Featured Application
Nanodiscs formed by styrene maleic acid copolymers allow separation of biologically intact protein complexes with native ligands for syst-OMICS analysis.
Abstract
The omics disciplines remain largely distinct sciences due to the necessity of separating molecular classes for different assays. For example, water-soluble and lipid bilayer-bound proteins and metabolites are usually studied separately. Nonetheless, it is at the interface between these sciences where biology happens. That is, lipid-interacting proteins typically recognize and transduce signals and regulate the flow of metabolites in the cell. Technologies are emerging to converge the omics. It is now possible to separate intact membrane:protein assemblies (memteins) directly from intact cells or cell membranes. Such complexes mediate complete metabolon, receptor, channel, and transporter functions. The use of poly(styrene-co-maleic acid) (SMA) copolymers has allowed their separation in a single step without any exposure to synthetic detergents or artificial lipids. This is a critical development as these agents typically strip away biological lipids, signals, and metabolites from their physiologically-relevant positions on proteins. The resulting SMA lipid particles (SMALPs) represent native nanodiscs that are suitable for elucidation of structures and interactions that occur in vivo. Compatible tools for resolving the contained memteins include X-ray diffraction (XRD), cryo-electron microscopy (cryoEM), mass spectrometry (MS), and nuclear magnetic resonance (NMR) spectroscopy. Recent progress shows that memteins are more representative than naked membrane proteins devoid of natural lipid and is driving the development of next generation polymers.
1. Advances and Challenges in Proteomics
Proteomics involves analysis of all the proteins from an organelle or cellular system. The data can include quantified protein levels, interactions, modifications, and turnover over a period of time or space. The information should be very accurate and reproducible given the high throughput data that is increasingly collected at multiple sites for clinical studies. Proteins can be classified as soluble, transmembrane, or peripheral membrane. Quantitative analyses of those that are membrane associated are particularly troublesome due to low abundance, hydrophobicity, and poor solubility issues, and hence, membrane proteome can be severely under-represented in proteomic studies [1,2]. This is a concern as membrane proteins are intricately involved in disease mechanisms and represent the majority of drug targets.
Proteomics analysis is usually carried out using mass spectrometry (MS) due to its high sensitivity and throughput. Both top-down and bottom-up approaches are employed. Top-down proteomics involves direct elucidation of intact proteins which are fractionated by gel electrophoresis or liquid chromatography (LC), and then fragmented and identified [3]. This approach is particularly useful for identifying isoforms and post-translational modifications (PTMs) of soluble proteins, less so for intact membrane proteins which are harder to separate in gels and columns. The technical challenges include generating minimally biased fractionation, ionization, and fragmentation of intact membrane protein complexes or memteins [4] for MS analysis in the gas phase.
Bottom-up proteomics improves protein solubilization through the use of a denaturing detergent followed by digestion with a protease such as trypsin. The resulting peptides are then fractionated and identified based on MS-based fragmentation patterns. This approach reduces the complexity of the original heterogeneous mixtures and bypasses the problems associated with analyzing intact proteins through ionization and MS characterization. However, the detergents, such as sodium dodecyl sulfate (SDS), that are commonly used to solubilize or unfold the proteins to enhance protease access are also problematic. In particular, they compromise protease function and produce MS peak clusters that obscure peptide signals. Moreover, the buffers and salts used in the biological mixtures lead to matrix effects that cause MS signal suppression or enhancement. Alternative proteomic-based protocols that use liposomes rather than detergents face challenges due to the heterogeneity and hydrophobic crevasses in vesicles which tend to trap nonspecific interactors. Hence, improved sample preparation methods are desirable, particularly for removal of detergents and salts that compromise data read-out and that would allow accurate and reproducible analysis of membrane proteins.
Proteomics protocols typically dissociate multimers and complexes with ligands including lipids, signaling molecules, and metabolites, particularly when detergents are used. Nonetheless, such interactions provide vital biological information, and ideally should be maintained during preparation. By analogy, one can count all the people in a room, but not know which ones are talking to one another let alone what they are saying. That is, proteomics can measure protein levels but not necessarily give insight into what the proteins are doing unless complexes can be kept intact to measure structures and kinetics. Hence, the preparative approaches should be compatible with parallel assays to characterize ligand interactions and high-resolution structures, thus yielding not only the players but also their partners and mechanisms.
Detergent-based protocols confront proteomics and structural biology experts with problems that need to be addressed. Amongst the developments are the use of native nanodiscs to separate intact memteins. Water-soluble nanodiscs surrounded by a membrane protein scaffold (MSP) can be used to solubilize memteins from, for example, the bacterial envelope for MS analysis and display lipid dependent protein-protein interactions [5]. Amphipols are polymers that have been designed to stabilize membrane proteins and can release them into the gas phase for MS analysis [6,7]. However, neither are not designed to extract memteins directly from native membranes without any exposure to detergent. A special class of synthetic polymers have been invented for detergent-free separation of native membrane:protein complexes from biological matter into nanodiscs along with their associated ligands and lipids for analysis by virtually any structural or biophysical assay.
2. Progress and Problems in Metabolomics and Lipidomics
Metabolomics involves the detection and quantification of all metabolites in a biological system, while lipidomics focuses on the subset of metabolites that are lipids [8]. The difference is pragmatic, with the lipids being extracted into chloroform and methanol or a similar nonpolar solvent mixture prior to analysis, while most other metabolites are water soluble. The challenge of full coverage is daunting, with over 200,000 structurally diverse metabolites of molecular weight <1500 Da at concentrations that span 12 orders of magnitude and fluctuate over many timescales [9]. Being able to capture rolling snapshots of this chemical diversity is critical for finding biomarkers that respond to stimulation or intervention and mirrors the challenges faced in proteomics.
The MS and nuclear magnetic resonance (NMR) methods play complementary roles in metabolomics and lipidomics analysis, providing high throughput and in-depth information, respectively. Both techniques have made gains in sensitivity although they still report on only a small fraction of the metabolome at a time. Both methods are used for either untargeted approaches to assess the broadest diversity of classes or for targeted approaches that employ protocols to separate metabolites into subgroups. The lipids can be divided into eight major groups; fatty acyls, glycerolipids, glycerophospholipids, polyketides, prenol lipids, saccharolipids, sphingolipids, and sterol lipids [10]. The universe of water-soluble metabolite is much larger and can be divided into over one hundred metabolic pathways. The stabilities of the compounds vary greatly. They change in concentration or composition during not only physiological processes but also purification and sampling protocols. As each can constitute a valuable biomarker if precisely measured, being able to accurately detect the structures and concentrations of a range of metabolites is crucial.
Measuring metabolites is nontrivial as their levels can be perturbed during each stage of sample preparation. The ionization efficiencies of various lipids for MS data collection depends on polar head and fatty acid structures. Due to the many isomeric and isobaric species, such perturbations can be hard to assess using NMR or MS. This necessitates the use of individual standards for each type of lipid as well as controls and detailed annotation of protocols for accurate quantitation and universal validation. Even so, accurately calibrated measurement of the absolute concentrations of pure lipid species in complex mixtures remains elusive.
