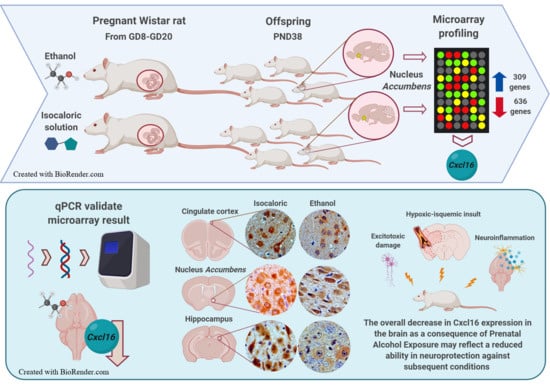
Prenatal Alcohol Exposure in Rats Diminishes Postnatal Cxcl16 Chemokine Ligand Brain Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Prenatal Alcohol Treatment
2.3. Sample Preparation
2.4. Extraction, Quantification, and Differential Expression of mRNA in Microarrays
2.5. Printing, Probe Preparation, Hybridization, Data Acquisition and Analysis of the Array
2.6. Functional Classification of Differentially Expressed Genes
2.7. qPCR mRNA Quantification
2.8. Immunohistochemistry
2.9. Data Analysis
3. Results
3.1. Changes in Patterns of Gene Expression in the Nucleus Accumbens after PAE
3.2. Functional Classification Analysis of Genes Affected by PAE
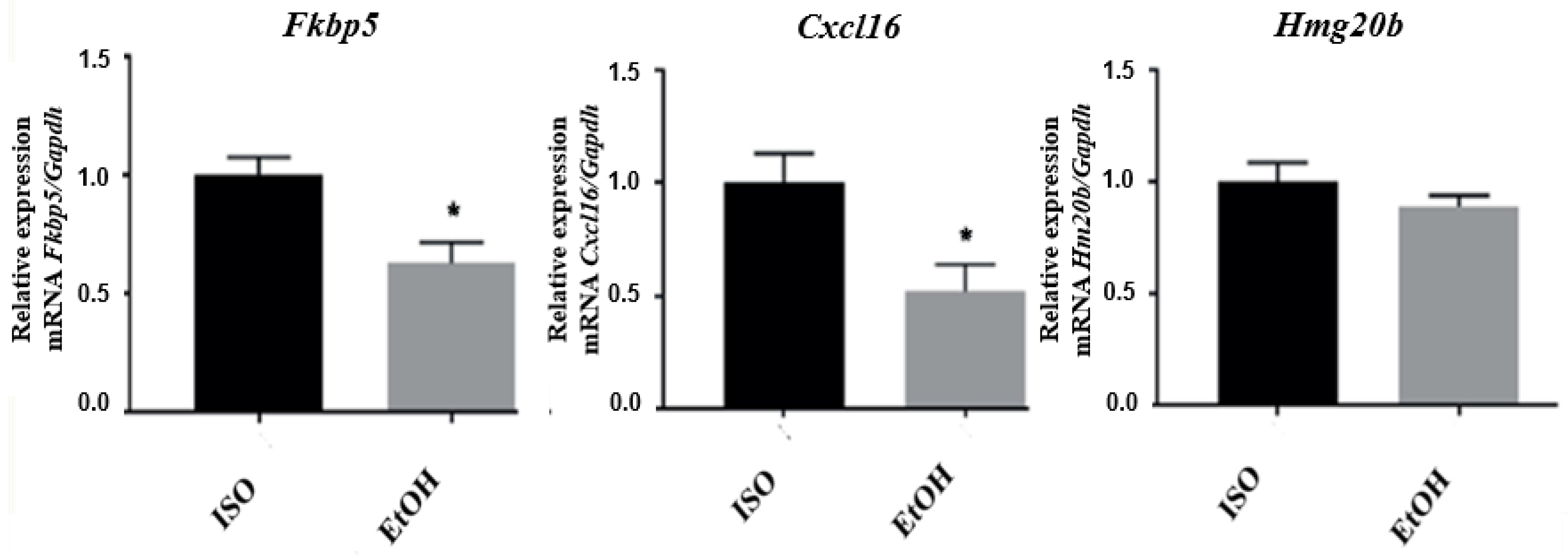
3.3. qPCR Validation of Microarray Results
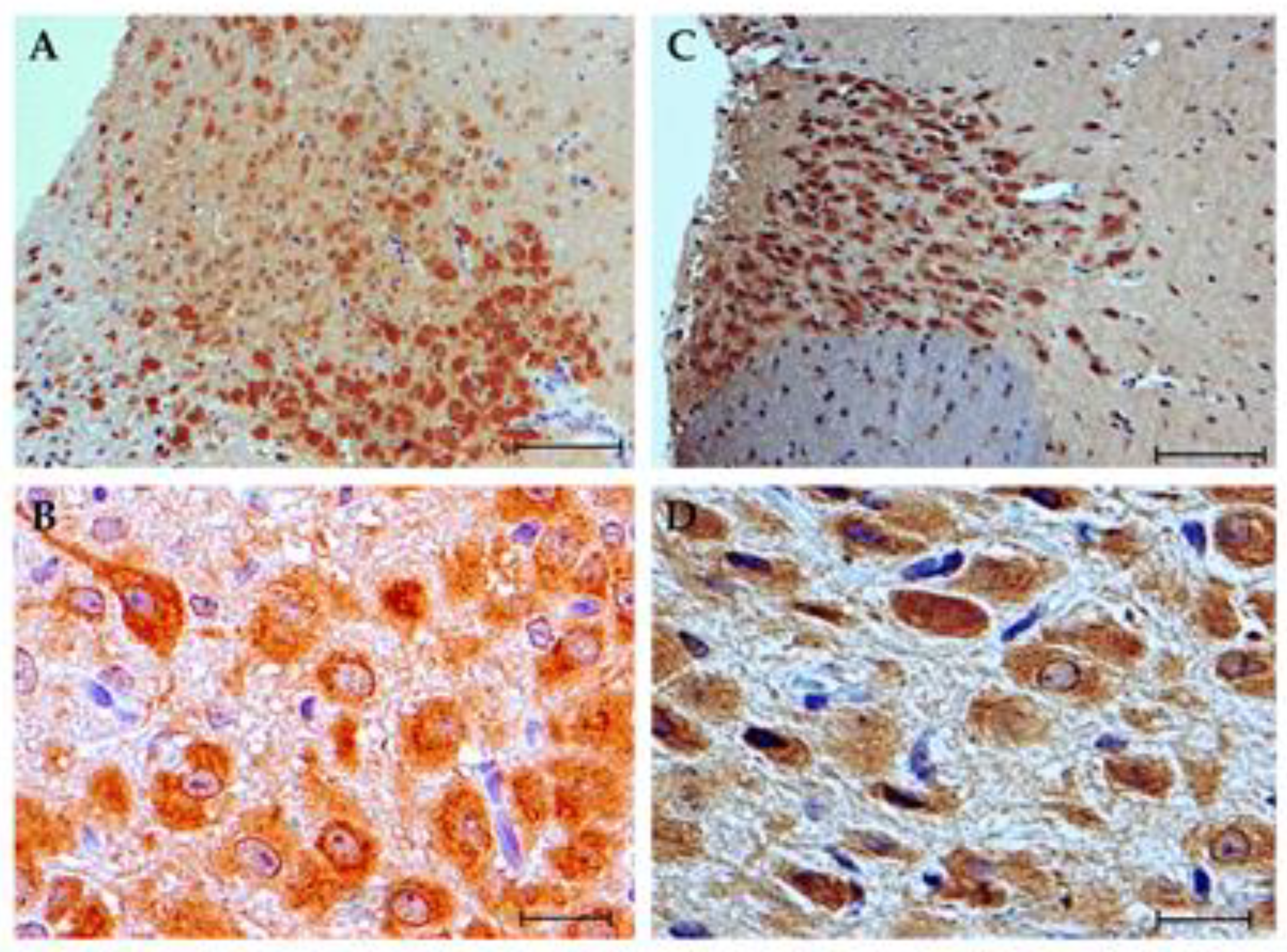
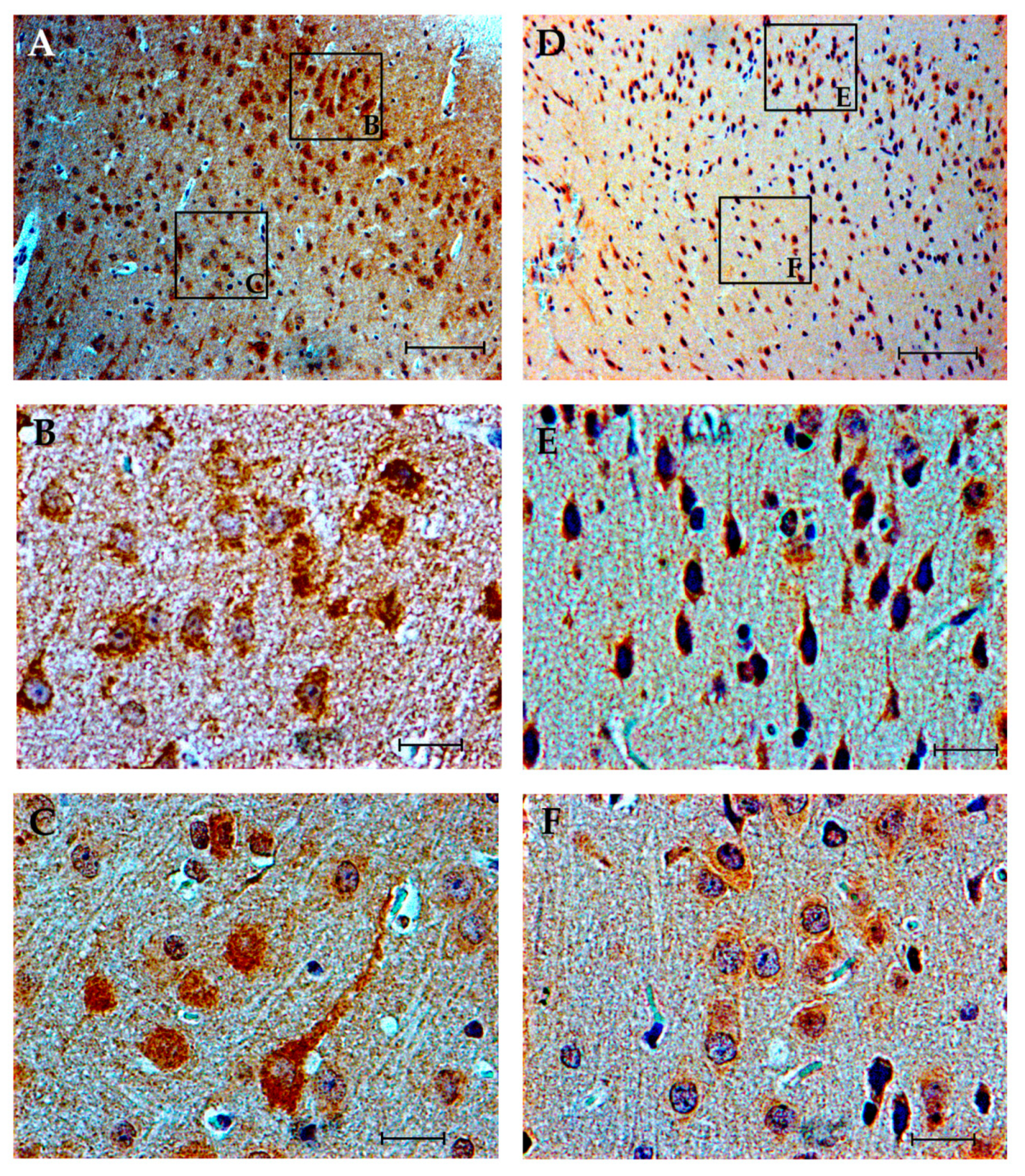
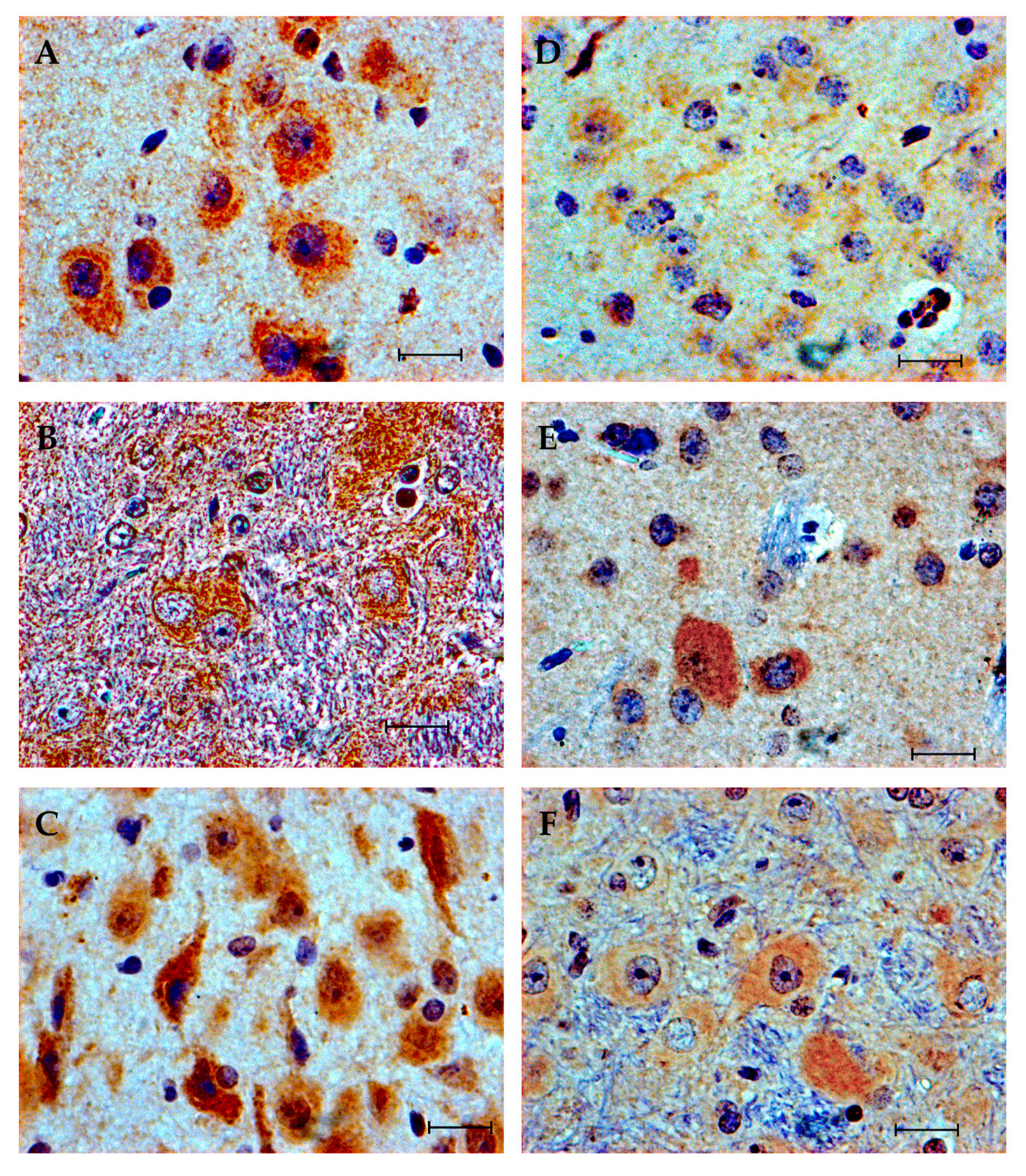
3.4. Immunohistochemistry
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- May, P.A.; Gossage, J.P.; Kalberg, W.O.; Robinson, L.K.; Buckley, D.; Manning, M.; Hoyme, H.E. Prevalence and epidemiologic characteristics of FASD from various research methods with an emphasis on recent in-school studies. Dev. Disabil. Res. Rev. 2009, 15, 176–192. [Google Scholar] [CrossRef]
