1. Introduction
Alzheimer’s disease (AD), a neurodegenerative brain disease caused by multiple factors, is one of the most common chronic diseases in old age [
1]. This disease usually causes progressive and disabling impairments of cognitive function, including memory, language, understanding and attention [
2]. In 2015, it was estimated that about 47 million people worldwide had AD, and the number is expected to reach 141 million by 2050 [
3]. At present, there is no practical method to cure AD [
4], so early diagnosis of AD is needed to obtain treatment time. Mild Cognitive Impairment (MCI) is an intermediate state between normal aging and dementia [
5], and one study showed that 32% of MCI converted to AD within five years [
6]. Therefore, early diagnosis and intervention of Alzheimer’s disease is very important.
In the past few decades, neuroimaging has been widely used to study brain diseases [
7,
8,
9]. Neuroimaging technology provides anatomical and functional images of the brain, such as Positron Emission Computed Tomography (PECT), Structural Magnetic Resonance Imaging (SMRI), Diffusion Magnetic Resonance Imaging (DMRI), Functional Magnetic Resonance Imaging (FMRI), Electroencephalogram (EEG), and Magnetoencephalography (MEG) [
10,
11,
12]. Among them, SMRI is often used for the characterization and prediction of AD due to its relatively low cost and good imaging quality. Previous studies have shown that the volume and thickness of the brain are closely related to AD [
13], the hippocampus region of AD patients is one third smaller than that of healthy subjects [
14], and the medial temporal lobe region is the most effective region of the brain for identifying patients with MCI [
15].
In recent years, machine learning and deep learning technologies have demonstrated revolutionary performance in many areas, such as action recognition, machine translation, image segmentation, and speech recognition [
16,
17,
18,
19]. Machine learning and deep learning have also achieved great success in the fields of medical image analysis and assisted diagnosis of brain diseases [
20,
21,
22,
23]. Unlike traditional methods based on manual feature extraction, deep learning can learn straightforward and low-level features from medical images, and construct complex high-level features in a hierarchical way [
24].
The methods of diagnosing AD through Magnetic Resonance Imaging (MRI) images can be roughly divided into two categories, including 1) methods based on two-dimensional (2D) view, and 2) methods based on three-dimensional (3D) view. In most studies based on 2D view, 2D slices are selected from each subject, and these coronal, sagittal, or axial brain images are considered as a whole to classify. Bi et al. [
25] manually selected brain images from three orthogonal panels of MRI data and performed unsupervised learning through PCA-Net. Neffati et al. [
26] extracted 2D discrete wavelet transform texture features from coronal slices to classify AD. Jain et al. [
27] selected the most informative set of 2D slices and used models trained on natural images to classify medical images through transfer learning. Mishra et al. [
28] extracted features through complete local binary pattern from 2D slices in three directions instead of a single direction. These methods based on 2D view have all achieved excellent classification performance.
In addition, there are many methods based on 3D view that can also classify AD well [
29,
30,
31,
32]. Zhang et al. [
33] discriminated between patients and healthy controls by using voxel-based morphometry (VBM) parameters from 3D images. Their results reported that the effect of classification using only 3D VBM parameters was better than the effect of classification using only 2D texture parameters. J. Liu et al. extracted 3D texture features through different divisions of the brain regions of interest (ROI) to construct multiple hierarchical networks [
34,
35]. Y. Wang et al. [
36] diagnosed AD through combining morphometric measures of 3D images and connectome measures of 3D ROI. M. Liu et al. [
37] used multivariate statistical tests to find discriminative 3D patch sets from the 3D rain images for the subsequent analysis. Basaia et al. [
38] adopted a convolutional neural network to distinguish mild cognitive impairment who will convert to AD (c-MCI) and stable MCI (s-MCI) through 3D MRI images with an accuracy of 75%.
Considering the excellent performance of methods based on 2D view and 3D view, researchers in different fields have combined 2D and 3D views for study. Nanni et al. [
39] combined texture features extracted from 2D slices with voxel features from 3D images and used multiple feature selection methods to improve the detection of early AD. In the field of action recognition and human pose estimation, Luvizon et al. [
40] used a multi-task learning framework to combine 2D still images with 3D videos to learn features and achieved the latest results at that time. In the field of tumor segmentation, Mlynarski et al. [
41] first divided 3D images into 2D images in three directions, and then fed them into the 2D model to generate 2D feature maps. Finally, they used 2D feature maps as the additional inputs of the 3D model to combine 2D and 3D information to achieve better segmentation results. The methods of combining 2D and 3D views in the different fields mentioned above have achieved excellent results. Therefore, we propose a multi-model learning framework based on multiple views to make full use of local and global information in this paper. We combine some 2D MRI images with the entire 3D MRI images, and adopt different deep learning models for different views to better distinguish AD and MCI.
