopez, C.; Vezina, G.; Limperopoulos, C. Global Network Organization of the Fetal Functional Connectome. Cereb. Cortex 2021, 31, 3034–3046. [Google Scholar] [CrossRef]
- Doria, V.; Beckmann, C.F.; Arichi, T.; Merchant, N.; Groppo, M.; Turkheimer, F.; Counsell, S.; Murgasova, M.; Aljabar, P.; Nunes, R.; et al. Emergence of resting state networks in the preterm human brain. Proc. Natl. Acad. Sci. USA 2010, 107, 20015–20020. [Google Scholar] [CrossRef]
- Thomason, M.E.; Dassanayake, M.T.; Shen, S.; Katkuri, Y.; Alexis, M.; Anderson, A.L.; Yeo, L.; Mody, S.; Hernandez-Andrade, E.; Hassan, S.S.; et al. Cross-Hemispheric Functional Connectivity in the Human Fetal Brain. Sci. Transl. Med. 2013, 5, 173ra24. [Google Scholar] [CrossRef]
- De Asis-Cruz, J.; Kapse, K.; Basu, S.; Said, M.; Scheinost, D.; Murnick, J.; Chang, T.; du Plessis, A.; Limperopoulos, C. Functional brain connectivity in ex utero premature infants compared to in utero fetuses. NeuroImage 2020, 219, 117043. [Google Scholar] [CrossRef]
- Thomason, M.E.; Grove, L.E.; Lozon, T.A., Jr.; Vila, A.M.; Ye, Y.; Nye, M.J.; Manning, J.H.; Pappas, A.; Hernandez-Andrade, E.; Yeo, L.; et al. Age-related increases in long-range connectivity in fetal functional neural connectivity networks in utero. Dev. Cogn. Neurosci. 2014, 11, 96–104. [Google Scholar] [CrossRef]
- Bullmore, E.T.; Sporns, O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009, 10, 186–198. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O.; Van Leeuwen, C.; Breakspear, M. Symbiotic relationship between brain structure and dynamics. BMC Neurosci. 2009, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Thomason, M.E.; Brown, J.A.; Dassanayake, M.T.; Shastri, R.; Marusak, H.A.; Hernandez-Andrade, E.; Yeo, L.; Mody, S.; Berman, S.; Hassan, S.S.; et al. Intrinsic Functional Brain Architecture Derived from Graph Theoretical Analysis in the Human Fetus. PLoS ONE 2014, 9, e94423. [Google Scholar] [CrossRef] [PubMed]
- Turk, E.; van den Heuvel, M.I.; Benders, M.J.; De Heus, R.; Franx, A.; Manning, J.H.; Hect, J.L.; Hernandez-Andrade, E.; Hassan, S.S.; Romero, R.; et al. Functional Connectome of the Fetal Brain. J. Neurosci. 2019, 39, 9716–9724. [Google Scholar] [CrossRef]
- Dosenbach, N.U.F.; Nardos, B.; Cohen, A.; Fair, D.A.; Power, J.D.; Church, J.; Nelson, S.M.; Wig, G.S.; Vogel, A.C.; Lessov-Schlaggar, C.N.; et al. Prediction of Individual Brain Maturity Using fMRI. Science 2010, 329, 1358–1361. [Google Scholar] [CrossRef]
- Van den Heuvel, M.P.; Kersbergen, K.J.; De Reus, M.A.; Keunen, K.; Kahn, R.S.; Groenendaal, F.; De Vries, L.S.; Benders, M.J. The Neonatal Connectome During Preterm Brain Development. Cereb. Cortex 2014, 25, 3000–3013. [Google Scholar] [CrossRef]