- Homberg, J.R.; Kyzar, E.J.; Nguyen, M.; Norton, W.H.; Pittman, J.; Poudel, M.K.; Gaikwad, S.; Nakamura, S.; Koshiba, M.; Yamanouchi, H.; et al. Understanding autism and other neurodevelopmental disorders through experimental translational neurobehavioral models. Neurosci. Biobehav. Rev. 2016, 65, 292–312. [Google Scholar] [CrossRef] [PubMed]
- Flak, A.L.; Su, S.; Bertrand, J.; Denny, C.H.; Kesmodel, U.S.; Cogswell, M.E. The association of mild, moderate, and binge prenatal alcohol exposure and child neuropsychological outcomes: A meta-analysis. Alcohol. Clin. Exp. Res. 2014, 38, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, D.A.; Barto, D.; Rodriguez, C.I.; Magcalas, C.M.; Fink, B.C.; Rice, J.P.; Bird, C.W.; Davies, S.; Savage, D.D. Effects of moderate prenatal ethanol exposure and age on social behavior, spatial response perseveration errors and motor behavior. Behav. Brain Res. 2014, 269, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Sarman, I. Review shows that early foetal alcohol exposure may cause adverse effects even when the mother consumes low levels. Acta Paediatr. 2018, 107, 938–941. [Google Scholar] [CrossRef] [PubMed]
- Scott-Goodwin, A.C.; Puerto, M.; Moreno, I. Toxic effects of prenatal exposure to alcohol, tobacco and other drugs. Reprod Toxicol. 2016, 61, 120–130. [Google Scholar] [CrossRef]