Clearly, there is an ongoing need for minimally perturbing methods that do not significantly alter metabolite or lipid levels and compositions. Factors to consider for protocols include coverage and recovery, simplicity, reproducibility, cost, and feasibility of scaling. For extremely low-abundance compounds, a method for enrichment is needed, and chromatography has often sufficed. The use of nanoparticles to collect low abundance metabolites for sensitive detection using NMR or MS represents a new opportunity [11] that could be exploited by styrene maleic acid lipid particles (SMALPs). As SMA binds lipids to form nanoparticles, it could potentially be used to obtain information about hydrophobic material in an organelle, biofluid, or environmental solutions. The SMALP system addresses many of the requirements in that no lipid bias is apparent; it provides low-cost, rapid 1-step solubilization into nanodiscs that are compatible with most chromatography systems. Because a 10–20 nm diameter section of membrane is typically captured intact, the original arrangement of contents within the membrane is at least partly retained. By displaying nanodisc arrays on a surface, the contents of each disc could be assessed using MS imaging (MSI) [12]. Moreover, the SMALP systems could potentially be automated for screening protocols, particularly with the tagged polymers that are in development.
An example of a metabolic system that has been studied in SMALPs is the dhurrin biosynthesis metabolon. Like many other processive enzymatic machines, it includes several components which are co-localized in membranes. Isolating the intact metabolon with all components in place is technically challenging, as demonstrated by a sorghum metabolon that produces the glucoside dhurrin [13]. This assembly includes multiple copies of three membrane proteins and a soluble glycotransferase protein. This memtein can be solubilized intact from microsomes using SMA(2:1) to yield 10–25 nm diameter nanodiscs. Conventional detergents such as cholate destroy the integrity of this assembly, while the soluble subunit binds to and modulates the SMALP’d memtein, the activity of which depends on retained negatively charged phospholipids.
3. Amphipathic Polymers and Nanodisc Formation
A growing family of SMA copolymers are being used to separate membranes into native nanodiscs that retain the original protein:lipid complexes found in cells. These polymers contain sequences of styrene (S) and maleic acid (MA) residues or their derivatives and are activated by hydrolysis of styrene maleimide (Figure 1). The most commonly used ratios of S:MA residues are between 2:1 and 3:1, which provides an optimal balance of hydrophobicity and polarity to allow insertion into bilayers while maintaining solubility and membrane fragmentation activity. The negatively charged MA sidechains provide solubility to the polymers and nanodiscs across the physiological pH range. Addition of the polymer at a critical concentration of around 1% to whole cells or membranes spontaneously yields water soluble nanodiscs that are stable and can be reconstituted after freeze-drying and storage while retaining protein activity [14]. A variety of SMA reagents are available commercially and more are in development. Polyscope (Polyscope, Geleen, Netherlands) offers XIRAN® 25010 and 30010 reagents, which have S:MA ratios of 3:1 and 2.3:1, respectively. Hydrolyzed SMA2000 and SMA3000 copolymers from Cray Valley (USA) contain S:MA ratios of 2:1 and 3:1, respectively, and are similar to Lipodisq® reagents. Thus far, the SMA2000 and the XIRAN reagents have been widely used for solubilizing bacterial, fungal, plant, and mammalian membrane proteins that contain anywhere between monomeric and multimeric proteins with 1 to 48 transmembrane α-helices [15,16,17].
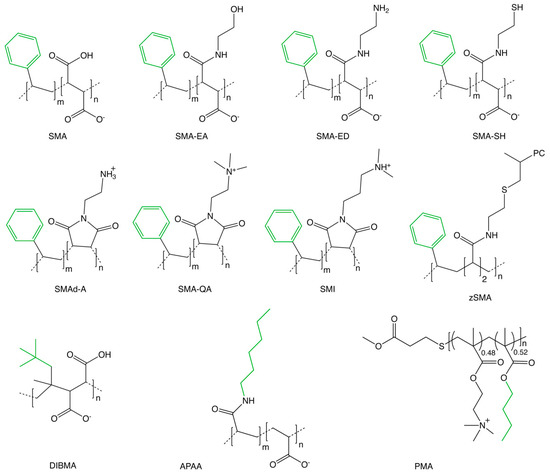
Figure 1.
Family of SMA copolymers used to solubilize memteins. The chemical structures of copolymers developed for solubilizing membrane:protein complexes from biological membranes into native nanodiscs include: SMA [20], SMA-EA [34], SMA-ED, SMAd-A [36], SMA-QA [37], SMI [38], SMA-SH [43], zSMA [39], DIBMA [40], APAA [42], and PMA [41]. The non-polar sidechains are colored green.
The nanodiscs formed spontaneously by adding activated SMA solutions have diameters of around 10 nm, although they can range from 6 to 30 nm depending on the protocol and SMA variant. For example, SMA(3:1) polymer solubilizes liposomes made of synthetic lipids and transfers their contents into 6–10 nm nanodiscs [18]. A fuzzy annulus of polydispersed polymers surrounds an inner patch of bilayer, as seen by small angle neutron scattering (SANS) experiments using hydrogenated and deuterated lipids [19]. The styrenes interact with the acyl tails of lipids as can be evidenced by intermolecular contacts observed using NMR [19], although the length and sequence polydispersity of current polymers as well as their non-specific interactions with memteins generally limit the experimental resolution.
The mechanism by which SMA forms nanodiscs involves at least two stages (Figure 2). The copolymer attaches to the membrane by nonspecific insertion of hydrophobic styrene rings between the lipid acyl tails while preserving conformations or the protein and directly associated lipid [20]. Above a critical concentration, the accumulated copolymer destabilizes and perforates the bilayer, which fragments to form nanodiscs surrounded by a boundary of multiple polymer molecules. The molecular processes involved can be modelled by molecular dynamics simulations, as illustrated by trajectories of SMA(2:1) and SMA(3:1) copolymers as they bind and perforate dimyristoylphosphatidylcholine (DMPC) bilayer and extract lipid patches into nanodiscs [21].
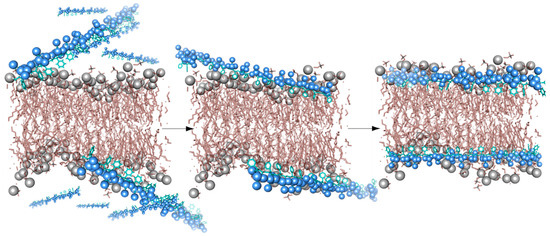
Figure 2.
A schematic model showing interactions between SMA polymers (blue) and a membrane bilayer (grey).