- Keunen, K.; Counsell, S.; Benders, M.J. The emergence of functional architecture during early brain development. NeuroImage 2017, 160, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Vasung, L.; Turk, E.A.; Ferradal, S.; Sutin, J.; Stout, J.N.; Ahtam, B.; Lin, P.-Y.; Grant, P.E. Exploring early human brain development with structural and physiological neuroimaging. NeuroImage 2019, 187, 226–254. [Google Scholar] [CrossRef] [PubMed]
- De Asis-Cruz, J.; Krishnamurthy, D.; Zhao, L.; Kapse, K.; Vezina, G.; Andescavage, N.; Quistorff, J.; Lopez, C.; Limperopoulos, C. Association of Prenatal Maternal Anxiety With Fetal Regional Brain Connectivity. JAMA Netw. Open 2020, 3, e2022349. [Google Scholar] [CrossRef]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef]
- Joshi, A.; Scheinost, D.; Okuda, H.; Belhachemi, D.; Murphy, I.; Staib, L.; Papademetris, X. Unified Framework for Development, Deployment and Robust Testing of Neuroimaging Algorithms. Neuroinformatics 2011, 9, 69–84. [Google Scholar] [CrossRef]
- Scheinost, D.; Onofrey, J.; Kwon, S.H.; Cross, S.N.; Sze, G.; Ment, L.R.; Papademetris, X. A fetal fMRI specific motion correction algorithm using 2nd order edge features. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1288–1292. [Google Scholar]
- Ojemann, J.G.; Akbudak, E.; Snyder, A.Z.; McKinstry, R.C.; Raichle, M.E.; Conturo, T.E. Anatomic Localization and Quantitative Analysis of Gradient Refocused Echo-Planar fMRI Susceptibility Artifacts. NeuroImage 1997, 6, 156–167. [Google Scholar] [CrossRef]
- Cox, R. AFNI: Software for Analysis and Visualization of Functional Magnetic Resonance Neuroimages. Comput. Biomed. Res. 1996, 29, 162–173. [Google Scholar] [CrossRef]
- Power, J.D.; Barnes, K.A.; Snyder, A.Z.; Schlaggar, B.L.; Petersen, S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 2012, 59, 2142–2154. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, M.; Hect, J.; Hernandez-Andrade, E.; Hassan, S.; Romero, R.; Eggebrecht, A.; Thomason, M. Sex differences in functional connectivity during fetal brain development. Dev. Cogn. Neurosci. 2019, 36, 100632. [Google Scholar] [CrossRef]
- Behzadi, Y.; Restom, K.; Liau, J.; Liu, T.T. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage 2007, 37, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Muschelli, J.; Nebel, M.B.; Caffo, B.S.; Barber, A.; Pekar, J.; Mostofsky, S.H. Reduction of motion-related artifacts in resting state fMRI using aCompCor. NeuroImage 2014, 96, 22–35. [Google Scholar] [CrossRef]
- Jo, H.J.; Gotts, S.J.; Reynolds, R.C.; Bandettini, P.A.; Martin, A.; Cox, R.; Saad, Z.S. Effective Preprocessing Procedures Virtually Eliminate Distance-Dependent Motion Artifacts in Resting State FMRI. J. Appl. Math. 2013, 2013, 935154. [Google Scholar] [CrossRef]
