3D U-Net for Skull Stripping in Brain MRI
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Volume Sampling
2.3. Network Architecture
2.4. Training
3. Experimental Results and Discussion
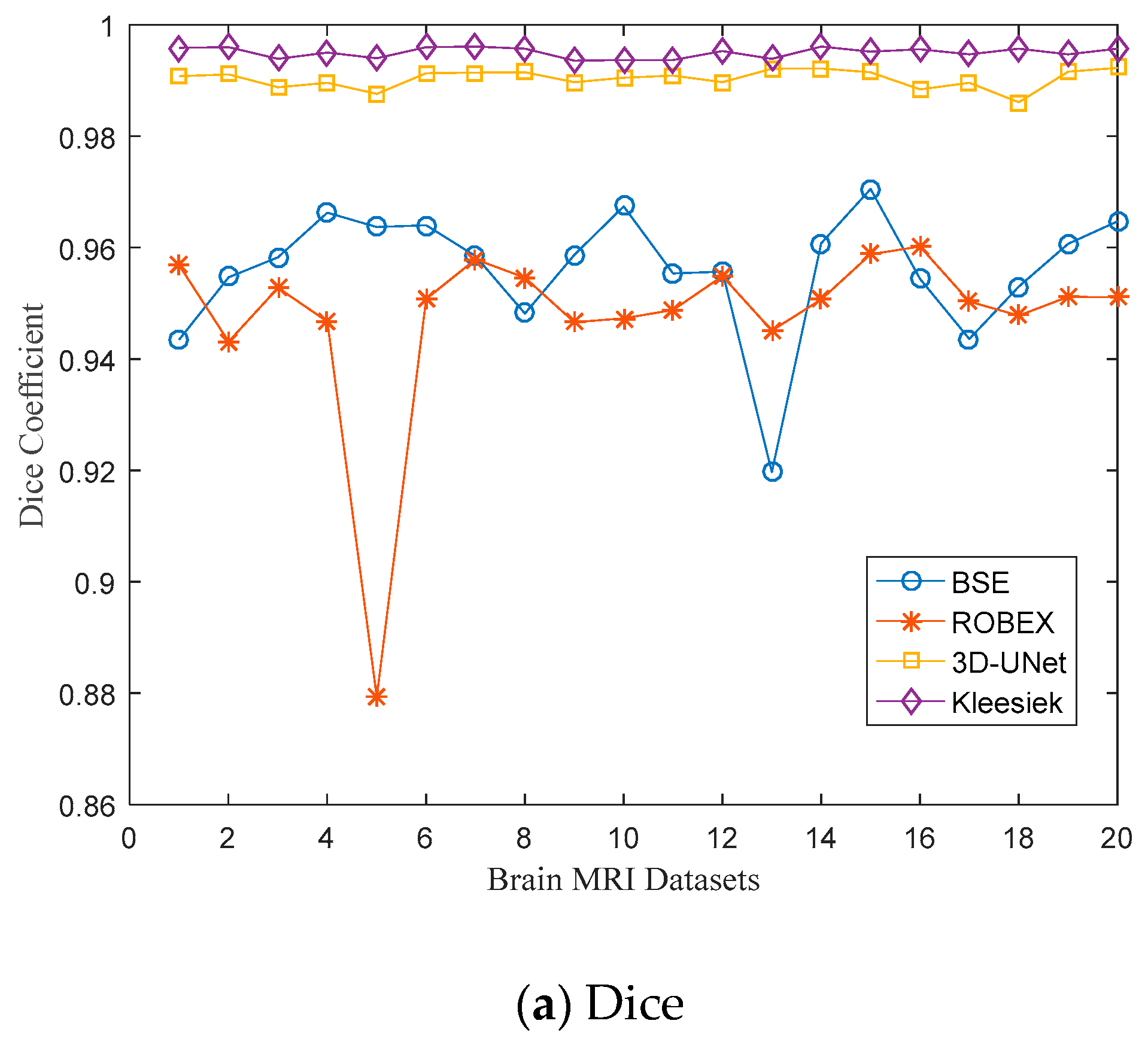
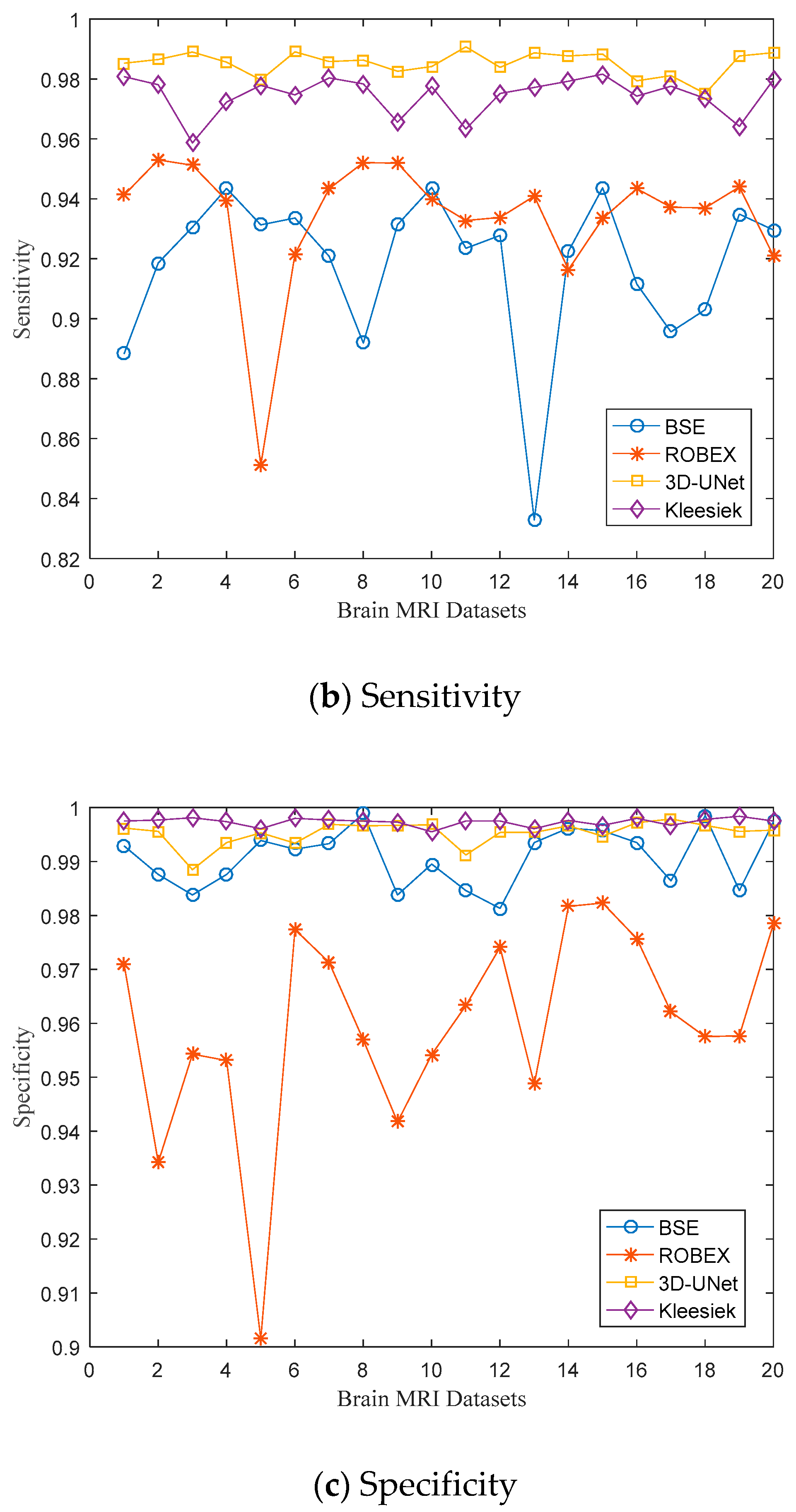