Unlike classical detergents, SMA polymers have very mild solubilizing activity and hence induce minimal alteration of memteins [22,23], allowing the position of protein-bound lipids to be kept intact including the packing of lipid acyl chains [19,24]. The optimal pH range at which SMA copolymers are effective at solubilizing membranes is between 7 and 9. Low salt levels can increase polymer solubility further [25], although higher salt may be necessary in the presence of millimolar divalent cations. Such cations otherwise bind the negatively charged MA groups and could cause polymer precipitation. A ratio of 2:1 for S:MA ratios appears most effective at solubilizing as this perturbs the lipid bilayer least [19], although other polymer types will behave differently. Comparison of the solubilizing activities of various SMAs indicated that molecular weights in the range of 10 kDa were most effective at solubilizing monomers, dimers, trimers, and tetramers of the Rhodobacter sphaeroides reaction center [26]. Higher yields of large oligomeric proteins in nanodiscs can be produced by adding lipids to membranes or employing longer SMA sequences. This can yield 50‒100 nm diameter discs depending on the nature of the protein and polymer used. Essentially, any molecules found within a native membrane can be incorporated into a SMALP. The lipids species can then be identified using MS or thin-layer chromatography, as described below. Exchange of lipid molecules between discs can occur through collisions [27], and exogenous lipids or proteins can be introduced or removed readily [28].
There is little or no apparent bias of SMA for solubilization of lipid species, as the styrene groups insert between the acyl tails in the bilayer through nonspecific hydrophobic interactions. However, SMA copolymers preferentially solubilize fluid bilayers and lipids with short or unsaturated acyl chains [29] over highly ordered membrane with tightly packed lipids and proteins [26]. These order specifies for dynamic regimes can be probed by incorporating spin labels [30]. The plasma and intracellular membranes in live cells can be seen to disintegrate with SMA treatment at different rates using fluorescence microscopy [31]. As SMA(2.3:1) encounters the outside of mammalian cells, the plasma membrane is fragmented first, followed by the inner membranes, as seen by the release of fluorescent proteins from cytosolic and organelle compartments. This is consistent with SMA binding initially to the liquid disordered regions before perforating the plasma membrane to allow cytosolic contents to be released. The intracellular membranes are bound and perforated next. Alterations in the length or concentration of polymer, temperature and inclusion of exogenous lipids such as DMPC provide ways to progressively liberate memteins from outer and inner membrane compartments into SMALPs [20]. These avenues could conceivably be exploited for progressive release of proteomes from compartmentalized intracellular layers or fragmentation and affinity-purification of distinct organelle or membrane microenvironments [32].
4. SMA Polymer Derivatives
The fields enabled by native nanodiscs are evolving through the emergence of new polymers to improve performance and enable new applications. A growing family of polymers that spontaneously fragment membranes into discs have been designed and tested (Figure 1). A systematic comparison of twelve SMA polymer types and conditions for solubilization of intact photosynthetic systems from spinach thylakoid membranes indicated those that best retain the activity and subunit composition based on chlorophyll fluorescence emission spectra, native gel electrophoresis, and immunoblotting. This narrowed the field to five that were most efficient in this case, including SMA 1440, XIRAN 25010, XIRAN 30010, SMA PRO10235, and SMA 1735, and suggests room for further optimization. Improvements to simplify separation and detection of the multiple transmembrane assemblies were found including detachment of peripheral membrane proteins by varying buffer conditions [33].
Specific derivatives have been synthesized to enhance functionality and operational range of polymers under a wider pH range and cation concentrations. This allows production of stable disks of different sizes, allowing larger memteins to be solubilized and stored. The maleic anhydride group can be modified, allowing addition of various moieties. An ethanolamine group was added to a SMA(1.3:1) polymer having an Mn of 1.6 kDa, the resulting SMA-EA polymer solubilizes liposomes over a wide range of pH, temperature, salt, and cation levels to produce nanodiscs of diameters between 10 and 50 nm [34,35]. The solution state NMR spectra of 15N labelled cytochrome b5 protein in SMA-EA discs display well-resolved signals indicating a folded state. When lanthanide ions such as Yb3+ are added the SMA-EA discs align with external magnetic fields, allowing transmembrane helix tilt angles to be measured using solid-state NMR. An ethylenediamine group was attached to the SMA(1.3:1) to produce the zwitterionic polymer SMA-ED, which solubilizes vesicles below pH 5 and over pH 7 [36]. This polymer can be dehydrated to yield the maleimide form SMAd-A, which also solubilizes DMPC vesicles at pH values below 6 where its amine group is positively charged. Both ethylenediamine-modified forms are effective solubilizers in high salt and divalent cation concentrations. These studies extend the range of conditions for solubilizing membranes by SMA-type polymers and allow virtually any membrane to be turned into nanodiscs.
Positively charged versions of SMA are also effective at solubilizing membranes into nanodiscs. The addition of a quaternary functional group to SMA(1.3:1) yields a cationic version that fragments vesicles into nanodiscs with diameters in the range of 10–30 nm depending on the ratio of added DMPC. This SMA-QA polymer is an effective solubilizer at pH values between 2 and 10 and tolerates high metal concentrations [37]. The incorporation of a positively charged group in styrene maleimide polymers yields nanodiscs from membrane at pH values below 7.8 and even in high concentrations of divalent cations [38]. Their discs have diameters of 6 nm and are stable up to 80 °C. The ZipA protein plays a role in bacterial cell division and purified well in SMI-based nanodiscs with yields that are only slightly reduced compared to those in the case of SMALPs. The human adenosine A2A receptor solubilized in SMI nanodiscs is able to bind ligand normally. The availability of both anionic SMA and cationic SMI copolymers to solubilize membranes free of detergent allows comparative proteomic studies of acidic or basic membranes. Moreover, the activities of these variants can be compared to minimize undesirable nonspecific electrostatic interactions.
The addition of a phosphatidylcholine (PC) lipid headgroup to form zwitterionic “zSMA” copolymers has been explored [39]. These lack MA groups and are not technically SMA polymers but still solubilize membranes into nanodiscs of diameters 10‒30 nm. The absence of carboxylic acid groups provides compatibility with a broad range of solution conditions such as low pH and high polyvalent cation concentrations.
An alternative copolymer with diisobutylene and maleic acid residues termed DIBMA lacks the styrene groups. It enables very mild fragmentation of membranes and is well suited to placing labile protein complexes and large memteins in nanodiscs with diameters of 12 to 29 nm [40]. An advantage of DIBMA over SMA polymers is their lack of a styrene-based signal in ultraviolet (UV) and circular dichroism spectra.
Similarly, a series of 25 polymethacrylate (PMA) copolymers, based on a distinct backbone with varying ratios of hydrophobic and cationic chains, were synthesized to replace the ultraviolet light-absorbing styrene group [41]. Nanodiscs with 17 nm diameter and 5.5 nm thickness could be formed from DMPC vesicles with members of this series, and their fragmentation of bacterial membranes yielded ~100 nm diameter particles based on electron microscopy analysis.
The effect of varying the hydrophobic group was further explored with a set of alkyl polyacrylic acid (APAA) copolymers [42]. Both the copolymer concentration and inclusion of butyl, pentyl, or hexyl sidechains influence the diameters of resulting nanoparticles, which range from 7 to 17 nm. All could solubilize DMPC vesicles at pH values over 6.5, with slight differences in divalent cation sensitivities and gel to liquid crystalline phase transition temperatures. A high yield of total membrane proteins comparable to SMA(3:1) could be obtained with the longest, hexyl chain variant. This augments the structure-activity relationship information available for these amphipathic copolymers, paving the way for further optimization of polymers for solubilizing membranes with specific protein targets or proteome-level analysis.
