Selection of Reliable Reference Genes for Analysis of Gene Expression in Spinal Cord during Rat Postnatal Development and after Injury
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
Spinal Cord Injury Models and Tissue Isolation
2.2. Analysis of Gene Expression
2.2.1. RNA Isolation and cDNA Synthesis
2.2.2. Primer Design and Gene Selection
2.2.3. Quantitative RT-PCR and Normalized Gene Expression
2.2.4. Reference Gene Stability Testing
2.3. Data Representation and Statistics
3. Results
3.1. Comparison of the Expressions of the Tested Genes
3.2. Stability of RGs Expression during Postnatal Development
3.3. Stability of RGs Expression after SCI
3.3.1. Stability of RGs’ Expression after SCI Examined en bloc (cSCI and mSCI Altogether)
3.3.2. Stability of RGs’ Expression after mSCI and cSCI Examined Separately
4. Discussions
4.1. Selection of Candidate RG Set
4.2. Stability of Expressed Genes during Rat Postnatal Development
4.3. Stability of RGs Expression after SCI
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vandesompele, J.; Kubista, M.; Pfaffl, M.W. Reference gene validation software for improved normalization. Real-Time PCR Curr. Technol. Appl. 2009, 4, 47–64. [Google Scholar]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Wyss, M.; Docquier, M.; Pfaffl, M.W.; Masanetz, S.; Borghi, L.; Verbrugghe, P.; Kalaydjieva, L.; Bleuler, S.; Laule, O.; et al. RefGenes: Identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genom. 2011, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Sturzenbaum, S.R.; Kille, P. Control genes in quantitative molecular biological techniques: The variability of invariance. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001, 130, 281–289. [Google Scholar] [CrossRef]
- Rydbirk, R.; Folke, J.; Winge, K.; Aznar, S.; Pakkenberg, B.; Brudek, T. Assessment of brain reference genes for RT-qPCR studies in neurodegenerative diseases. Sci. Rep. 2016, 6, 37116. [Google Scholar] [CrossRef]
- Bemeur, C.; Ste-Marie, L.; Desjardins, P.; Hazell, A.S.; Vachon, L.; Butterworth, R.; Montgomery, J. Decreased beta-actin mRNA expression in hyperglycemic focal cerebral ischemia in the rat. Neurosci. Lett. 2004, 357, 211–214. [Google Scholar] [CrossRef]
- He, Y.; Lv, B.; Huan, Y.; Liu, B.; Li, Y.; Jia, L.; Qu, C.; Wang, D.; Yu, H.; Yuan, H. Zhenbao pill protects against acute spinal cord injury via miR-146a-5p regulating the expression of GPR17. Biosci. Rep. 2018, 38, BSR20171132. [Google Scholar] [CrossRef]
- Ko, W.K.; Kim, S.J.; Jo, M.J.; Choi, H.; Lee, D.; Kwon, I.K.; Lee, S.H.; Han, I.B.; Sohn, S. Ursodeoxycholic Acid Inhibits Inflammatory Responses and Promotes Functional Recovery After Spinal Cord Injury in Rats. Mol. Neurobiol. 2019, 56, 267–277. [Google Scholar] [CrossRef]