- Parkinson, J.A.; Dalley, J.W.; Cardinal, R.N.; Bamford, A.; Fehnert, B.; Lachenal, G.; Rudarakanchana, N.; Halkerston, K.M.; Robbins, T.W.; Everitt, B.J. Nucleus accumbens dopamine depletion impairs both acquisition and performance of appetitive Pavlovian approach behaviour: Implications for mesoaccumbens dopamine function. Behav. Brain Res. 2002, 137, 149–163. [Google Scholar] [CrossRef]
- Basar, K.; Sesia, T.; Groenewegen, H.; Steinbusch, H.W.; Visser-Vandewalle, V.; Temel, Y. Nucleus accumbens and impulsivity. Prog. Neurobiol. 2010, 92, 533–557. [Google Scholar] [CrossRef]
- Burd, L. FASD and ADHD: Are they related and How? BMC Psychiatry 2016, 16, 325. [Google Scholar] [CrossRef]
- Atalar, E.G.; Uzbay, T.; Karakaş, S. Modeling Symptoms of Attention-Deficit Hyperactivity Disorder in a Rat Model of Fetal Alcohol Syndrome. Alcohol Alcohol. 2016, 51, 684–690. [Google Scholar] [CrossRef]
- Rojas-Mayorquín, A.E.; Padilla-Velarde, E.; Ortuño-Sahagún, D. Prenatal Alcohol Exposure in Rodents as a Promising Model for the Study of ADHD Molecular Basis. Front. Neurosci. 2016, 10, 565. [Google Scholar] [CrossRef] [PubMed]
- Hattermann, K.; Ludwig, A.; Gieselmann, V.; Held-Feindt, J.; Mentlein, R. The chemokine CXCL16 induces migration and invasion of glial precursor cells via its receptor CXCR6. Mol. Cell Neurosci. 2008, 39, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Di Castro, M.A.; Trettel, F.; Milior, G.; Maggi, L.; Ragozzino, D.; Limatola, C. The chemokine CXCL16 modulates neurotransmitter release in hippocampal CA1 area. Sci. Rep. 2016, 6, 34633. [Google Scholar] [CrossRef] [PubMed]