The functionalization of SMALPs has been demonstrated by addition of thiol groups to the MA sidechains. In particular, the SMA-SH polymer contains an amino-mercapto-ethane group to which fluorescent dyes or molecular tags can be conjugated [43]. This allows affinity purification and fluorescence detection of the nanodiscs once biotin tags or Alexa Fluor 488 groups are attached here. This offers methods to readily purify and observe nanodiscs containing endogenous proteins.
5. Membrane Proteins in SMALPs
At least 40 different membrane proteins expressed in bacterial, yeast, insect, and mammalian cells have been solubilized using SMA for detailed biochemical analysis. Entire lipid rafts can be solubilized intact using SMA with no detergent exposure, allowing the protein:lipid bilayer patches to be examined. The solubilization of T cell membranes by addition of SMA(3:1) at 1% (w/v) yields >250 nm sized fragments that can be immunoprecipitated to isolate those with Src family kinases and glycosylphosphatidylinositol-anchored proteins [44]. These “raft” nanodiscs are the largest observed to date and contain ordered lipids including an excess of cholesterol, phosphatidylserines, sphingomyelins, ceramides, and monohexosylceramides compared to ~20 nm nanodiscs, which contain relatively more phosphatidylinositol, phosphatidylglycerol (PG), phosphatidylcholine (PC), and phosphatidylethanolamine (PE) lipids. Thus, SMA provides a detergent-free approach to solubilize both large lipid rafts and smaller nanodomains with distinct groups of lipids and compare those associated with specific transmembrane proteins.
Structures of diverse memteins in SMALPs have been solved using various high-resolution methods, indicating the potential feasibility of the SMALP approach for membrane proteomics. Several notable recent reports are discussed below to illustrate the potential of the technique and build on dozens of earlier studies of expressed and endogenous proteins solubilized in SMA for structure-function analysis.
The preparation of the multidrug exporter AcrB in SMA2000 yielded a structure of the asymmetric trimer at 3.0 Å resolution [45]. Its detergent-free purification from Escherichia coli membrane involved single-step metal affinity chromatography of the SMALPs which could then be directly applied to grids to collect cryo-EM data. Unlike detergent approaches, this reveals the intact memtein, which is 12 nm in diameter. It contains 31 visible lipids around the protein trimer, which is 9 nm in diameter. The native lipid bilayer is highly organized with 24 lipid molecules engaging protein residues and being packed into hexagonal arrays within the inner leaflet and an ordered irregular array in the outer leaflet (Figure 3). Although the polymers and lipid headgroup identities are not discernible, the development of more homogeneous polymers is likely to improve resolution further.
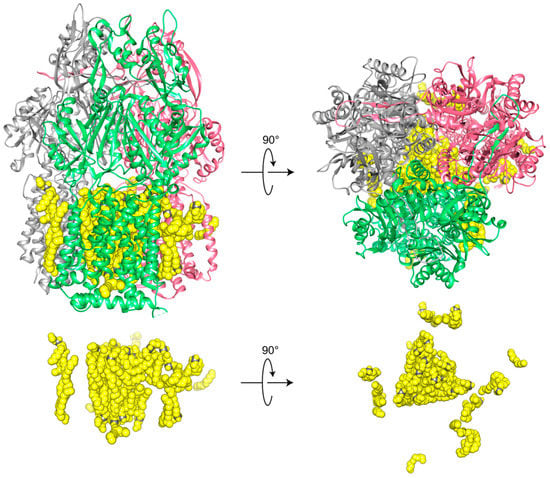
Figure 3.
Top and side views of high resolution cryo-EM structure of trimeric wild-type AcrB (gray, green, pink subunits) and associated lipids (yellow) in SMA2000 nanodiscs (EMD-7074, PDB ID 6BAJ) [45]. The central cavity is filled with 24 triangular-like core lipids.
The largest SMALP-based structure is that of the Alternative Complex III (ACIII) supercomplex, which was resolved at 3.4 Å resolution using cryoEM [17]. This study used SMA3000 as well as XIRAN 25010 to prepare functional complexes of this 464 kDa memtein and revealed the positions of acylated residues mediating photosynthetic electron transport as well as multiple phospholipids haem groups and iron-sulfur clusters. The SMALP contains this entire 9 × 13 nm assembly with its 6 protein subunits, 48 transmembrane helices, and accessory cytochrome c oxidase within a shell of native lipid that were extracted together from the membrane. This demonstrates the adaptability of the SMA nanoparticles, which is shown to conform to the irregular dimensions of membrane protein multimers.
The G protein-coupled receptor (GPCR) superfamily includes many valuable targets for drug discovery. The first 3D structure of a seven transmembrane helix protein that had been extracted with SMA(3:1) was elucidated using XRD at 2.0 Å resolution, which is better than that of detergent-extracted protein in crystals [28]. The seven transmembrane helices of this microbial rhodopsin protein are arranged in a trimer that binds trans-retinal as well as monoolein molecules which were used to prepare the lipidic cubic phase for in meso crystallization. Bound bacterial lipids could not be resolved, presumably due to their displacement by the excess monoolein molecules. A heterotetrameric GPCR complex that recognizes the growth hormone secretagogue and dopamine can be solubilized using SMA (2:1) and binds to G proteins [46]. The adenosine receptor expressed in Pichia pastoris or HEK 293T cells retains ligand binding and stability after extraction with SMA(2:1). Moreover, after storage and freeze-thawing in SMALPs it remains stable seven times longer than in detergent preparations [14]. Solubilization of the melatonin and ghrelin receptors from Pichia pastoris membranes or liposomes with SMA(2:1) or SMA(3:1) yields 13 nm diameter nanodiscs that retain ligand binding and signal transduction activity [47]. The cannabinoid receptor 1 expressed in insect cells retains its native fold once solubilized with SMA(2:1) based on recognition using conformationally specific antibody in surface plasmon resonance experiments [48]. Moreover, this SMALP’d GPCR is stable and active in yeast and mammalian display formats using fluorescence-activated cell sorting (FACS) even after repeated freeze-thaws. It could be used for screening of libraries of proteins to select for those with enhanced ligand affinity, stability, or potential for targeting membrane protein, demonstrating the potential of the approach for screening for novel biologics.
Full length ion channels have been solubilized using SMA(3:1) to reveal the intact pores and symmetric tetramers using negative stain EM [49]. The human hKCNH5 and hKCNQ1 α-subunits of the human neuronal and cardiac voltage-gated potassium (Kv) channels as well as the regulatory transmembrane KCNE1 β-subunit were SMALP’d from mammalian COS-1 cells and showed better behavior than detergent solubilized forms, being more stable and easier to concentrate without aggregation. The channels are oriented randomly on EM grids, displaying orientations with dimensions in the range of 12–15 nm within the discs. Electron paramagnetic resonance (EPR) studies of a spin-labeled KCNE1 protein solubilized in SMA(3:1) nanodiscs show that sidechains in the aqueous phase are more flexible than those inside the bilayer [50]. This report showed that use of SMA can increase the signal to noise and phase memory time in double electron–electron resonance spectra to yield improved distance measurements.