- Yang, P.; Chen, A.; Qin, Y.; Yin, J.; Cai, X.; Fan, Y.J.; Li, L.; Huang, H.Y. Buyang huanwu decoction combined with BMSCs transplantation promotes recovery after spinal cord injury by rescuing axotomized red nucleus neurons. J. Ethnopharmacol. 2019, 228, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yang, Y.Y.; Zhou, M.W.; Liu, N.; Xing, H.Y.; Liu, X.X.; Li, F. Ketogenic diet attenuates oxidative stress and inflammation after spinal cord injury by activating Nrf2 and suppressing the NF-kappaB signaling pathways. Neurosci. Lett. 2018, 683, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Mukhamedshina, Y.O.; Akhmetzyanova, E.R.; Kostennikov, A.A.; Zakirova, E.Y.; Galieva, L.R.; Garanina, E.E.; Rogozin, A.A.; Kiassov, A.P.; Rizvanov, A.A. Adipose-Derived Mesenchymal Stem Cell Application Combined With Fibrin Matrix Promotes Structural and Functional Recovery Following Spinal Cord Injury in Rats. Front. Pharmacol. 2018, 9, 343. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Decker, J.T.; Smith, D.R.; Cummings, B.J.; Anderson, A.J.; Shea, L.D. Reducing inflammation through delivery of lentivirus encoding for anti-inflammatory cytokines attenuates neuropathic pain after spinal cord injury. J. Control. Release Off. J. Control. Release Soc. 2018, 290, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, V.K.; Sampathkumar, N.K.; Massaad, C.; Grenier, J. Optimal use of statistical methods to validate reference gene stability in longitudinal studies. PLoS ONE 2019, 14, e0219440. [Google Scholar] [CrossRef] [PubMed]
- Piller, N.; Decosterd, I.; Suter, M.R. Reverse transcription quantitative real-time polymerase chain reaction reference genes in the spared nerve injury model of neuropathic pain: Validation and literature search. BMC Res. Notes 2013, 6, 266. [Google Scholar] [CrossRef] [PubMed]
- Sandercock, D.A.; Coe, J.E.; Di Giminiani, P.; Edwards, S.A. Determination of stable reference genes for RT-qPCR expression data in mechanistic pain studies on pig dorsal root ganglia and spinal cord. Res. Vet. Sci. 2017, 114, 493–501. [Google Scholar] [CrossRef]
- Walder, R.Y.; Wattiez, A.S.; White, S.R.; Marquez de Prado, B.; Hamity, M.V.; Hammond, D.L. Validation of four reference genes for quantitative mRNA expression studies in a rat model of inflammatory injury. Mol. Pain 2014, 10, 55. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, Research0034. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics (Oxf. UK) 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Bangaru, M.L.; Park, F.; Hudmon, A.; McCallum, J.B.; Hogan, Q.H. Quantification of gene expression after painful nerve injury: Validation of optimal reference genes. J. Mol. Neurosci. MN 2012, 46, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Galea, E.; Dupouey, P.; Feinstein, D.L. Glial fibrillary acidic protein mRNA isotypes: Expression in vitro and in vivo. J. Neurosci. Res. 1995, 41, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Dean, B.; Udawela, M.; Scarr, E. Validating reference genes using minimally transformed qpcr data: Findings in human cortex and outcomes in schizophrenia. BMC Psychiatry 2016, 16, 154. [Google Scholar] [CrossRef]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Kutna, V.; Sevc, J.; Gombalova, Z.; Matiasova, A.; Daxnerova, Z. Enigmatic cerebrospinal fluid-contacting neurons arise even after the termination of neurogenesis in the rat spinal cord during embryonic development and retain their immature-like characteristics until adulthood. Acta Histochem. 2014, 116, 278–285. [Google Scholar] [CrossRef]