- Trettel, F.; Di Castro, M.A.; Limatola, C. Chemokines: Key Molecules that Orchestrate Communication among Neurons, Microglia and Astrocytes to Preserve Brain Function. Neuroscience 2020, 439, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Rosito, M.; Lauro, C.; Chece, G.; Porzia, A.; Monaco, L.; Mainiero, F.; Catalano, M.; Limatola, C.; Trettel, F. Trasmembrane chemokines CX3CL1 and CXCL16 drive interplay between neurons, microglia and astrocytes to counteract pMCAO and excitotoxic neuronal death. Front. Cell Neurosci. 2014, 8, 193. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.E.S.; Goodkey, K.; Footz, T.; Voronova, A. Regulation of CNS precursor function by neuronal chemokines. Neurosci. Lett. 2020, 715, 134533. [Google Scholar] [CrossRef]
- Zhang, K.; Luo, J. Role of MCP-1 and CCR2 in alcohol neurotoxicity. Pharmacol. Res. 2019, 139, 360–366. [Google Scholar] [CrossRef]
- Choong, K.; Shen, R. Prenatal ethanol exposure alters the postnatal development of the spontaneous electrical activity of dopamine neurons in the ventral tegmental area. Neuroscience 2004, 126, 1083–1091. [Google Scholar] [CrossRef]
- Choong, K.C.; Shen, R.Y. Methylphenidate restores ventral tegmental area dopamine neuron activity in prenatal ethanol-exposed rats by augmenting dopamine neurotransmission. J. Pharmacol. Exp. Ther. 2004, 309, 444–451. [Google Scholar] [CrossRef]