The main contributions are listed as follows:
- (1)
We employ entropy to select 2D slices and fuse them to learn 2D local features.
- (2)
We propose a combination of 2D and 3D images of MRI to diagnose MCI, rather than a single view.
- (3)
We propose a multi-model learning framework that uses different models to train data from different views.
2. Materials and Methods
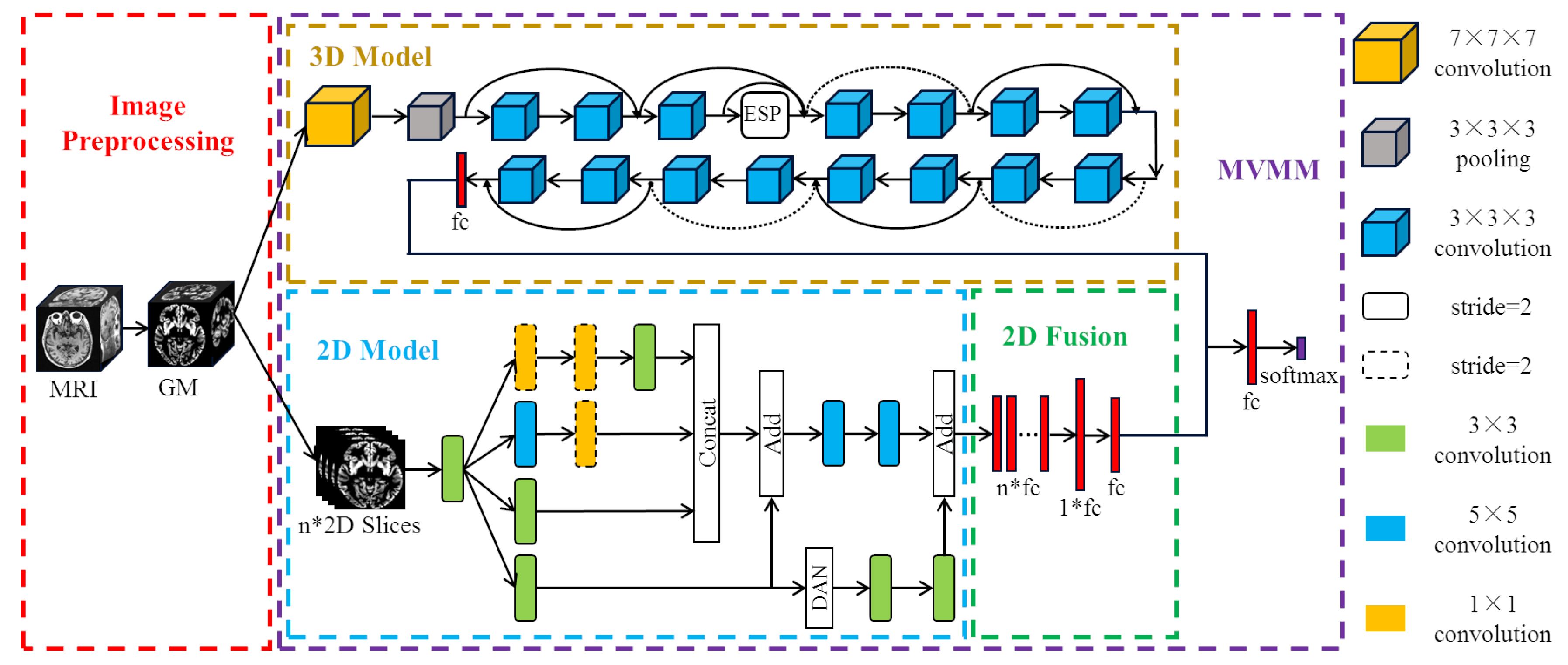
We propose a new method of MCI diagnosis based on multi-view based multi-model (MVMM) framework. The MVMM framework mainly includes a 3D model for extracting global features and a 2D model for extracting local features. The 3D model we use is the Dilated Residual Network (DRN), which adds an Efficient Space Pyramid (ESP) module. The 2D model is the Dual Attention Inception Network (DAIN), which adds a dual attention mechanism to the Inception network. The flow of our MVMM framework is shown in
Figure 1. Firstly, the gray matter (GM) images of subjects are divided into whole-brain gray matter images and some selected two-dimensional slices, and then they are respectively input into the corresponding different models. Finally, the local features and global features are concatenated together for integration training.