- Alexovic Matiasova, A.; Sevc, J.; Tomori, Z.; Gombalova, Z.; Gedrova, S.; Daxnerova, Z. Quantitative analyses of cellularity and proliferative activity reveals the dynamics of the central canal lining during postnatal development of the rat. J. Comp. Neurol. 2017, 525, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Sevc, J.; Matiasova, A.; Kutna, V.; Daxnerova, Z. Evidence that the central canal lining of the spinal cord contributes to oligodendrogenesis during postnatal development and adulthood in intact rats. J. Comp. Neurol. 2014, 522, 3194–3207. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.M.; Yaksh, T.L. Neuraxial analgesia in neonates and infants: A review of clinical and preclinical strategies for the development of safety and efficacy data. Anesth. Analg. 2012, 115, 638–662. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Shaydurov, V.A.; Kasianov, A.; Bolshakov, A.P. Analysis of Housekeeping Genes for Accurate Normalization of qPCR Data During Early Postnatal Brain Development. J. Mol. Neurosci. MN 2018, 64, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, N.; Neubert, G.; Issa, L.; Ninnemann, O.; Seiler, A.E.; Kaindl, A.M. Reference genes in the developing murine brain and in differentiating embryonic stem cells. Neurol. Res. 2012, 34, 664–668. [Google Scholar] [CrossRef]
- Tanic, N.; Perovic, M.; Mladenovic, A.; Ruzdijic, S.; Kanazir, S. Effects of aging, dietary restriction and glucocorticoid treatment on housekeeping gene expression in rat cortex and hippocampus-evaluation by real time RT-PCR. J. Mol. Neurosci. MN 2007, 32, 38–46. [Google Scholar] [CrossRef]
- Boda, E.; Pini, A.; Hoxha, E.; Parolisi, R.; Tempia, F. Selection of reference genes for quantitative real-time RT-PCR studies in mouse brain. J. Mol. Neurosci. MN 2009, 37, 238–253. [Google Scholar] [CrossRef]
- Wan, G.; Yang, K.; Lim, Q.; Zhou, L.; He, B.P.; Wong, H.K.; Too, H.P. Identification and validation of reference genes for expression studies in a rat model of neuropathic pain. Biochem. Biophys. Res. Commun. 2010, 400, 575–580. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Luo, D.; Liao, D.J. Pseudogenes as weaknesses of ACTB (Actb) and GAPDH (Gapdh) used as reference genes in reverse transcription and polymerase chain reactions. PLoS ONE 2012, 7, e41659. [Google Scholar] [CrossRef]
- Sevc, J.; Daxnerova, Z.; Miklosova, M. Role of radial glia in transformation of the primitive lumen to the central canal in the developing rat spinal cord. Cell. Mol. Neurobiol. 2009, 29, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shan, Q.; Meng, Y.; Pan, J.; Yi, S. Mrpl10 and Tbp Are Suitable Reference Genes for Peripheral Nerve Crush Injury. Int. J. Mol. Sci. 2017, 18, 263. [Google Scholar] [CrossRef] [PubMed]