The tetraspanin superfamily represents a technically challenging group of membrane proteins to study because of their small size, oligomerization, multiple disulfides, and glycosylation and palmitoylation groups, as well as the lack of many biochemical assays. Fifteen human tetraspanins were expressed in S. cerevisiae, and the five highest yield proteins were solubilized by three different SMA copolymers. The yield of TSPAN7 was comparable to conventional detergents, while CO-029, TSPAN12, and TSPAN18 yields were higher in detergents, and CD63 was resistant to any approach [51]. The variable expression levels and organized superstructure of such proteins emphasize the need for further development of tools for extraction and purification of low abundance memteins and efficient dispersal of very large, ordered networks.
NMR studies of SMALPs reveal the structures, dynamics, and interactions of contained transmembrane proteins. The Pf1 bacteriophage coat protein forms stable helical structures in magnetically-oriented 30 nm nanodiscs that are formed with SMA(3:1) [52]. They display sharper NMR signals than those which are obtained in bicelles or peptide-based nanodiscs. The NMR signals of cytochrome b5 protein within magnetically aligned ~50 nm nanodiscs made of SMA-EA reveal a stable transmembrane helix [34]. The 34 kDa zinc diffusion facilitator CzcD solubilized with XIRAN 25010 or XIRAN 30010 into 10–15 nm nanodiscs maintains a boundary of bound lipid [53]. The SMALPs display resolvable NMR signals for amide or methyl groups, and NMR assignments can be transferred between solid and solution states. Solving NMR structures of novel proteins in SMALPs remains difficult due to the polydispersity of the non-alternating SMA polymers, which could be addressed by the synthesis of more homogeneous polymers.
The mitochondrial cytochrome c oxidase expressed in Saccharomyces cerevisiae has been solubilized using SMA(3:1) to form 12 nm nanodiscs that also contain biological source lipids and display the native ligand binding and reaction kinetic properties [54]. The SMALP’d memtein include two additional respiratory supercomplex factors which are weakly bound and dissociate upon detergent extraction [55]. The entire assembly has dimensions of 11 by 14 nm, including the entire supercomplex of 11 protein subunits and mitochondrial lipids that together pump protons across membranes.
The holo-translocon assembly that secretes proteins through bacterial inner membranes includes SecYEG, SecDF, YajC, and YidC subunits. These are solubilized along with bound lipids from membrane using SMA(2:1) treatment and can be recognized by specific antibodies [56]. The native SecYEG translocon complex solubilizes with the motor protein SecA and essential biological lipids upon SMA(3:1) exposure, while conventional detergents do not maintain the complex [57]. This assembly was moved into proteoliposomes using Bio-Beads and displayed its abilities to translocate transmembrane proteins and engage the ribosome. These examples illustrate some of the successful characterizations of memteins in nanodiscs. However, not all are as easy to purify and maintain in active forms, motivating further studies on maximally efficient polymers that are minimally interactive with proteins.
6. Lipids in SMALPs
The design of SMA as an effective mild solubilizer of virtually any lipid bilayer or memtein relies on the short styrene sidechains which insert into hydrophobic material [20]. The specific lipid species bound to proteins in SMALPs can most easily be identified using MS. This was demonstrated for SMALP’d insect cell membranes containing the equilibrative nucleoside transporter-1 (hENT1). This protein solubilizes with 16 PC and 2 PE molecules but no polyunsaturated lipids based on electrospray ionization (ESI) MS data [58]. To prevent protein degradation and inactivation, the XIRAN 30010 polymer had been added at a relatively low concentration (0.25% w/v) along with cholesteryl hemisuccinate at low temperature. The resulting protein in nanodiscs shows the predicted inhibitor binding activity, in contrast to protein solubilized in decyl maltoside.
The phospholipids which co-extract with three bacterial membrane proteins can be determined using LC-MS/MS methods [59]. SMA copolymer does not preferentially solubilize any specific types of phospholipids from the membrane, and thus is suitable for unbiased detection of protein-bound phospholipids. The ZipA and PgpB proteins contain 1 and 6 transmembrane helices, respectively, and bind similar ratios of various phospholipids, with higher levels of monounsaturated PE and PG species. In contrast the membrane associated protein FtsA bound preferentially to a distinct profile of PE and PG lipids, presumably as it only inserts part of the way into the inner leaflet of the cytoplasmic membrane.
The GlpG rhomboid proteases from Escherichia coli and Vibrio cholerae have been solubilized using DIBMA, SMA(3:1), and SMA(2.3:1), and their yields and activities compared [60]. Native-like protease activities and high stabilities were found after purification with SMA(3:1) and DIBMA but not with conventional detergents. GlpG binds a variable number of about 50 phospholipids of different chain lengths and degrees of saturation depending on the cell type and temperature based on electrospray ionization and collision-induced dissociation MS data [61].
While the polydispersity of synthetic polymers such as SMA allows a large range of lipids and proteins to be solubilized effectively, this also poses technical issues for MS analysis of SMALPs and multimeric proteins. In particular, the broad range of polymer masses, stoichiometries, and nanodisc diameters can lead to a peak broadening of the assemblies when analyzed using MS. Moveover, the voltages in ESI-MS experiments may be insufficient to release the contained protein from strong interactions with lipid sheets inside the SMALP. This is being addressed by laser-induced liquid bead ion desorption (LILBID) MS, which allows nanodiscs to be transferred within 50 µm diameter droplets that experience soft or hard laser pulses. This results in detection of memteins that can be progressively dissociated in their component lipid-bound subunits, as demonstrated by the GlpG monomers, KtrB potassium channel dimers, and AcrB trimers [62]. This approach allows novel multimeric states to be resolved, including the monomeric and dimeric states of the sodium–solute symporter and KimA proteins, suggesting wider utility.
7. Conclusions
These studies and many others document the attractive option that SMA polymers present for solubilizing a wide diversity of memteins, including those monomers and multimers that are unstable, low abundance, or lipid-dependent. The advantages include the solubilization of intact complexes of proteins and lipids as found in vivo, a feat not afforded by other methods. The breadth of solution conditions including wide ranges of pH, salt, and metals allows new targets to be solubilized. The challenges include nonspecific binding to some surfaces and proteins, which can compromise yield and activity in some cases. The polydispersity is helpful for extracting diverse arrays of memteins but is a challenge for high resolution structural analysis. The heterogeneity of disc sizes also poses challenges, with structure-function studies emerging to help design discs of defined dimensions and dynamics. Quantitating polymer concentrations remains challenging, although fluorescent tags are being developed. Polymer affinity tags and columns that allow endogenous memteins to be purified rapidly and easily are needed. Such tags could also be used in high throughput systems and biophysical assays. The tools with which SMALPs can already be studied include size exclusion chromatography, cryoEM, EPR, MS, NMR, SANS, SPR, and XRD, suggesting applicability to proteomics and lipidomics analysis. A growing body of experimental data on memteins combined with structure-activity relationships and modelling of nanodiscs is accumulating. This is informing the design of next generation polymers to increase resolution, broaden conditions, and incorporate fluorescent and affinity tags to allow detection and purification of endogenous complexes. Together, this promises to open up an even wider range of applications and mechanistic insights into biology in its many dimensions.