2.1. Data and Pre-Processing
2.1.1. Data Acquisition
In this work, the MRI images we use are obtained from Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (
http://adni.loni.usc.edu/) [
42]. ADNI started in 2004 under the leadership of Dr. Michael W. Weiner. ADNI is a longitudinal multicenter study with the primary goal of early detection of AD and the use of biomarkers to track disease progression. ADNI has already begun three phases, namely ADNI1 (2004-2009), ADNI2/GO (2010-2016) and ADNI3. At each phase of ADNI, new participants are recruited and agreed to complete various imaging acquisitions and clinical evaluations. Later phases include follow-up scans of some previously scanned subjects and scans of new subjects. In this paper, we select the SMRI data acquired at 1.5 Tesla [
43]. The data we use are obtained from 649 subjects, which include 175 scans of AD, 214 scans of healthy controls (HC), and 260 scans of MCI. The demographic and clinical characteristics of all subjects are reported in
Table 1.
2.1.2. Data Pre-Processing
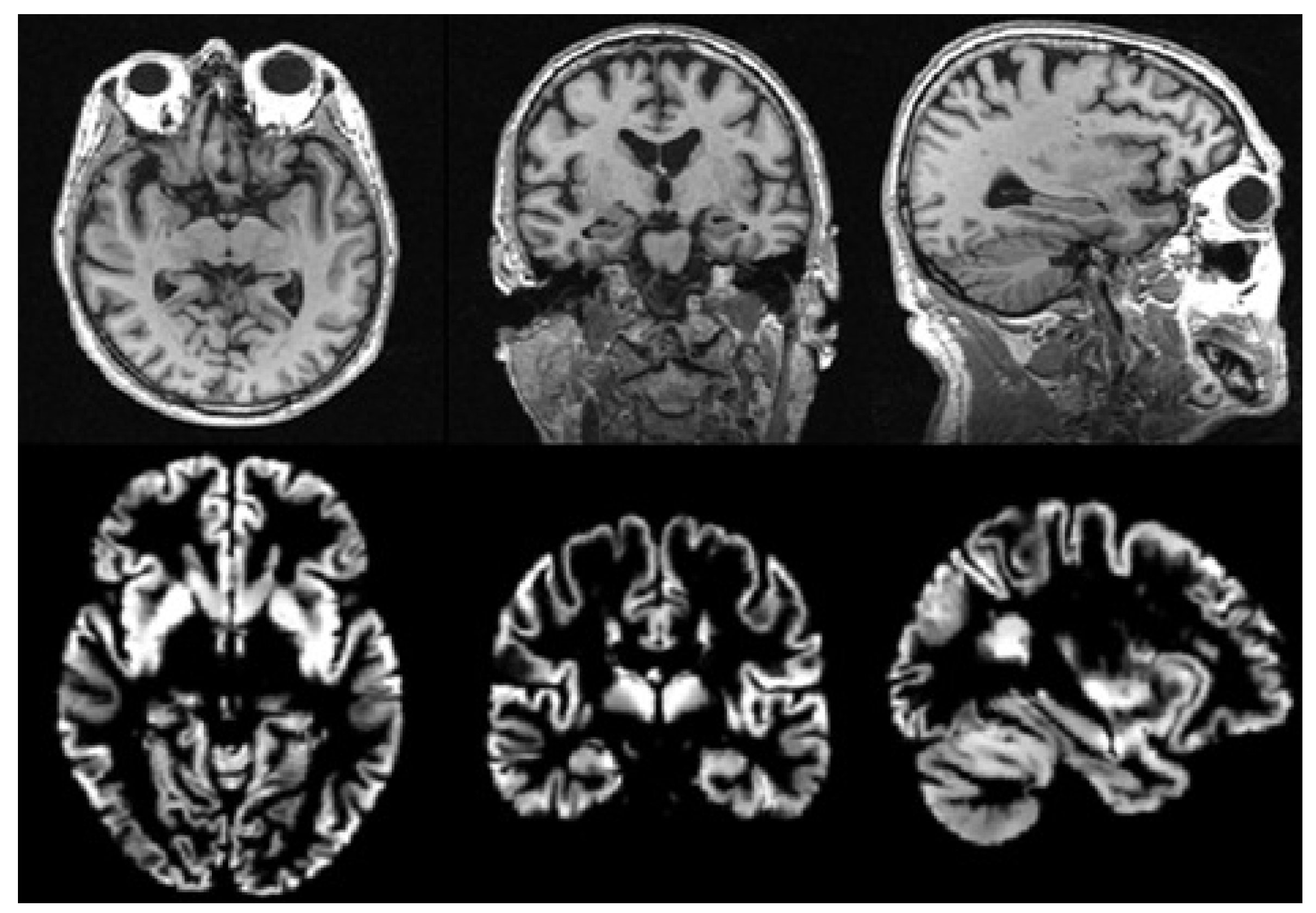
There is usually much noise in the raw data, so we need to preprocess the MRI data first. In this paper, we use the voxel-based morphological preprocessing method. Specifically, we use the CAT12 toolbox which is an extension to SPM12 [
44] to provide computational anatomy. First, we register the MRI images to the standard space through DARTEL (Diffeomorphic Anatomical Registration Through Exponentiated Liealgebra) algorithm [
45]. Second, we use the maximum a posteriori and partial volume estimation segmentation techniques [
46] to segment the image into gray matter, white matter, and cerebrospinal fluid. Then, the Jacobian determinant is used to modulate the gray matter image nonlinearly. Finally, the gray matter image is spatially smoothed with the 8mm Gaussian smoothing kernel. The size of each gray matter image we get in the standard space is