- Rhinn, H.; Marchand-Leroux, C.; Croci, N.; Plotkine, M.; Scherman, D.; Escriou, V. Housekeeping while brain's storming Validation of normalizing factors for gene expression studies in a murine model of traumatic brain injury. BMC Mol. Biol. 2008, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.L.; Vink, R.; Donkin, J.J.; van den Heuvel, C. Validation of reference genes for normalization of real-time quantitative RT-PCR data in traumatic brain injury. J. Neurosci. Res. 2009, 87, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.L.; Reeves, T.M.; Phillips, L.L. Injury modality, survival interval, and sample region are critical determinants of qRT-PCR reference gene selection during long-term recovery from brain trauma. J. Neurotrauma 2009, 26, 1669–1681. [Google Scholar] [CrossRef]
- Timaru-Kast, R.; Herbig, E.L.; Luh, C.; Engelhard, K.; Thal, S.C. Influence of Age on Cerebral Housekeeping Gene Expression for Normalization of Quantitative Polymerase Chain Reaction after Acute Brain Injury in Mice. J. Neurotrauma 2015, 32, 1777–1788. [Google Scholar] [CrossRef]
- Plemel, J.R.; Wee Yong, V.; Stirling, D.P. Immune modulatory therapies for spinal cord injury--past, present and future. Exp. Neurol. 2014, 258, 91–104. [Google Scholar] [CrossRef]
- Grube, S.; Gottig, T.; Freitag, D.; Ewald, C.; Kalff, R.; Walter, J. Selection of suitable reference genes for expression analysis in human glioma using RT-qPCR. J. Neuro-Oncol. 2015, 123, 35–42. [Google Scholar] [CrossRef]
- Rohn, G.; Koch, A.; Krischek, B.; Stavrinou, P.; Goldbrunner, R.; Timmer, M. ACTB and SDHA Are Suitable Endogenous Reference Genes for Gene Expression Studies in Human Astrocytomas Using Quantitative RT-PCR. Technol. Cancer Res. Treat. 2018, 17, 1533033818802318. [Google Scholar] [CrossRef]
- Nelissen, K.; Smeets, K.; Mulder, M.; Hendriks, J.J.; Ameloot, M. Selection of reference genes for gene expression studies in rat oligodendrocytes using quantitative real time PCR. J. Neurosci. Methods 2010, 187, 78–83. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, S.; Zhang, L.; You, H.; Huang, R.; Sun, L.; He, P.; Chen, S.; Zhang, H.; Xie, P. Real-time qPCR identifies suitable reference genes for Borna disease virus-infected rat cortical neurons. Int. J. Mol. Sci. 2014, 15, 21825–21839. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.R.; Vieira, H.L.; Duarte, C.B. Effect of carbon monoxide on gene expression in cerebrocortical astrocytes: Validation of reference genes for quantitative real-time PCR. Nitric Oxide Biol. Chem. 2015, 49, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Wierschke, S.; Gigout, S.; Horn, P.; Lehmann, T.N.; Dehnicke, C.; Brauer, A.U.; Deisz, R.A. Evaluating reference genes to normalize gene expression in human epileptogenic brain tissues. Biochem. Biophys. Res. Commun. 2010, 403, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Durrenberger, P.F.; Fernando, F.S.; Magliozzi, R.; Kashefi, S.N.; Bonnert, T.P.; Ferrer, I.; Seilhean, D.; Nait-Oumesmar, B.; Schmitt, A.; Gebicke-Haerter, P.J.; et al. Selection of novel reference genes for use in the human central nervous system: A BrainNet Europe Study. Acta Neuropathol. 2012, 124, 893–903. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, M.; Li, J.; Xiao, H.; Wu, D.; Guo, Q.; Zhang, Y.; Wang, H.; Li, S.; Liao, S. Selection and Validation of Reference Genes for RT-PCR Expression Analysis of Candidate Genes Involved in Morphine-Induced Conditioned Place Preference Mice. J. Mol. Neurosci. 2018, 66, 587–594. [Google Scholar] [CrossRef]