- Friston, K.J.; Williams, S.; Howard, R.; Frackowiak, R.; Turner, R. Movement-Related effects in fMRI time-series. Magn. Reson. Med. 1996, 35, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Craddock, R.C.; James, G.A.; Holtzheimer, P.E.; Hu, X.P.; Mayberg, H.S. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 2012, 33, 1914–1928. [Google Scholar] [CrossRef]
- Peer, M.; Abboud, S.; Hertz, U.; Amedi, A.; Arzy, S. Intensity-based masking: A tool to improve functional connectivity results of resting-state fMRI. Hum. Brain Mapp. 2016, 37, 2407–2418. [Google Scholar] [CrossRef][Green Version]
- De Asis-Cruz, J.; Donofrio, M.T.; Vezina, G.; Limperopoulos, C. Aberrant brain functional connectivity in newborns with congenital heart disease before cardiac surgery. NeuroImage Clin. 2018, 17, 31–42. [Google Scholar] [CrossRef]
- Achard, S.; Salvador, R.; Whitcher, B.; Suckling, J.; Bullmore, E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J. Neurosci. 2006, 26, 63–72. [Google Scholar] [CrossRef]
- Bassett, D.S.; Meyer-Lindenberg, A.; Achard, S.; Duke, T.; Bullmore, E. Adaptive reconfiguration of fractal small-world human brain functional networks. Proc. Natl. Acad. Sci. USA 2006, 103, 19518–19523. [Google Scholar] [CrossRef]
- Lynall, M.-E.; Bassett, D.S.; Kerwin, R.; McKenna, P.; Kitzbichler, M.; Muller, U.; Bullmore, E. Functional Connectivity and Brain Networks in Schizophrenia. J. Neurosci. 2010, 30, 9477–9487. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Humphries, M.D.; Gurney, K. Network ‘Small-World-Ness’: A Quantitative Method for Determining Canonical Network Equivalence. PLoS ONE 2008, 3, e0002051. [Google Scholar] [CrossRef]
- Crucitti, P.; Latora, V.; Marchiori, M.; Rapisarda, A. Efficiency of scale-free networks: Error and attack tolerance. Phys. A Stat. Mech. Its Appl. 2003, 320, 622–642. [Google Scholar] [CrossRef]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef]
- Stiles, J.; Jernigan, T.L. The Basics of Brain Development. Neuropsychol. Rev. 2010, 20, 327–348. [Google Scholar] [CrossRef]
- Kostović, I.; Sedmak, G.; Judaš, M. Neural histology and neurogenesis of the human fetal and infant brain. NeuroImage 2019, 188, 743–773. [Google Scholar] [CrossRef]
- Hoerder-Suabedissen, A.; Molnár, Z. Development, evolution and pathology of neocortical subplate neurons. Nat. Rev. Neurosci. 2015, 16, 133–146. [Google Scholar] [CrossRef]
- Judaš, M.; Sedmak, G.; Kostović, I. The significance of the subplate for evolution and developmental plasticity of the human brain. Front. Hum. Neurosci. 2013, 7, 423. [Google Scholar] [CrossRef]
- Kostovic, I.; Rakic, P. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J. Comp. Neurol. 1990, 297, 441–470. [Google Scholar] [CrossRef] [PubMed]
- Kostović, I.; Judaš, M. Embryonic and Fetal Development of the Human Cerebral Cortex. Brain Mapp. 2015, 167–175. [Google Scholar] [CrossRef]