, then we use scikit-image package to resample it to a size of
. It is noted that gray matter loss in the medial temporal lobe is characteristic of MCI [
47], so we use gray matter images to analyze in this paper. The MRI images before and after preprocessing are shown in
Figure 2.
2.2. DRN Model Based on a 3D View
For the whole brain three-dimensional view, we take the preprocessed whole gray matter image directly as input. In order to learn the 3D global information more comprehensively, we use 3D convolutional neural networks to perform global feature extraction related to AD. The 3D gray matter image contains the entire brain, which is very informative. Therefore, how to comprehensively learn useful features is a challenge. Convolutional neural networks have developed rapidly. Since the birth of AlexNet in 2012 [
48], the depth of subsequent advanced convolutional neural network models has grown deeper. But as the depth increases, the problem of gradient disappearance during training becomes more serious. In order to avoid the problem of gradient disappearance caused by the network being too deep, the residual network (ResNet) [
49] introduces an identity shortcut connection and skips one or more layers directly. Assuming that the layer
is connected to the layer
l, the output
of the layer
l is:
represents a non-linear transformation function, including batch normalization (BN), rectified linear unit (ReLu), and convolution operation. ResNet reduces the difficulty of training deep networks by adding shortcut connections. For AD-related information, ResNet uses linear activation to obtain identity mapping. In contrast, ResNet uses non-linear activation for redundant information not related to AD. Since the non-linear activation is for redundant information, less useful information is lost. ResNet effectively solves the problem of network degradation caused by too deep depth, so that the model can learn more powerful advanced features.