- He, Y.; Yu, S.; Bae, E.; Shen, H.; Wang, Y. Methamphetamine alters reference gene expression in nigra and striatum of adult rat brain. Neurotoxicology 2013, 39, 138–145. [Google Scholar] [CrossRef]
- Taki, F.A.; Abdel-Rahman, A.A.; Zhang, B. A comprehensive approach to identify reliable reference gene candidates to investigate the link between alcoholism and endocrinology in Sprague-Dawley rats. PLoS ONE 2014, 9, e94311. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, X.; Ren, J.; Wang, P.; Zhang, J.; Wei, Z.; Tian, Y. Validation of reference genes for quantitative real-time PCR in valproic acid rat models of autism. Mol. Biol. Rep. 2016, 43, 837–847. [Google Scholar] [CrossRef]
- Gholami, K.; Loh, S.Y.; Salleh, N.; Lam, S.K.; Hoe, S.Z. Selection of suitable endogenous reference genes for qPCR in kidney and hypothalamus of rats under testosterone influence. PLoS ONE 2017, 12, e0176368. [Google Scholar] [CrossRef]
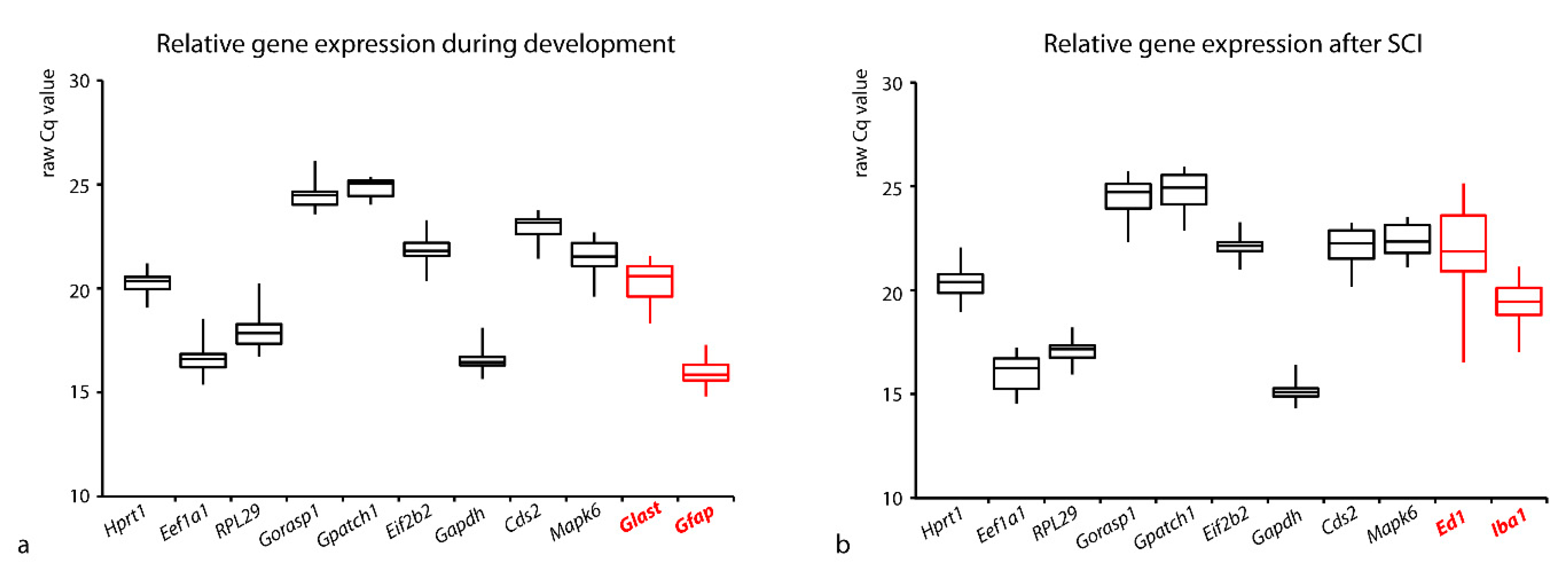
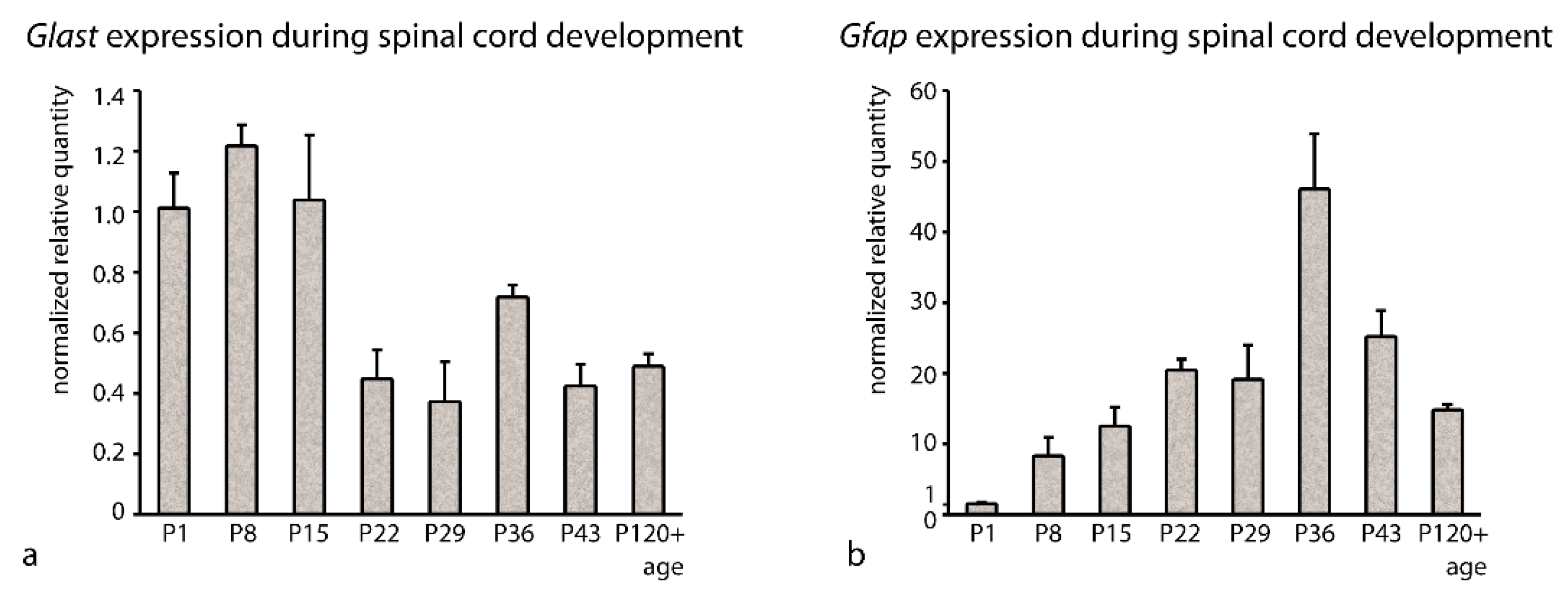
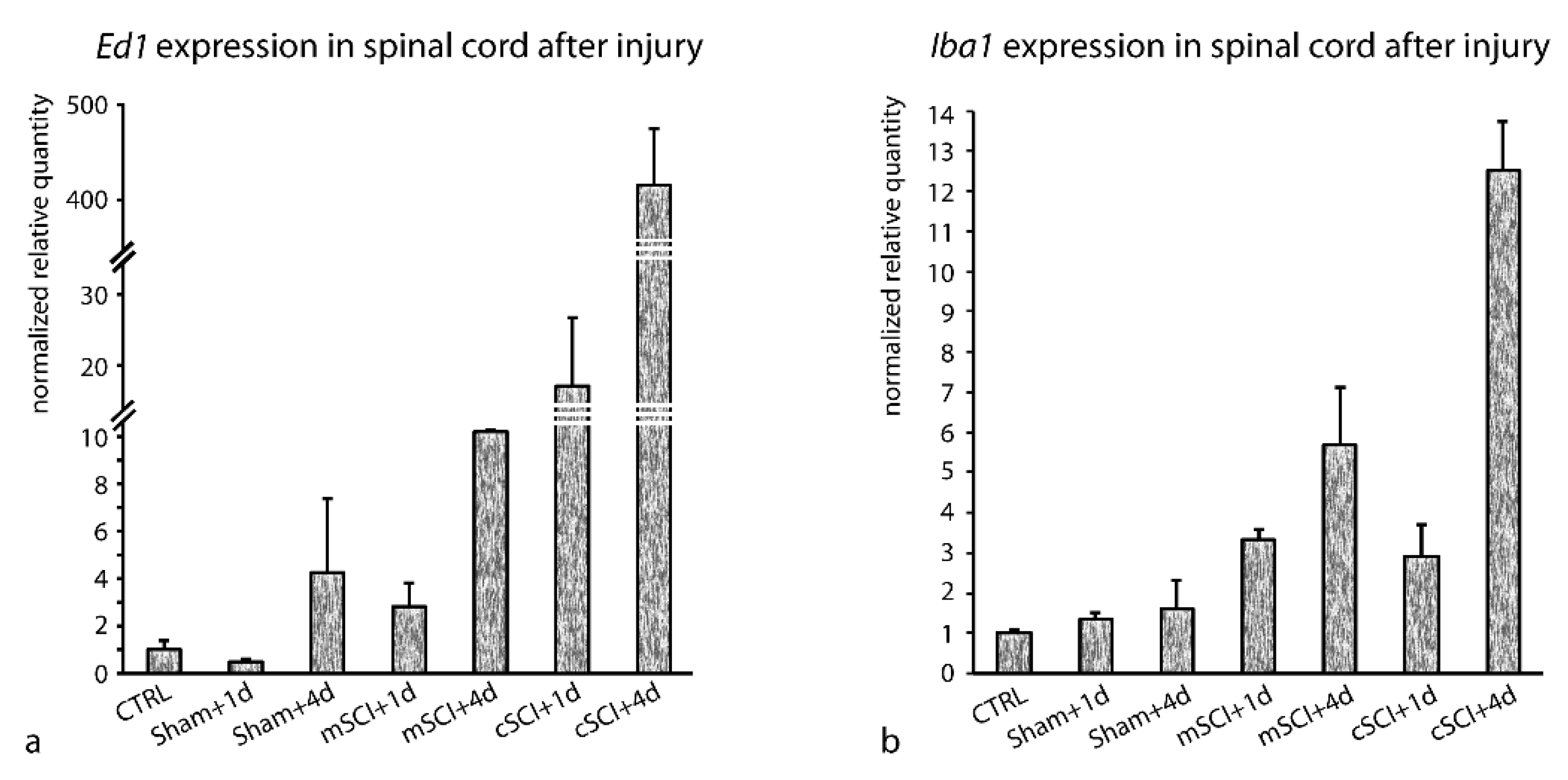
| Gene Symbol | Gene Name | Main Function |
|---|---|---|
| Cds2 | CDP-diacylglycerol synthase 2 | Glycerophospholipid metabolism and phosphatidylinositol signaling system |
| Eef1a1 | eukaryotic translation elongation factor 1 alpha 1 | Protein synthesis |
| Eif2b2 | eukaryotic translation initiation factor 2B subunit beta | Protein synthesis |
| Gapdh | glyceraldehyde-3-phosphate dehydrogenase | Glycolysis |
| Gorasp1 | golgi reassembly stacking protein 1 | Golgi apparatus structure. |
| Gpatch1 | G patch domain containing 1 | Nucleic acid-binding protein |
| Hprt1 | hypoxanthine phosphoribosyltransferase 1 | Metabolic salvage of purines |
| Mapk6 | mitogen-activated protein kinase 6 | Protein kinase |
| Rpl29 | ribosomal protein L29 | Component of ribosomes (60S subunit) |
| Gene Symbol | Acc. No | Sequence for | Sequence Rev | Length (bp) |
|---|---|---|---|---|
| Reference Genes | ||||
| Cds2 | NM_053643 | TGGCTGGAAAACCATGAGGAT | GGTACTGGCAGTCAAAGCGA | 185 |
| Eef1a1 | NM_175838 | TGCTGGAGCCAAGTGCTAAT | GTGCCAATGCCGCCAATTTT | 181 |
| Eif2b2 | NM_032058 | ATCCGCAGAGAGGGTAGGAG | GTGCCTTCCAGTTCCACTAGC | 260 |
| Gapdh1 | NM_017008 | AGACAGCCGCATCTTCTTGT | TGATGGCAACAATGTCCACT | 142 |
| Gorasp1 | NM_019385 | CTGAAGGCTAATGTGGAGAAG | CCAACAATGTAGTCTGTGTAAGG | 239 |
| Gpatch1 | NM_001106246 | GGACCAGCCATCTTCTTGGA | TCTCTCTCGGGTTCTTTGTGA | 226 |
| Hprt1 | NM_012583 | TGCTGGTGAAAAGGACCTCTC | AGATTCAACTTGCCGCTGTCT | 192 |
| Mapk62 | NM_031622 | TAAAGCCATTGACATGTGGG | TCGTGCACAACAGGGATAGA | 129 |