- Vanhatalo, S.; Kaila, K.; Lagercrantz, H. Emergence of spontaneous and evoked electroencephalographic activity in the human brain. In The Newborn Brain; Lagercrantz, H., Hanson, M.A., Ment, L.R., Peebles, D.M., Eds.; Cambridge University Press (CUP): Cambridge, UK, 2011; pp. 229–244. [Google Scholar]
- Tolonen, M.; Palva, J.M.; Andersson, S.; Vanhatalo, S. Development of the spontaneous activity transients and ongoing cortical activity in human preterm babies. Neuroscience 2007, 145, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Khazipov, R.; Luhmann, H.J. Early patterns of electrical activity in the developing cerebral cortex of humans and rodents. Trends Neurosci. 2006, 29, 414–418. [Google Scholar] [CrossRef]
- Luhmann, H.J.; Sinning, A.; Yang, J.-W.; Reyes-Puerta, V.; Stüttgen, M.C.; Kirischuk, S.; Kilb, W. Spontaneous neuronal activity in developing neocortical networks: From single cells to large-scale interactions. Front. Neural Circuits 2016, 10, 40. [Google Scholar] [CrossRef]
| Metric | GA at Scan = 28 | GA at Scan = 29 | GA at Scan = 30 | GA at Scan = 31 | GA at Scan = 32 | GA at Scan = 33 |
|---|---|---|---|---|---|---|
| Q | −0.12 | −0.1243 | −0.1286 | −0.1328 | −0.1371 | −0.1414 |
| (−0.3497–0.0477) | (−0.3622−0.0494) | (−0.3747−0.0511) | (−0.3871−0.0528) | (−0.3996−0.0545) | (−0.4121−0.0562) | |
| GE | 0.0056 | 0.0058 | 0.006 | 0.0062 | 0.0064 | 0.0066 |
| (−0.0102−0.0196) | (−0.0105−0.0203) | (−0.0109−0.0210) | (−0.0113−0.0217) | (−0.0116−0.0224) | (−0.0120−0.0231) | |
| LE | −0.0179 | −0.0185 | −0.0192 | −0.0198 | −0.0205 | −0.0211 |
| (−0.0466−0.0315) | (−0.0483−0.0326) | (−0.0500−0.0337) | (−0.0516−0.0349) | (−0.0533−0.0360) | (−0.0550−0.0371) | |
| γ | −0.2449 | −0.2537 | −0.2624 | −0.2712 | −0.2799 | −0.2887 |
| (−0.5488−0.4955) | (−0.5684−0.5132) | (−0.5880−0.5309) | (−0.6076−0.5486) | (−0.6272−0.5662) | (−0.6468−0.5839) | |
| λ | −0.0283 | −0.0293 | −0.0303 | −0.0313 | −0.0323 | −0.0333 |
| (−0.0788−0.0447) | (−0.0816−0.0463) | (−0.0845−0.0479) | (−0.0873−0.0495) | (−0.0901−0.0511) | (−0.0929−0.0527) | |
| σ | −0.2027 | −0.2099 | −0.2171 | −0.2244 | −0.2316 | −0.2389 |
| (−0.4537−0.3698) | (−0.4699−0.3830) | (−0.4861−0.3962) | (−0.5023−0.4095) | (−0.5185−0.4227 | (−0.5347−0.4359) |
| Metric | Exceed | GA at Scan = 28 | GA at Scan = 29 | GA at Scan = 30 | GA at Scan = 31 | GA at Scan = 32 | GA at Scan = 33 |
|---|---|---|---|---|---|---|---|
| Cut Point | N = 13/97 | N = 14/96 | N = 20/90 | N = 24/86 | N = 30/80 | N = 34/76 | |
| Q | No | −0.0061 | −0.0064 | −0.0048 | −0.0063 | −0.0048 | −0.0055 |
| (−0.0144–−0.0010) | (−0.0139–−0.0017) | (−0.0095− −0.0011) | −0.0072 | (−0.0077–−0.0024) | (−0.0082–−0.0031) | ||
| Yes | −0.0014 | −0.0014 | 0.0013 | 0.0008 | 0.0027 | 0.0019 | |