Author Contributions
M.O. wrote the manuscript. M.E. prepared the figures and edited the manuscript.
Funding
This research was funded by NSERC RGPIN-2018-04994, the Campus Alberta Innovation Program (RCP-12-002C) and Alberta Prion Research Institute/Alberta Innovates Bio Solutions (201600018) grants awarded to M.O. and The Metabolomics Innovation Centre (TMIC) funders the Canada Foundation for Innovation and Genome Canada.
Acknowledgments
M.O. thanks the SMALP network members including Stefan Scheidelaar for discussions and for sharing polymers and methods.
Conflicts of Interest
M.O. is a co-inventor of patent WO2011004158A1 entitled “Solubilisation of Membrane Proteins”, which is owned by the University of Birmingham, and co-directs the SMALP network.
Abbreviations
The following abbreviations are used in this manuscript:
| DIBMA | poly(diisobutylene-alt-maleic acid) |
| cryoEM | cryo-electron microscopy |
| EM | electron microscopy |
| GPCR | G protein-coupled receptor |
| MA | maleic acid |
| MI | maleimide |
| NMR | nuclear magnetic resonance spectroscopy |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| PG | phosphatidylglycerol |
| S | styrene |
| SMA | poly(styrene-co-maleic acid) |
| SMAd-A | dehydrated styrene maleic acid ethylenediamine |
| SMA-EA | styrene maleic acid ethanolamine |
| SMA-ED | styrene maleic acid ethylenediamine |
| SMALP | styrene maleic acid lipid particle |
| SMA-QA | styrene maleimide quaternary ammonium |
| SMA-SH | SMA with sulfhydrils |
| SMI | poly(styrene-co-maleimide) |
| zSMA | zwitterionic SMA |
| TM | transmembrane |
| XRD | X-ray diffraction |
References
- Vuckovic, D.; Dagley, L.F.; Purcell, A.W.; Emili, A. Membrane proteomics by high performance liquid chromatography-tandem mass spectrometry: Analytical approaches and challenges. Proteomics 2013, 13, 404–423. [Google Scholar] [CrossRef]
- Wehner, A.; Geiger, T.; Cox, J.; Schaab, C.; Mann, M. Comparative Proteomic Analysis of Eleven Common Cell Lines Reveals Ubiquitous but Varying Expression of Most Proteins. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef]
- Speers, A.E.; Wu, C.C. Proteomics of Integral Membrane ProteinsTheory and Application. Chem. Rev. 2007, 107, 3687–3714. [Google Scholar] [CrossRef]
- Overduin, M.; Esmaili, M. Memtein: The fundamental unit of membrane-protein structure and function. Chem. Phys. Lipids 2019, 218, 73–84. [Google Scholar] [CrossRef]
- Zhang, X.X.; Chan, C.S.; Bao, H.; Fang, Y.; Foster, L.J.; Duong, F. Nanodiscs and SILAC-based mass spectrometry to identify a membrane protein interactome. J. Proteome Res. 2012, 11, 1454–1459. [Google Scholar] [CrossRef] [PubMed]
- Watkinson, T.G.; Calabrese, A.N.; Giusti, F.; Zoonens, M.; Radford, S.E.; Ashcroft, A.E. Systematic analysis of the use of amphipathic polymers for studies of outer membrane proteins using mass spectrometry. Int. J. Mass Spectrom. 2015, 391, 54–61. [Google Scholar] [CrossRef]
- Hopper, J.T.S.; Yu, Y.T.C.; Li, D.; Raymond, A.; Bostock, M.; Liko, I.; Mikhailov, V.; Laganowsky, A.; Benesch, J.L.P.; Caffrey, M.; et al. Detergent-free mass spectrometry of membrane protein complexes. Nat. Methods 2013, 10, 1206–1208. [Google Scholar] [CrossRef] [PubMed]
- Brügger, B. Lipidomics: Analysis of the Lipid Composition of Cells and Subcellular Organelles by Electrospray Ionization Mass Spectrometry. Annu. Rev. Biochem. 2014, 83, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Kurland, I.J.; Jones, M.R.; Dunn, W.B. How close are we to complete annotation of metabolomes? Curr. Opin. Chem. Biol. 2017, 36, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Simons, K. Lipidomics: Coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010, 11, 593–598. [Google Scholar] [CrossRef]
- Zhang, B.; Xie, M.; Bruschweiler-Li, L.; Brüschweiler, R. Nanoparticle-assisted metabolomics. Metabolites 2018, 88, 21. [Google Scholar] [CrossRef]
- Gross, R.W. The evolution of lipidomics through space and time. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 731–739. [Google Scholar] [CrossRef]
- Laursen, T.; Borch, J.; Knudsen, C.; Bavishi, K.; Torta, F.; Martens, H.J.; Silvestro, D.; Hatzakis, N.S.; Wenk, M.R.; Dafforn, T.R.; et al. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science 2016, 354, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Jamshad, M.; Charlton, J.; Lin, Y.-P.; Routledge, S.J.; Bawa, Z.; Knowles, T.J.; Overduin, M.; Dekker, N.; Dafforn, T.R.; Bill, R.M.; et al. G-protein coupled receptor solubilization and purification for biophysical analysis and functional studies, in the total absence of detergent. Biosci. Rep. 2015, 35, e00188. [Google Scholar] [CrossRef] [PubMed]
- Gulamhussein, A.; Meah, D.; Fenner, S.; Saidani, Z.; Akram, A.; Lallie, S.; Mathews, A.; Painter, C.; Liddar, M.; Mohammed, Z.; et al. Examining the stability of membrane proteins within SMALPs. Eur. Polym. J. 2019, 112, 120–125. [Google Scholar] [CrossRef]
- Paulin, S.; Jamshad, M.; Dafforn, T.R.; Garcia-Lara, J.; Foster, S.J.; Galley, N.F.; Roper, D.I.; Rosado, H.; Taylor, P.W. Surfactant-free purification of membrane protein complexes from bacteria: Application to the staphylococcal penicillin-binding protein complex PBP2/PBP2a. Nanotechnology 2014, 25, 285101. [Google Scholar] [CrossRef]
- Sun, C.; Benlekbir, S.; Venkatakrishnan, P.; Wang, Y.; Hong, S.; Hosler, J.; Tajkhorshid, E.; Rubinstein, J.; Gennis, R. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 2018, 557, 123–126. [Google Scholar] [CrossRef]