| Rpl29 | NM_017150 | AGTCCAAGAACCACACCACA | ATTCGTATCTTTGTGACCGGGG | 84 |
| Target Genes | ||||
| Aif1 (Iba1) | NM_017196 | CCTCATCGTCATCTCCCCAC | CTCCATGTACTTCGTCTTGAAGG | 214 |
| Cd68 (Ed1) | NM_001031638 | TGGTTCCCAGCCATGTGTTC | TCTGATGTCGGTCCTGTTTG | 209 |
| Gfap | NM_017009 | CACTCAGTACGAGGCAGTGG | ACTCAAGGTCGCAGGTCAAG | 176 |
| Slc1a3 (Glast) | NM_019225 | GACCTCCTCAAGTTCTGCCA | ATCTGGTGATGCGTTTGTCC | 83 |
| geNorm | NormFinder | BestKeeper | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene Rank | Gene | geNorm M | geNorm V | Gene | S Value | Gene | Coeff of Corr. [r] | Std Dev [±Cq] |
| 1 | Eef1a1/Gapdh | 0.378 | --- | Gapdh | 0.196 | Eef1a1 | 0.935 | 0.57 |
| 2 | Eef1a1 | 0.222 | Gapdh | 0.891 | 0.49 | |||
| 3 | Rpl29 | 0.431 | 0.141 | Gpatch1 | 0.231 | Rpl29 | 0.889 | 0.58 |
| 4 | Hprt1 | 0.474 | 0.113 | Hprt1 | 0.270 | Hprt1 | 0.789 | 0.36 |
| 5 | Gpatch1 | 0.502 | 0.095 | Rpl29 | 0.290 | Gorasp1 | 0.767 | 0.56 |
| 6 | Gorasp1 | 0.537 | 0.088 | Gorasp1 | 0.312 | Mapk6 | 0.703 | 0.65 |
| 7 | Eif2b2 | 0.573 | 0.082 | Cds2 | 0.355 | Gpatch1 | 0.669 | 0.33 |
| 8 | Cds2 | 0.610 | 0.079 | Mapk6 | 0.400 | Eif2b2 | 0.646 | 0.58 |
| 9 | Mapk6 | 0.643 | 0.073 | Eif2b2 | 0.407 | Cds2 | 0.590 | 0.49 |
| geNorm | NormFinder | BestKeeper | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene Rank | Gene | geNorm M | geNorm V | Gene | S Value | Gene | Coeff of Corr. [r] | Std Dev [±Cq] |
| 1 | Eif2b2/Gapdh | 0.340 | --- | Eif2b2 | 0.199 | Gapdh | 0.881 | 0.36 |
| 2 | Gapdh | 0.201 | Eif2b2 | 0.879 | 0.38 | |||
| 3 | Gorasp1 | 0.453 | 0.164 | Gorasp1 | 0.313 | Gorasp1 | 0.825 | 0.67 |
| 4 | Hprt1 | 0.498 | 0.118 | Gpatch1 | 0.313 | Gpatch1 | 0.813 | 0.70 |
| 5 | Gpatch1 | 0.538 | 0.104 | Hprt1 | 0.319 | Cds2 | 0.720 | 0.63 |
| 6 | Mapk6 | 0.555 | 0.080 | Mapk6 | 0.352 | Mapk6 | 0.717 | 0.67 |
| 7 | Cds2 | 0.565 | 0.069 | Cds2 | 0.379 | eEF1a1 | 0.690 | 0.78 |
| 8 | Eef1a1 | 0.625 | 0.091 | Eef1a1 | 0.421 | Hprt1 | 0.658 | 0.53 |
| 9 | Rpl29 | 0.689 | 0.094 | Rpl29 | 0.457 | Rpl29 | 0.212 | 0.55 |
| geNorm | NormFinder | BestKeeper | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene Rank | Gene | geNorm M | geNorm V | Gene | S Value | Gene | Coeff of Corr. [r] | Std Dev [±Cq] |
| 1 | Eif2b2/Gapdh | 0.261 | --- | Eif2b2 | 0.162 | Gorasp1 | 0.951 | 0.81 |
| 2 | Eef1a1 | 0.187 | Eif2b2 | 0.945 | 0.46 | |||
| 3 | Eef1a1 | 0.281 | 0.087 | Gapdh | 0.211 | eEF1a1 | 0.916 | 0.52 |
| 4 | Hprt1 | 0.361 | 0.104 | Mapk6 | 0.266 | Gpatch1 | 0.915 | 0.80 |
| 5 | Mapk6 | 0.410 | 0.085 | Hprt1 | 0.277 | Gapdh | 0.888 | 0.37 |
| 6 | Gorasp1 | 0.454 | 0.080 | Gorasp1 | 0.299 | Mapk6 | 0.845 | 0.59 |
| 7 | Gpatch1 | 0.485 | 0.070 | Gpatch1 | 0.307 | Cds2 | 0.844 | 0.67 |
| 8 | Cds2 | 0.508 | 0.061 | Cds2 | 0.328 | Hprt1 | 0.821 | 0.49 |
| 9 | Rpl29 | 0.576 | 0.084 | Rpl29 | 0.428 | Rpl29 | 0.151 | 0.40 |
| geNorm | NormFinder | BestKeeper | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene Rank | Gene | geNorm M | geNorm V | Gene | S Value | Gene | Coeff of Corr. [r] | Std Dev [±Cq] |