- Shen, R.Y.; Hannigan, J.H.; Kapatos, G. Prenatal ethanol reduces the activity of adult midbrain dopamine neurons. Alcohol. Clin. Exp. Res. 1999, 23, 1801–1807. [Google Scholar] [CrossRef]
- Wang, R.; Shen, Y.L.; Hausknecht, K.A.; Chang, L.; Haj-Dahmane, S.; Vezina, P.; Shen, R.Y. Prenatal ethanol exposure increases risk of psychostimulant addiction. Behav. Brain Res. 2019, 356, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Wiss, D.A.; Avena, N.; Rada, P. Sugar Addiction: From Evolution to Revolution. Front. Psychiatry 2018, 9, 545. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.Q.; Karatayev, O.; Halkina, V.; Edelstien, J.; Ramirez, E.; Leibowitz, S.F. Hypothalamic CCL2/CCR2 Chemokine System: Role in Sexually Dimorphic Effects of Maternal Ethanol Exposure on Melanin-Concentrating Hormone and Behavior in Adolescent Offspring. J. Neurosci. 2018, 38, 9072–9090. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.Q.; Karatayev, O.; Boorgu, D.S.S.K.; Leibowitz, S.F. CCL2/CCR2 Chemokine System in Embryonic Hypothalamus: Involvement in Sexually Dimorphic Stimulatory Effects of Prenatal Ethanol Exposure on Peptide-Expressing Neurons. Neuroscience 2020, 424, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Wolraich, M.L.; Hagan, J.F., Jr.; Allan, C.; Chan, E.; Davison, D.; Earls, M.; Evans, S.W.; Flinn, S.K.; Froehlich, T.; Frost, J.; et al. Clinical Practice Guideline for the Diagnosis, Evaluation, and Treatment of Attention-Deficit/Hyperactivity Disorder in Children and Adolescents. Pediatrics 2019, 144, e20192528. [Google Scholar] [CrossRef] [PubMed]