In this paper, ResNet-18 is used as the basic model to train gray matter images from 3D view to obtain disease characteristics related to AD. However, the traditional ResNet model only uses
convolution kernels, and the receptive field of a single convolution kernel can only reach 27. In order to enable the convolution kernel to obtain a larger receptive field, and to allow the model to learn 3D features more comprehensively, we add the dilated convolution [
50] on the basis of the ResNet network. Assuming that the size of the convolution kernel is
k and the dilation rate is
r, the receptive field (RF) of the dilated convolution is:
When
, it is ordinary convolution, and the receptive field is
. When
, for the same
convolution, the dilated convolution not only increases the receptive field (from 9 to 25), but also does not increase the parameters like ordinary convolution (the weight in the dilated part is 0). Although dilated convolutions increase the receptive field, the mesh effect is prone to occur if misused. Therefore, we use an efficient space pyramid (ESP) [
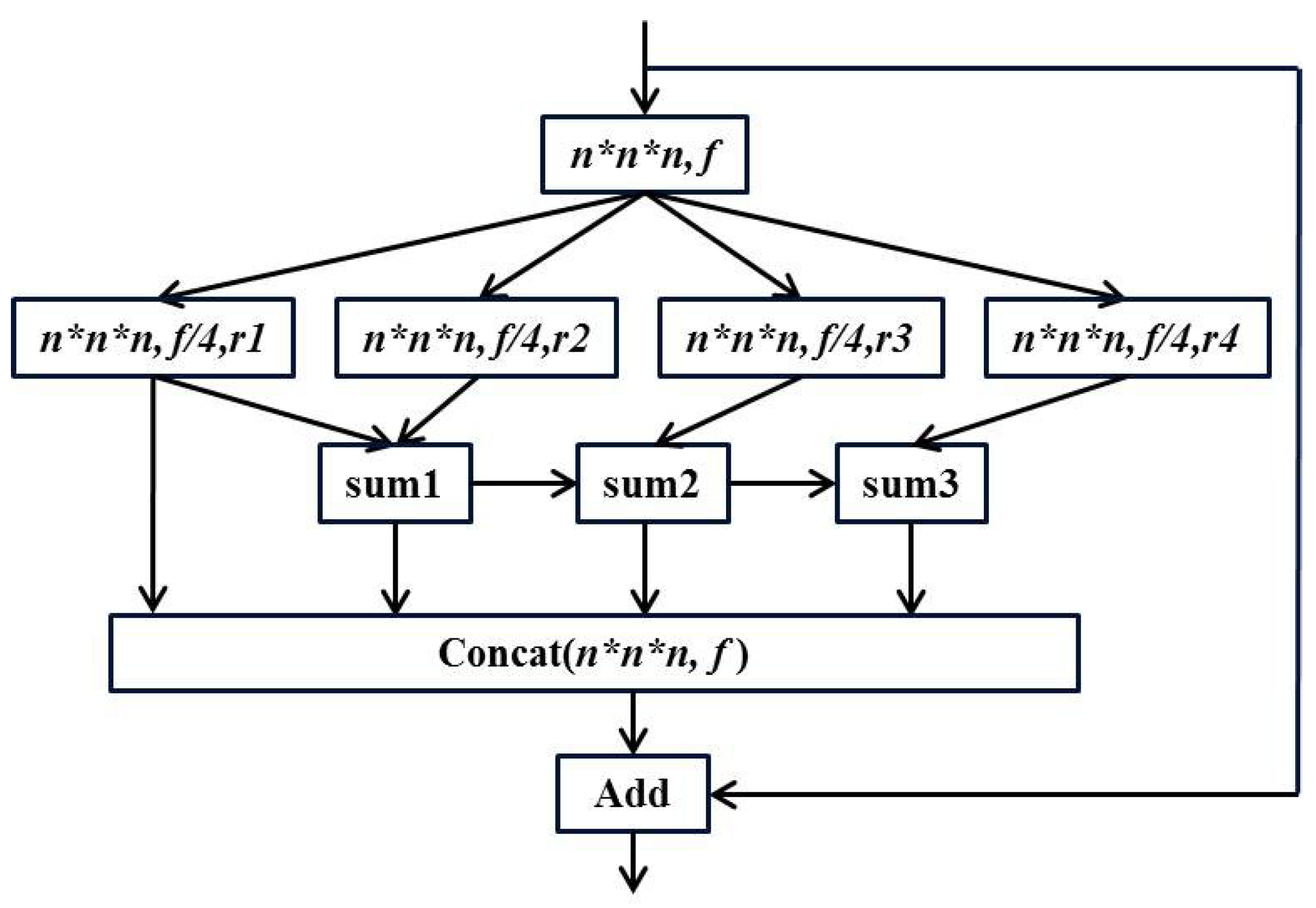
51] module to avoid the mesh effect. Specifically, a
convolution is performed on the input to obtain a feature map of
, and then four parallel dilated convolutions are used. Finally, the hierarchical features are merged to obtain the same size feature map. The structure of the ESP module is shown in
Figure 3.