| 1 | Hprt1/Gorasp1 | 0.358 | --- | Gapdh | 0.202 | Gapdh | 0.845 | 0.27 |
| 2 | Eif2b2 | 0.260 | Mapk6 | 0.838 | 0.78 | |||
| 3 | Eif2b2 | 0.405 | 0.131 | Hprt1 | 0.268 | Gpatch1 | 0.817 | 0.81 |
| 4 | Gpatch1 | 0.491 | 0.133 | Gpatch1 | 0.334 | Cds2 | 0.787 | 0.81 |
| 5 | Mapk6 | 0.531 | 0.104 | Mapk6 | 0.358 | Eif2b2 | 0.779 | 0.35 |
| 6 | Gapdh | 0.555 | 0.086 | Gorasp1 | 0.373 | Gorasp1 | 0.773 | 0.71 |
| 7 | Cds2 | 0.585 | 0.083 | Eef1a1 | 0.475 | Hprt1 | 0.768 | 0.53 |
| 8 | Eef1a1 | 0.660 | 0.102 | Cds2 | 0.484 | eEF1a1 | 0.496 | 0.67 |
| 9 | Rpl29 | 0.744 | 0.110 | Rpl29 | 0.494 | Rpl29 | −0.112 | 0.60 |
| Experimental Condition | Tissue | Species | Tested Genes | Best Rated RGs | Reference |
|---|---|---|---|---|---|
| development | spinal cord, brain (cerebellum) | mouse, C57BL/6J | Actb, Gapdh, Hsp60, Mrpl10, Pgk1, Ppia, Rpl13a, Rps26, Sdha, Tbp | Mrpl10, Ppia | [15] |
| development | brain (somatosensory cortex, visual cortex) | rat, Wistar | Gapdh, Hprt1, Kif5c, Ospb, Rn18s, Rps18, Tfr1, Uqcrfs1, Ywhaz | Ywhaz, Uqcrfs1 Gapdh, Tfr1 Osbp | [35] |
| development, in vitro differentiation | brain (neocortex), cell culture—mESC | mouse, C57BL/6 | 18S rRNA, Actb, Gapdh, Hprt1, RpII | Gapdh Hprt1 | [36] |
| development | brain (different parts) | mouse, CD-1 | 18s rRNA, B2m, Gapdh, Gusb, Pgk1, Tfrc | Pgk1 | [38] |
| aging, dietary restriction, glucocorticoid treatment | brain (cortex, hippocampus) | rat, Wistar | 18S rRNA, Actb, Gapdh, Cypb | Actb, Gapdh | [37] |
| PNI | dorsal root ganglia | rat, Sprague-Dawley | 18s rRNA, Act, Gapdh, Hprt1, Mapk6, Tubb3, Tubb5 | Mapk6, Gapdh | [24] |
| PNI | sciatic nerve, dorsal root ganglia | rat, Sprague-Dawley | 18S rRNA, Actb, Ankrd27, CypA, Gapdh, Hprt1, Mrpl10, Pgk1, Rictor, Tbp, Ubc, Ubxn11, Ywhaz | Mrpl10, Tbp | [42] |
| SNI | spinal cord, dorsal root ganglia | rat, Sprague-Dawley | 18S rRNA, Actb, Gapdh, Hmbs, Hprt1, Rpl13a, Rpl29 | Rpl29, Rpl13a Hprt1, Actb | [16] |
| neuropathic pain | dorsal root ganglia | rat, Sprague-Dawley | Actb, Gapdh, Hmbs, Rpl3, Rpl19, Rpl29 | Rpl29, Rpl3 | [39] |
| inflammatory injury | spinal cord | rat, Sprague-Dawley | Actb, B2m, Hprt1, Mapk6 | Actb, B2m, Hprt1, Mapk6 | [18] |
| TBI | brain | mouse, CD-1 | 18S rRNA, Actb, B2m, Gapd, S100b | 18S rRNA, Gapdh | [43] |
| TBI | brain (cortex, hippocampus) | rat, Sprague-Dawley | B2m, Gapdh, Gusb, Hprt1, Tbp, Sdha | Hprt1, Sdha, Gusb B2m, Tbp, Gapdh | [44] |
| TBI | brain (hippocampus, parietotemporal cortex) | rat, Sprague-Dawley | 18S rRNA, Actb, Cyca, Gapdh | Actb, Ppia | [45] |
| TBI, aging | brain (hemispheres) | mouse, C57BL/6N | 18S rRNA, Actb, B2m, Gapdh, Hprt1, Pbgd, Ppia, S100b | Hprt1, Ppia | [46] |
| cancer (astrocytoma) | cell culture—astrocytoma | human | B2M, CYC1, GAPDH, HMBS, HPRT1, RPL13a, SDHA, TBA, YWHAZ | GAPDH, RPL13A, CYC1 | [48] |
| cancer (gliomas) | cell culture—glioma | human | ACTB, GAPDH, POLR2A, RPL13A, SDHA, TBP | ACTB, SDHA | [49] |
| in vitro (differentiation0 | cell culture—Oligodendrocytes | rat, Wistar | 18S rRNA, Actb, Cyca, Gapdh, Hmbs, Hprt1, Pgk1, Rpl13A, Tbp, Ywhaz | Cyca, Pgk1, Rpl13A, Ywhaz | [50] |