| (−0.0120–0.0075) | (−0.0120–0.0075) | (−0.0066–0.0088) | (−0.0036–0.0203) | (−0.0033–0.0204) | (−0.0047–0.0204) | ||
| GE | No | 0.0005 | 0.0006 | 0.0004 | 0.0007 | 0.0008 | 0.0007 |
| (−0.0015–0.0032) | (−0.0012–0.0030) | (−0.0007–0.0018) | (−0.0003–0.0018) | (0.0000–0.0016) | (0.0001–0.0015) | ||
| Yes | −0.0001 | −0.0001 | −0.0004 | −0.0006 | −0.0003 | −0.0002 | |
| (−0.0010–0.0005) | (−0.0010–0.0005) | (−0.0020–0.0004) | −0.0027–0.0002) | (−0.0010–0.0005) | (−0.0011–0.0007) | ||
| LE | No | −0.0016 | −0.0019 | −0.0015 | −0.0019 | −0.0017 | −0.0018 |
| (−0.0060–0.0018) | (−0.0059–0.0012 | (−0.0041–0.0006) | (−0.0041–−0.0001) | −0.0033–−0.0002) | (−0.0031–−0.0005) | ||
| Yes | 0 | −0.0001 | 0.0001 | 0.0006 | 0.0003 | 0 | |
| (−0.0016–0.0020) | (−0.0016–0.0020) | (−0.0026–0.0031) | (−0.0008–0.0031) | (−0.0012–0.0015) | (−0.0057–0.0014) | ||
| γ | No | −0.0312 | −0.0348 | −0.0243 | −0.0263 | −0.0176 | −0.0208 |
| (−0.0510–−0.0149) | (−0.0543–−0.0172) | (−0.0406–−0.0064) | (−0.0386–−0.0134) | (−0.0266–−0.0054) | −0.0298–−0.0083) | ||
| Yes | 0.0048 | 0.0041 | 0.0021 | 0.0056 | 0.0144 | 0.012 | |
| (−0.0111–0.0341) | (−0.0128–0.0341) | (−0.0102–0.0384) | (−0.0161–0.1078) | (−0.0147–0.1081) | (−0.0167–0.1082) | ||
| λ | No | −0.0044 | −0.0044 | −0.0026 | −0.003 | −0.003 | −0.0029 |
| (−0.0130–0.0022) | (−0.0123–0.0016) | (−0.0079–0.0021) | (−0.0075–0.0009) | (−0.0064–0.0001) | (−0.0056–−0.0001) | ||
| Yes | −0.0004 | −0.0005 | 0.0004 | 0.0007 | −0.0005 | −0.0009 | |
| −0.0036–0.0023) | (−0.0038–0.0022) | (−0.0019–0.0060) | (−0.0020–0.0085) | (−0.0021–0.0015) | (−0.0028–0.0015) | ||
| σ | No | −0.0281 | −0.0295 | −0.0186 | −0.0188 | −0.0102 | −0.0128 |
| (−0.0492–−0.0139) | (−0.0484–−0.0163) | (−0.0296–−0.0072) | (−0.0265–− 0.0106) | (−0.0166–−0.0000) | (−0.0202–−0.0035) | ||
| Yes | 0.0006 | −0.0007 | −0.0015 | 0.001 | 0.0103 | 0.0079 | |
| (−0.0148–0.0344) | (−0.0167–0.0344) | (−0.0153–0.0344) | (−0.0199–0.1047) | (−0.0177–0.1049) | (−0.0231–0.1050) |
| Metric | Exceed | GA at Scan = 28 | GA at Scan = 29 | GA at Scan = 30 | GA at Scan = 31 | GA at Scan = 32 | GA at Scan = 33 |
|---|---|---|---|---|---|---|---|
| Cut Point | N = 13/97 | N = 14/96 | N = 20/90 | N = 24/86 | N = 30/80 | N = 34/76 | |
| Q | No | 11.867 | 12.634 | 17.145 | 20.132 | 24.664 | 27.635 |
| (9.0000−14.0000) | (9.5000−15.0000) | (14.0000−20.0000) | (16.0000−24.0000) | (21.0000−28.0000) | (23.0000−32.0000) | ||
| Yes | 75.1168 | 74.3619 | 69.7688 | 66.7001 | 62.0553 | 59.2125 | |
| (72.0004−78.0001) | (74.0000−76.5054) | (66.0000−73.0002) | (62.0002−71.0000) | (57.0405−66.0000) | (55.0000−63.0004) | ||
| GE | No | 11.867 | 12.634 | 17.145 | 20.132 | 24.664 | 27.635 |
| (9.0000−14.0000) | (9.5000−15.0000) | (14.0000−20.0000) | (16.0000−24.0000) | (21.0000−28.0000) | 23.0000−32.0000) | ||