- Semple, B.D.; Blomgren, K.; Gimlin, K.; Ferriero, D.M.; Noble-Haeusslein, L.J. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013, 106–107, 1–16. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Eddy, S.F.; Storey, K.B. Comparative molecular physiological genomics. Heterologous probing of cDNA arrays. Methods Mol. Biol. 2008, 410, 81–110. [Google Scholar] [CrossRef]
- Ortuño-Sahagún, D.; Rivera-Cervantes, M.C.; Gudiño-Cabrera, G.; Junyent, F.; Verdaguer, E.; Auladell, C.; Pallàs, M.; Camins, A.; Beas-Zárate, C. Microarray analysis of rat hippocampus exposed to excitotoxicity: Reversal Na(+)/Ca(2+) exchanger NCX3 is overexpressed in glial cells. Hippocampus 2012, 22, 128–140. [Google Scholar] [CrossRef]
- González-Castillo, C.; Ortuño-Sahagún, D.; Guzmán-Brambila, C.; Márquez-Aguirre, A.L.; Raisman-Vozari, R.; Pallás, M.; Rojas-Mayorquín, A.E. The absence of pleiotrophin modulates gene expression in the hippocampus in vivo and in cerebellar granule cells in vitro. Mol. Cell Neurosci. 2016, 75, 113–121. [Google Scholar] [CrossRef]
- Linares, P.S.; Carvajal, M.A.; Coello, G. Package genArise2. 2010. Available online: http://www.ifc.unam.mx/genarise/ (accessed on 1 August 2020).