3.1. Evaluation Metrics
3.2. Quantitative Results
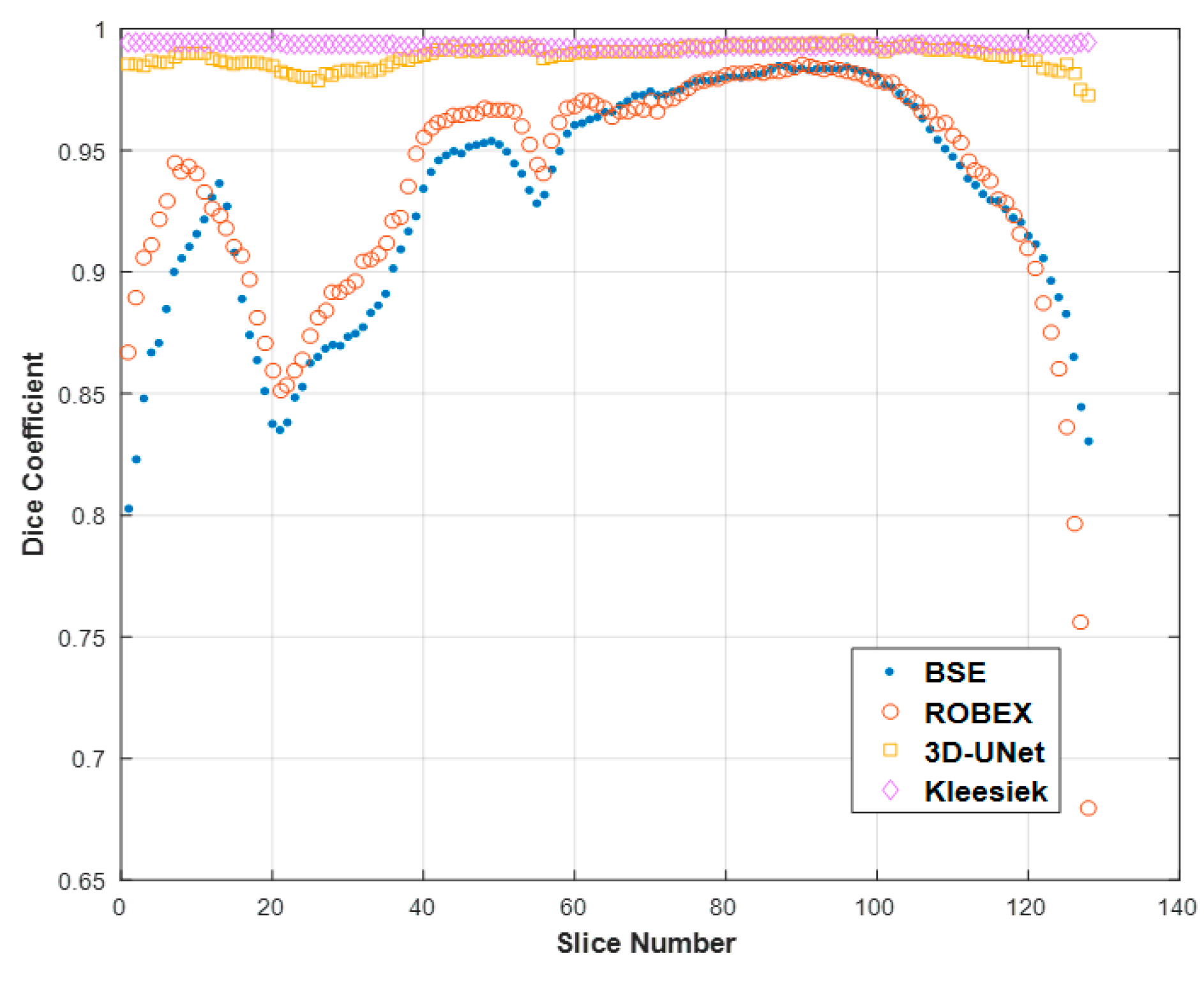