| Yes | 75.1168 | 74.4058 | 69.7883 | 66.6606 | 62.2288 | 59.4004 | |
| (72.0014−78.0001) | (71.0013−78.0000) | (65.5393−74.0000) | (62.0000−71.0000) | (57.9407−67.0256) | (55.0000−64.5115) | ||
| LE | No | 11.867 | 12.634 | 17.145 | 20.132 | 24.664 | 27.635 |
| (9.0000−14.0000) | (9.5000−15.0000) | (14.0000−20.0000) | (16.0000−24.0000) | (21.0000−28.0000) | (23.0000−32.0000) | ||
| Yes | 74.9755 | 74.1004 | 68.6799 | 64.0572 | 57.8721 | 57.0111 | |
| (72.0000−78.0037) | (71.10003−78.0000) | (26.1065−74.0000) | (25.8099−71.0012) | (26.0766−71.9152) | (29.0706−95.2327) | ||
| γ | No | 11.867 | 12.634 | 17.145 | 20.132 | 24.664 | 27.635 |
| (98.0000−14.0000) | (9.5000−15.0000) | (14.0000−20.0000) | (16.0000−24.0000) | (21.0000−28.0000) | (23.0000−32.0000) | ||
| Yes | 75.1457 | 74.3887 | 69.8371 | 66.8617 | 62.2667 | 59.3355 | |
| (72.0003−78.0002) | (71.0000−78.0000) | (66.0000−73.0003) | (63.0000−71.0000) | (58.1335−66.0000) | (55.0000−63.4042) | ||
| λ | No | 11.867 | 12.634 | 17.145 | 20.132 | 24.664 | 27.635 |
| (9.0000−14.0000) | (9.5000−15.0000) | (14.0000−20.0000) | (16.0000−24.0000) | (21.0000−28.0000) | (23.0000−32.0000) | ||
| Yes | 74.9952 | 74.228 | 70.154 | 67.6423 | 63.6002 | 59.8447 | |
| (72.0004−78.0003) | (71.0007−77.0000) | (65.9967−74.0195) | (62.0000−74.8341) | (57.6592−69.9663) | (55.0000−64.0018) | ||
| σ | No | 11.867 | 12.634 | 17.145 | 20.132 | 24.664 | 27.635 |
| (9.0000−14.0000) | (9.5000−15.0000) | (14.0000−20.0000) | (16.0000−24.0000) | (21.0000−28.0000) | (23.0000−32.0000) | ||
| Yes | 75.2052 | 74.3849 | 69.8515 | 66.8673 | 62.3361 | 59.3695 | |
| (72.0002−78.0002) | (71.5005−78.0000) | (66.4253−73.0002) | (63.0000−71.0000) | (59.0000−66.0004) | (55.0000−64.0000) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Asis-Cruz, J.; Barnett, S.D.; Kim, J.-H.; Limperopoulos, C. Functional Connectivity-Derived Optimal Gestational-Age Cut Points for Fetal Brain Network Maturity. Brain Sci. 2021, 11, 921. https://doi.org/10.3390/brainsci11070921
De Asis-Cruz J, Barnett SD, Kim J-H, Limperopoulos C. Functional Connectivity-Derived Optimal Gestational-Age Cut Points for Fetal Brain Network Maturity. Brain Sciences. 2021; 11(7):921. https://doi.org/10.3390/brainsci11070921
Chicago/Turabian StyleDe Asis-Cruz, Josepheen, Scott Douglas Barnett, Jung-Hoon Kim, and Catherine Limperopoulos. 2021. "Functional Connectivity-Derived Optimal Gestational-Age Cut Points for Fetal Brain Network Maturity" Brain Sciences 11, no. 7: 921. https://doi.org/10.3390/brainsci11070921
APA StyleDe Asis-Cruz, J., Barnett, S. D., Kim, J.-H., & Limperopoulos, C. (2021). Functional Connectivity-Derived Optimal Gestational-Age Cut Points for Fetal Brain Network Maturity. Brain Sciences, 11(7), 921. https://doi.org/10.3390/brainsci11070921