- Dominguez Pardo, J.J.; Dorr, J.M.; Iyer, A.; Cox, R.C.; Scheidelaar, S.; Koorengevel, M.C.; Subramaniam, V.; Killian, J.A. Solubilization of lipids and lipid phases by the styrene maleic acid copolymer. Eur. Biophys. J. 2017, 46, 91–101. [Google Scholar] [CrossRef]
- Jamshad, M.; Grimard, V.; Idini, I.; Knowles, T.J.; Dowle, M.R.; Schofield, N.; Sridhar, P.; Lin, Y.; Finka, R.; Wheatley, M.; et al. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 2015, 8, 774–789. [Google Scholar] [CrossRef]
- Knowles, T.J.; Finka, R.; Smith, C.; Lin, Y.-P.; Dafforn, T.; Overduin, M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009, 131, 7484–7485. [Google Scholar] [CrossRef] [PubMed]
- Orekhov, P.S.; Bozdaganyan, M.; Voskoboynikova, N.; Mulkidjanian, A.Y.; Steinhoff, H.J.; Shaitan, K.V. Styrene-maleic acid copolymers form SMALPs by pulling lipid patches out of the lipid bilayer. Langmuir 2019, 35, 3748–3758. [Google Scholar] [CrossRef]
- Cuevas Arenas, R.; Klingler, J.; Vargas, C.; Keller, S. Influence of lipid bilayer properties on nanodisc formation mediated by styrene/maleic acid copolymers. Nanoscale 2016, 8, 15016–15026. [Google Scholar] [CrossRef]
- Vargas, C.; Cuevas Arenas, R.; Frotscher, E.; Keller, S. Nanoparticle self-assembly in mixtures of phospholipids with styrene/maleic acid copolymers or fluorinated surfactants. Nanoscale 2015, 7, 20685–20696. [Google Scholar] [CrossRef] [PubMed]
- Orwick, M.C.; Judge, P.J.; Procek, J.; Lindholm, L.; Graziadei, A.; Engel, A.; Gröbner, G.; Watts, A. Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: Lipodisq. Angew. Chem. Int. Ed. 2012, 51, 4653–4657. [Google Scholar] [CrossRef] [PubMed]
- Scheidelaar, S.; Koorengevel, M.C.; van Walree, C.A.; Dominguez, J.J.; Dörr, J.M.; Killian, J.A. Effect of Polymer Composition and pH on Membrane Solubilization by Styrene-Maleic Acid Copolymers. Biophys. J. 2016, 111, 1974–1986. [Google Scholar] [CrossRef]
- Swainsbury, D.J.K.; Scheidelaar, S.; Foster, N.; van Grondelle, R.; Antoinette Killian, J.; Jones, M.R. The effectiveness of styrene–maleic acid (SMA) copolymers for solubilisation of integral membrane proteins from SMA-accessible and SMA-resistant membranes. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2133–2143. [Google Scholar] [CrossRef] [PubMed]
- Cuevas Arenas, R.; Danielczak, B.; Martel, A.; Porcar, L.; Breyton, C.; Ebel, C.; Keller, S. Fast Collisional Lipid Transfer Among Polymer-Bounded Nanodiscs. Sci. Rep. 2017, 7, 45875. [Google Scholar] [CrossRef]
- Broecker, J.; Eger, B.T.; Ernst, O.P. Crystallogenesis of Membrane Proteins Mediated by Polymer-Bounded Lipid Nanodiscs. Structure 2017, 25, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Scheidelaar, S.; Koorengevel, M.C.; Pardo, J.D.; Meeldijk, J.D.; Breukink, E.; Killian, J.A. Molecular Model for the solubilization of membranes into nanodisks by styrene maleic acid copolymers. Biophys. J. 2015, 108, 279–290. [Google Scholar] [CrossRef]
- Bali, A.P.; Sahu, I.D.; Craig, A.F.; Clark, E.E.; Burridge, K.M.; Dolan, M.T.; Dabney-Smith, C.; Konkolewicz, D.; Lorigan, G.A. Structural characterization of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using EPR spectroscopy. Chem. Phys. Lipids 2019, 220, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Dörr, J.M.; van Coevorden-Hameete, M.H.; Hoogenraad, C.C.; Killian, J.A. Solubilization of human cells by the styrene-maleic acid copolymer: Insights from fluorescence microscopy. Biochim. Biophys. Acta 2017, 2736, 30260–30262. [Google Scholar] [CrossRef]
- Satori, C.P.; Kostal, V.; Arriaga, E.A. Review on recent advances in the analysis of isolated organelles. Anal. Chim. Acta 2012, 753, 8–18. [Google Scholar] [CrossRef]
- Korotych, O.; Mondal, J.; Asfura, K.G.; Hendricks, J.; Bruce, B. Evaluation of commercially available styrene-co-maleic acid polymers in extraction of membrane proteins from chloroplast thylakoids. Eur. Polym. J. 2018, in press. [Google Scholar] [CrossRef]
- Ravula, T.; Ramadugu, S.K.; Di Mauro, G.; Ramamoorthy, A. Bioinspired, Size-Tunable Self-Assembly of Polymer–Lipid Bilayer Nanodiscs. Angew. Chem. Int. Ed. Engl. 2017, 56, 11466–11470. [Google Scholar] [CrossRef]
- Ravula, T.; Hardin, N.; Di Mauro, G.; Ramamoorthy, A. Styrene maleic acid derivates to enhance the applications of bio-inspired polymer based lipid-nanodiscs. Eur. Polym. J. 2018, 108, 597–602. [Google Scholar] [CrossRef]
- Ravula, T.; Hardin, N.Z.; Ramadugu, S.K.; Ramamoorthy, A. pH Tunable and Divalent Metal Ion Tolerant Polymer Lipid Nanodiscs. Langmuir 2017, 33, 10655–10662. [Google Scholar] [CrossRef] [PubMed]
- Ravula, T.; Hardin, N.Z.; Ramadugu, S.K.; Cox, S.J.; Ramamoorthy, A. Formation of pH-Resistant Monodispersed Polymer–Lipid Nanodiscs. Angew. Chem. Int. Ed. 2018, 57, 1342–1345. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.; Tognoloni, C.; Charlton, J.; Bragginton, É.; Rothnie, A.; Sridhar, P.; Wheatley, M.; Knowles, T.; Arnold, T.; Edler, K.; et al. An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles. Nanoscale 2018, 10, 10609–10619. [Google Scholar] [CrossRef]
- Fiori, M.C.; Jiang, Y.; Altenberg, G.A.; Liang, H. Polymer-encased nanodiscs with improved buffer compatibility. Sci. Rep. 2017, 7, 7432. [Google Scholar] [CrossRef]
- Oluwole, A.O.; Danielczak, B.; Meister, A.; Babalola, J.O.; Vargas, C.; Keller, S. Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angew. Chem. Int. Ed. 2017, 56, 1919–1924. [Google Scholar] [CrossRef]
- Yasuhara, K.; Arakida, J.; Ravula, T.; Ramadugu, S.K.; Sahoo, B.; Kikuchi, J.I.; Ramamoorthy, A. Spontaneous Lipid Nanodisc Fomation by Amphiphilic Polymethacrylate Copolymers. J. Am. Chem. Soc. 2017, 139, 18657–18663. [Google Scholar] [CrossRef] [PubMed]