In the figure above, the
are different dilation rates. It can be seen that the residual operation is also used in the ESP module, and the information of different receptive fields is concatenated before the residual operation to ensure the output quality. We replace one layer in the original ResNet-18 network with an ESP module to learn the 3D global features of the brain image more effectively. The overall structure of the final three-dimensional DRN model is shown in
Figure 1. The pre-processed whole-brain gray matter image contains much information. In order to effectively learn useful information, we choose 3D deep convolutional neural network for training. We use ResNet-18 with shortcut connections as our base model. At the same time, we also use dilated convolution to expand the receptive field in order to learn AD-related features more comprehensively.
2.3. DAIN Model Based on 2D View
For the 2D local view, we select some slices from the preprocessed gray matter image for training. We can select a large number of 2D slices from the 3D gray matter image. How to choose the best training data is very important for the success of the entire method. In this paper, we select 2D images based on image entropy [
52] and extract the most informative slices to train the network. Generally, for a set of M gray values with probabilities
, the entropy can be calculated as follows:
The higher the entropy, the more information the image contains. The subsequent problem is that MRI images generally contain much noise. Blindly selecting the image with a large amount of information may lead to the selection of some useless images with much noise. The images we study are preprocessed by CAT12. Compared with the original MRI image, the image we use is standardized and smoothed, and the skull of the image is removed. Therefore, we sort the slices in descending order of entropy and select the first 32 images for training to provide robustness according to previous research [
53].
After obtaining the selected 2D slices from the 3D MR image, we use them as the inputs of 2D model to learn AD features. Assume that the image set of each subject is
, the input of the two-dimensional model can be expressed as:
Thirty-two slices are selected from each subject through the axial brain image. In order to enhance the representation ability of the model, we use the Inception structure [
54] to obtain information on different scales and fuse the features learned by convolution kernels of different sizes. In the 2D model, we pay more attention to the local information as the selected pictures are comparatively informative. How to find the local information that can distinguish MCI from other categories is the focus of our research. The attentional mechanism solves this problem by enabling the model to think globally and focus on more critical local information. We use a dual attention network (DAN) proposed by Fu et al. [
55] that combines channel attention and spatial attention. The spatial attention module (PAM) in DAN first performs three
convolutions on the feature map
A to obtain three feature maps
B,
C, and
D of the same size
. Moreover, these three feature maps are converted to the size of
. Then the spatial attention map is calculated from
B and
C. We multiply the spatial attention map by
D, and then multiply the result by the scale coefficient
(initialized to 0). Finally, the output of PAM is obtained by adding the original feature map
A:
The channel attention module (CAM) in DAN first converts the feature map
A into
, and multiplies
and
to obtain the channel attention map with the size of
. We multiply the channel attention map by
and then multiply it by the scale coefficient
(initialized to 0). The result of the product is converted to the size of
. Finally, the output of CAM is obtained by adding to the original feature map
A:
Finally, the results of the two attention modules are added together to form the output of DAN:
We combine DAN with Inception to form the final two-dimensional model. The structure of our proposed DAIN model is shown in
Figure 1. The proposed DAIN model first obtains multi-scale AD information through the Inception structure. Then, the dual attention mechanism is used to obtain more significant local information. Finally, the important local information is combined with the fused multi-scale information to obtain the AD features that represent the 2D view.
2.4. Combination of 2D and 3D Views
Before constructing the multi-view model, we first integrate the output of the DAIN Model. Because in the previous DAIN model, we selected 32 slices for each subject and treated each slice as one subject. In this study, we use the above DAIN model as the pre-trained model of the final 2D model. In the final 2D model, the features extracted from each of the 32 images are concatenated together for classification. The final full connection is the MCI feature representing the local information extracted from the 2D slices.
After integrating the 2D features, we combine the features extracted from the 2D model with the features extracted from the 3D model. Specifically, 32 full connections of size 32 are obtained from every MRI image after the DAIN model. Then, through the final 2D model, the 32 full connections of the same subject are concatenated together to obtain the feature of size . After this layer of full connection, we add another full connection of size 32. That is, the information learned by the same person from the DAIN model is nonlinearly integrated, and then the integrated features are taken as the final two-dimensional AD features. The 2D model and the 3D model are trained separately. Finally, the full connections of the 2D model and the full connections of the 3D model are concatenated together for training. The method of combining 2D with 3D models is our proposed MVMM framework. Our MVMM framework can learn both global information of 3D images and local information of 2D slices.