| in vitro (Borna disease virus infection) | cell culture—primary cortical neurons | rat, Sprague-Dawley | 18S rRNA, Actb, Arbp, B2m, Gapdh, Hprt1, Ppia, Rpl13a, Tpp, Ywhaz | Arbp, Actb | [51] |
| in vitro (treatment with carbon monoxide) | cell culture—cortical astrocytes | mouse, C57BL/6 | Actg1, Gapdh, Hprt1, Pgk1, Ppia, Rn18s, Sdha, Tbp | Gapdh, Ppia | [52] |
| Disease—neurodegenerative diseases | brain (prefrontal cortex, cerebellum) | human | ACTB, ATP5B, B2M, CYC1, EIF4A2, GAPDH, HMBS, HPRT1, PPIA, PUM1, RPL13, SDHA, TBP, TOP1, UBE2D2, UBC | UBE2D2, CYC1, RPL13 | [7] |
| Disease—epilepsy | brain (neocortex temporal lobe) | human | ACTB, B2M, CYPA, GAPDH, HPRT1, MAP-2, MRPL, NNE, SDHA, SYP, TBP, UBC | SYP, NSE, MRPLl | [53] |
| Disease—neurodegenerative disorders | CNS (brain, spinal cord) | human | AARS, ATP5E, BECN1, CSNK2B, DCTN2, GAPDH, GAPVD1, OSBP, QARS, NAT5, TUBB, XPNPEP1 | XPNPEP1 | [54] |
| Neuroplasticity—morphine addiction | brain (caudate putamen, hippocampus) | mouse, C57BL/6J | Actb, B2m, Gapdh, Hmbs, Hprt1, Oaz1, Rps6, Tbp | Tbp Tbp, Oaz1 | [55] |
| Neuroplasticity—methamphetamine | brain (striatum, substantia nigra) | rat, Sprague-Dawley | 18S rRNA, B2m, Actb, Gapdh, Hmbs, Hprt1, Oaz1, Rps6, Tbp, Ubc | Actb,, Gapdh, Hprt1, Rps6 | [56] |
| Neuroplasticity—alcoholism, estrogen | brain, hearth | rat, Sprague-Dawley | U2, U5a, U6, U87, Z39, 5S rRNA, 18S rRNA, Actb, B2m, Gadd45af, Gapdh, Hprt1, Tbp, Tnks, Ubc | U87, 5S rRNA, Gapdh, U5a | [57] |
| autism | brain (prefrontal cortex, hippocampus) | rat, Sprague-Dawley | Actb, Gapdh, Hmbs, Hprt1, Ppia, Rpl13a, Rps18, Tbp, Ywhaz | Hprt1 Hmbs, Tbp | [58] |
| testosterone treatment | brain (hypothalamus), kidney | rat, Sprague-Dawley | Actb, B2m, Gapdh, Hmbs, Hprt1, Ppia | Hmbs, Ppia Hmbs, Gapdh | [59] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Košuth, J.; Farkašovská, M.; Mochnacký, F.; Daxnerová, Z.; Ševc, J. Selection of Reliable Reference Genes for Analysis of Gene Expression in Spinal Cord during Rat Postnatal Development and after Injury. Brain Sci. 2020, 10, 6. https://doi.org/10.3390/brainsci10010006
Košuth J, Farkašovská M, Mochnacký F, Daxnerová Z, Ševc J. Selection of Reliable Reference Genes for Analysis of Gene Expression in Spinal Cord during Rat Postnatal Development and after Injury. Brain Sciences. 2020; 10(1):6. https://doi.org/10.3390/brainsci10010006
Chicago/Turabian StyleKošuth, Ján, Martina Farkašovská, Filip Mochnacký, Zuzana Daxnerová, and Juraj Ševc. 2020. "Selection of Reliable Reference Genes for Analysis of Gene Expression in Spinal Cord during Rat Postnatal Development and after Injury" Brain Sciences 10, no. 1: 6. https://doi.org/10.3390/brainsci10010006
APA StyleKošuth, J., Farkašovská, M., Mochnacký, F., Daxnerová, Z., & Ševc, J. (2020). Selection of Reliable Reference Genes for Analysis of Gene Expression in Spinal Cord during Rat Postnatal Development and after Injury. Brain Sciences, 10(1), 6. https://doi.org/10.3390/brainsci10010006