- Rojas-Mayorquín, A.E.; Torres-Ruíz, N.M.; Ortuño-Sahagún, D.; Gudiño-Cabrera, G. Microarray analysis of striatal embryonic stem cells induced to differentiate by ensheathing cell conditioned media. Dev. Dyn. 2008, 237, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Fabián, S.; Ortuño-Sahagún, D.; Vázquez-Carrera, M.; López-Roa, R.I. Alliin, a garlic (Allium sativum) compound, prevents LPS-induced inflammation in 3T3-L1 adipocytes. Mediat. Inflamm. 2013, 2013, 381815. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Chong, T.T.-J.; Husain, M. The role of dopamine in the pathophysiology and treatment of apathy. Prog. Brain Res. 2016, 229, 389–426. [Google Scholar] [CrossRef]
- Caballero, J.P.; Scarpa, G.B.; Remage-Healey, L.; Moorman, D.E. Differential Effects of Dorsal and Ventral Medial Prefrontal Cortex Inactivation during Natural Reward Seeking, Extinction, and Cue-Induced Reinstatement. eNeuro 2019, 6, 0296-19. [Google Scholar] [CrossRef]
- Hammes, J.; Theis, H.; Giehl, K.; Hoenig, M.C.; Greuel, A.; Tittgemeyer, M.; Timmermann, L.; Fink, G.R.; Drzezga, A.; Eggers, C.; et al. Dopamine metabolism of the nucleus accumbens and fronto-striatal connectivity modulate impulse control. Brain 2019, 142, 733–743. [Google Scholar] [CrossRef]
- Caldwell, K.K.; Goggin, S.L.; Tyler, C.R.; Allan, A.M. Prenatal alcohol exposure is associated with altered subcellular distribution of glucocorticoid and mineralocorticoid receptors in the adolescent mouse hippocampal formation. Alcohol. Clin. Exp. Res. 2014, 38, 392–400. [Google Scholar] [CrossRef]
- Petrelli, B.; Weinberg, J.; Hicks, G.G. Effects of prenatal alcohol exposure (PAE): Insights into FASD using mouse models of PAE. Biochem. Cell Biol. 2018, 96, 131–147. [Google Scholar] [CrossRef]
- Tunc-Ozcan, E.; Ullmann, T.M.; Shukla, P.K.; Redei, E.E. Low-dose thyroxine attenuates autism-associated adverse effects of fetal alcohol in male offspring’s social behavior and hippocampal gene expression. Alcohol. Clin. Exp. Res. 2013, 37, 1986–1995. [Google Scholar] [CrossRef]
- Ignacio, C.; Mooney, S.M.; Middleton, F.A. Effects of Acute Prenatal Exposure to Ethanol on microRNA Expression are Ameliorated by Social Enrichment. Front. Pediatr. 2014, 2, 103. [Google Scholar] [CrossRef] [PubMed]
- Kleiber, M.L.; Mantha, K.; Stringer, R.L.; Singh, S.M. Neurodevelopmental alcohol exposure elicits long-term changes to gene expression that alter distinct molecular pathways dependent on timing of exposure. J. Neurodev. Disord. 2013, 5, 6. [Google Scholar] [CrossRef]
- Mandal, C.; Halder, D.; Chai, J.C.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. Profiling ethanol-targeted transcription factors in human carcinoma cell-derived embryoid bodies. Gene 2016, 576 Pt 1, 119–125. [Google Scholar] [CrossRef]
- Zhou, F.C.; Zhao, Q.; Liu, Y.; Goodlett, C.R.; Liang, T.; McClintick, J.N.; Edenberg, H.J.; Li, L. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genom. 2011, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Mandal, C.; Park, K.S.; Jung, K.H.; Chai, Y.G. Ethanol-related alterations in gene expression patterns in the developing murine hippocampus. Acta Biochim. Biophys. Sin. 2015, 47, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Lussier, A.A.; Weinberg, J.; Kobor, M.S. Epigenetics studies of fetal alcohol spectrum disorder: Where are we now? Epigenomics 2017, 9, 291–311. [Google Scholar] [CrossRef] [PubMed]
- Wochnik, G.M.; Rüegg, J.; Abel, G.A.; Schmidt, U.; Holsboer, F.; Rein, T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 2005, 280, 4609–4616. [Google Scholar] [CrossRef] [PubMed]
- McClintick, J.N.; Xuei, X.; Tischfield, J.A.; Goate, A.; Foroud, T.; Wetherill, L.; Ehringer, M.A.; Edenberg, H.J. Stress-response pathways are altered in the hippocampus of chronic alcoholics. Alcohol 2013, 47, 505–515. [Google Scholar] [CrossRef]
- Alfonso-Loeches, S.; Guerri, C. Molecular and behavioral aspects of the actions of alcohol on the adult and developing brain. Crit. Rev. Clin. Lab. Sci. 2011, 48, 19–47. [Google Scholar] [CrossRef]