- Hardin, N.Z.; Ravula, T.; Di Mauro, G.; Ramamoorthy, A. Hydrophobic Functionalization of Polyacrylic Acid as a Versatile Platform for the Development of Polymer Lipid Nanodisks. Small 2019, 15, 1804813. [Google Scholar] [CrossRef] [PubMed]
- Lindhoud, S.; Carvalho, V.; Pronk, J.W.; Aubin-Tam, M.E. SMA-SH: Modified Styrene-Maleic Acid Copolymer for Functionalization of Lipid Nanodiscs. Biomacromolecules 2016, 17, 1516–1522. [Google Scholar] [CrossRef] [PubMed]
- Angelisová, P.; Ballek, O.; Sýkora, J.; Benada, O.; Čajka, T.; Pokorná, J.; Pinkas, D.; Hořejší, V. The use of styrene-maleic acid copolymer (SMA) for studies on T cell membrane rafts. Biochim. Biophys. Acta Biomembr. 2018, 1861, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Fu, Z.; Xu, G.G.; Grassucci, R.A.; Zhang, Y.; Frank, J.; Hendrickson, W.A.; Guo, Y. Structure and activity of lipid bilayer within a membrane-protein transporter. Proc. Natl. Acad. Sci. USA 2018, 115, 12985–12990. [Google Scholar] [CrossRef] [PubMed]
- Damian, M.; Pons, V.; Renault, P.; M’Kadmi, C.; Delort, B.; Hartmann, L.; Kaya, A.I.; Louet, M.; Gagne, D.; Ben Haj Salah, K.; et al. GHSR-D2R heteromerization modulates dopamine signaling through an effect on G protein conformation. Proc. Natl. Acad. Sci. USA 2018, 115, 4501–4506. [Google Scholar] [CrossRef] [PubMed]
- Logez, C.; Damian, M.; Legros, C.; Dupré, C.; Guéry, M.; Mary, S.; Wagner, R.; M’Kadmi, C.; Nosjean, O.; Fould, B.; et al. Detergent-free Isolation of Functional G Protein-Coupled Receptors into Nanometric Lipid Particles. Biochemistry 2016, 55, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Luna, V.M.; Vazir, M.; Vaish, A.; Chong, S.; Chen, I.; Yamane, H. Generation of Membrane Proteins in Polymer-based Lipoparticles as Flow Cytometry Antigens. Eur. Polym. J. 2018, 109, 483–488. [Google Scholar] [CrossRef]
- Karlova, M.G.; Voskoboynikova, N.; Gluhov, G.S.; Bramochkin, D.; Malak, O.A.; Mulkidzhanyan, A.; Loussouarn, G.; Steinhoff, H.J.; Shaitan, K.V.; Sokolova, O.S. Detergent-free solubilization of human Kv channels expressed in mammalian cells. Chem. Phys. Lipids 2019, 219, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.D.; Zhang, R.; Dunagan, M.M.; Craig, A.F.; Lorigan, G.A. Characterization of KCNE1 inside Lipodisq Nanoparticles for EPR Spectroscopic Studies of Membrane Proteins. J. Phys. Chem. B 2017, 121, 5312–5321. [Google Scholar] [CrossRef]
- Skaar, K.; Korza, H.J.; Tarry, M.; Sekyrova, P.; Högbom, M. Expression and subcellular distribution of GFP-Tagged human tetraspanin proteins in saccharomyces cerevisiae. PLoS ONE 2015, 10, e0134041. [Google Scholar] [CrossRef] [PubMed]
- Radoicic, J.; Park, S.H.; Opella, S.J. Macrodiscs Comprising SMALPs for Oriented Sample Solid-State NMR Spectroscopy of Membrane Proteins. Biophys. J. 2018, 115, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Bersch, B.; Dörr, J.M.; Hessel, A.; Killian, J.A.; Schanda, P. Proton-Detected Solid-State NMR Spectroscopy of a Zinc Diffusion Facilitator Protein in Native Nanodiscs. Angew. Chem. Int. Ed. 2017, 56, 2508–2512. [Google Scholar] [CrossRef] [PubMed]
- Long, A.R.; O’Brien, C.C.; Malhotra, K.; Schwall, C.T.; Albert, A.D.; Watts, A.; Alder, N.N. A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs. BMC Biotechnol. 2013, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, I.A.; Sjostrand, D.; Li, F.; Bjorck, M.; Schafer, J.; Ostbye, H.; Hogbom, M.; von Ballmoos, C.; Lander, G.C.; Adelroth, P.; et al. Isolation of yeast complex IV in native lipid nanodiscs. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2984–2992. [Google Scholar] [CrossRef] [PubMed]
- Komar, J.; Alvira, S.; Schulze, R.J.; Martin, R.; Lycklama a Nijeholt, J.A.; Lee, S.C.; Dafforn, T.R.; Deckers-Hebestreit, G.; Berger, I.; Schaffitzel, C.; et al. Membrane protein insertion and assembly by the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Biochem. J. 2016, 473, 3341–3354. [Google Scholar] [CrossRef] [PubMed]
- Prabudiansyah, I.; Kusters, I.; Caforio, A.; Driessen, A.J.M. Characterization of the annular lipid shell of the Sec translocon. Biochim. Biophys. Acta Biomembr. 2015, 1848, 2050–2056. [Google Scholar] [CrossRef]
- Rehan, S.; Paavilainen, V.O.; Jaakola, V.P. Functional reconstitution of human equilibrative nucleoside transporter-1 into styrene maleic acid co-polymer lipid particles. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Teo, A.C.K.; Lee, S.C.; Pollock, N.L.; Stroud, Z.; Hall, S.; Thakker, A.; Pitt, A.R.; Dafforn, T.R.; Spickett, C.M.; Roper, D.I. Analysis of SMALP co-extracted phospholipids shows distinct membrane environments for three classes of bacterial membrane protein. Sci. Rep. 2019, 9, 1813. [Google Scholar] [CrossRef]
- Barniol-Xicota, M.; Verhelst, S.H.L. Stable and Functional Rhomboid Proteases in Lipid Nanodiscs by Using Diisobutylene/Maleic Acid Copolymers. J. Am. Chem. Soc. 2018, 140, 14557–14561. [Google Scholar] [CrossRef]
- Reading, E.; Hall, Z.; Martens, C.; Haghighi, T.; Findlay, H.; Ahdash, Z.; Politis, A.; Booth, P.J. Interrogating Membrane Protein Conformational Dynamics within Native Lipid Compositions. Angew. Chem. Int. Ed. 2017, 56, 15654–15657. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, N.; Peetz, O.; Ahdash, Z.; Tascón, I.; Booth, P.J.; Mikusevic, V.; Diskowski, M.; Politis, A.; Hellmich, Y.; Hänelt, I.; et al. Native mass spectrometry goes more native: Investigation of membrane protein complexes directly from SMALPs. Chem. Commun. 2018, 54, 13702–13705. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).