- Ludwig, A.; Schulte, A.; Schnack, C.; Hundhausen, C.; Reiss, K.; Brodway, N.; Held-Feindt, J.; Mentlein, R. Enhanced expression and shedding of the transmembrane chemokine CXCL16 by reactive astrocytes and glioma cells. J. Neurochem. 2005, 93, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.; Sinha, R. The neurobiology of alcohol craving and relapse. Handb. Clin. Neurol. 2014, 125, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Rosito, M.; Deflorio, C.; Limatola, C.; Trettel, F. CXCL16 orchestrates adenosine A3 receptor and MCP-1/CCL2 activity to protect neurons from excitotoxic cell death in the CNS. J. Neurosci. 2012, 32, 3154–3163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xiong, W.; Jackson, M.F.; Parkinson, F.E. Ethanol Tolerance Affects Endogenous Adenosine Signaling in Mouse Hippocampus. J. Pharmacol. Exp. Ther. 2016, 358, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, R. CXCL16 protects against oxygen and glucose deprivation-induced injury in human microvascular endothelial cells-1: Potential role in ischemic stroke. J. Cell Physiol. 2019, 234, 20149–20160. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Gai, H.; Su, R.; Deji, C.; Cui, J.; Lai, J.; Zhu, Y. PI3K-AKT-GSK3β-CREB signaling pathway regulates anxiety-like behavior in rats following alcohol withdrawal. J. Affect Disord. 2018, 235, 96–104. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, J.H.; Song, G.G. Pathway analysis of a genome-wide association study in schizophrenia. Gene 2013, 525, 107–115. [Google Scholar] [CrossRef]
- Chang, G.Q.; Collier, A.D.; Karatayev, O.; Gulati, G.; Boorgu, D.S.S.K.; Leibowitz, S.F. Moderate Prenatal Ethanol Exposure Stimulates CXCL12/CXCR4 Chemokine System in Radial Glia Progenitor Cells in Hypothalamic Neuroepithelium and Peptide Neurons in Lateral Hypothalamus of the Embryo and Postnatal Offspring. Alcohol Clin. Exp. Res. 2020, 44, 866–879. [Google Scholar] [CrossRef]
- Lepore, F.; D’Alessandro, G.; Antonangeli, F.; Santoro, A.; Esposito, V.; Limatola, C.; Trettel, F. CXCL16/CXCR6 Axis Drives Microglia/Macrophages Phenotype in Physiological Conditions and Plays a Crucial Role in Glioma. Front. Immunol. 2018, 9, 2750. [Google Scholar] [CrossRef]

| Functional Annotation | No. of Genes | Enrichment Score | Benjamini |
|---|---|---|---|
| Transcription regulation | 104 | 6.69 | 5.9 × 10−3 |
| Homeobox | 19 | 2.04 | 7.2 × 10−2 |
| Transcription factor fork head | 6 | 2.00 | 1.02 × 100 |
| Nuclear hormone receptor | 7 | 1.87 | 1.0 × 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juárez-Rodríguez, P.; Godínez-Rubí, M.; Guzmán-Brambila, C.; Padilla-Velarde, E.; Orozco-Barocio, A.; Ortuño-Sahagún, D.; Rojas-Mayorquín, A.E. Prenatal Alcohol Exposure in Rats Diminishes Postnatal Cxcl16 Chemokine Ligand Brain Expression. Brain Sci. 2020, 10, 987. https://doi.org/10.3390/brainsci10120987
Juárez-Rodríguez P, Godínez-Rubí M, Guzmán-Brambila C, Padilla-Velarde E, Orozco-Barocio A, Ortuño-Sahagún D, Rojas-Mayorquín AE. Prenatal Alcohol Exposure in Rats Diminishes Postnatal Cxcl16 Chemokine Ligand Brain Expression. Brain Sciences. 2020; 10(12):987. https://doi.org/10.3390/brainsci10120987
Chicago/Turabian StyleJuárez-Rodríguez, Pedro, Marisol Godínez-Rubí, Carolina Guzmán-Brambila, Edgar Padilla-Velarde, Arturo Orozco-Barocio, Daniel Ortuño-Sahagún, and Argelia E. Rojas-Mayorquín. 2020. "Prenatal Alcohol Exposure in Rats Diminishes Postnatal Cxcl16 Chemokine Ligand Brain Expression" Brain Sciences 10, no. 12: 987. https://doi.org/10.3390/brainsci10120987
APA StyleJuárez-Rodríguez, P., Godínez-Rubí, M., Guzmán-Brambila, C., Padilla-Velarde, E., Orozco-Barocio, A., Ortuño-Sahagún, D., & Rojas-Mayorquín, A. E. (2020). Prenatal Alcohol Exposure in Rats Diminishes Postnatal Cxcl16 Chemokine Ligand Brain Expression. Brain Sciences, 10(12), 987. https://doi.org/10.3390/brainsci10120987