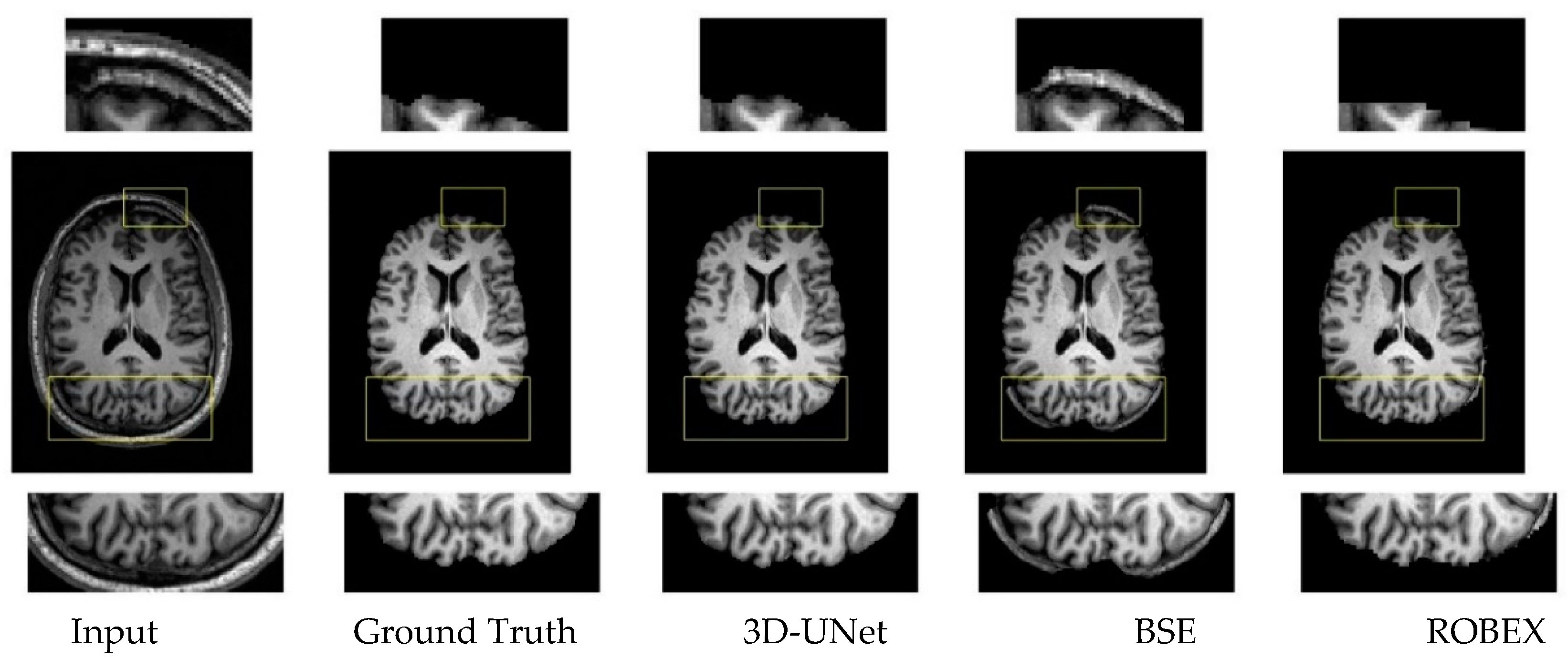
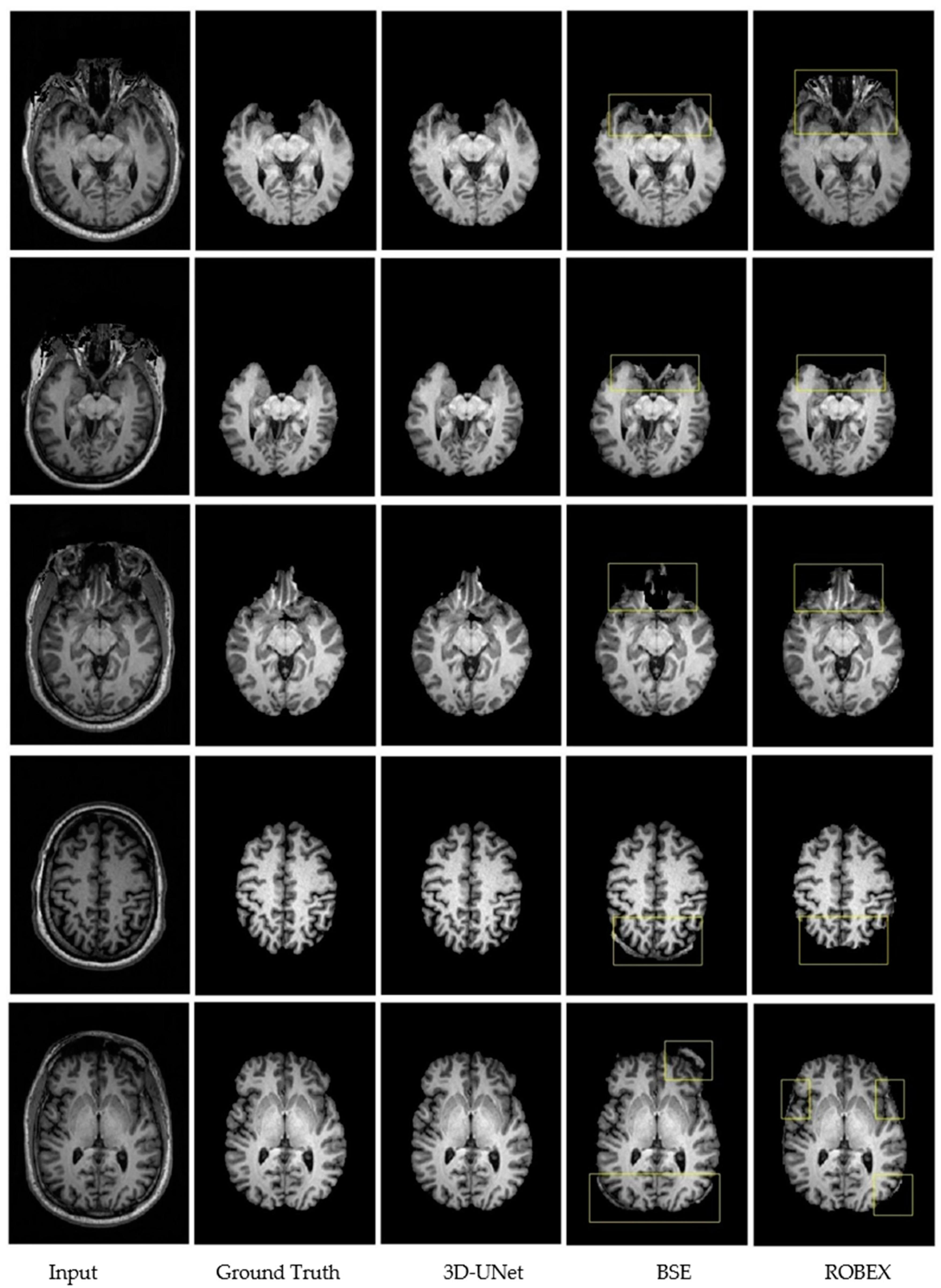
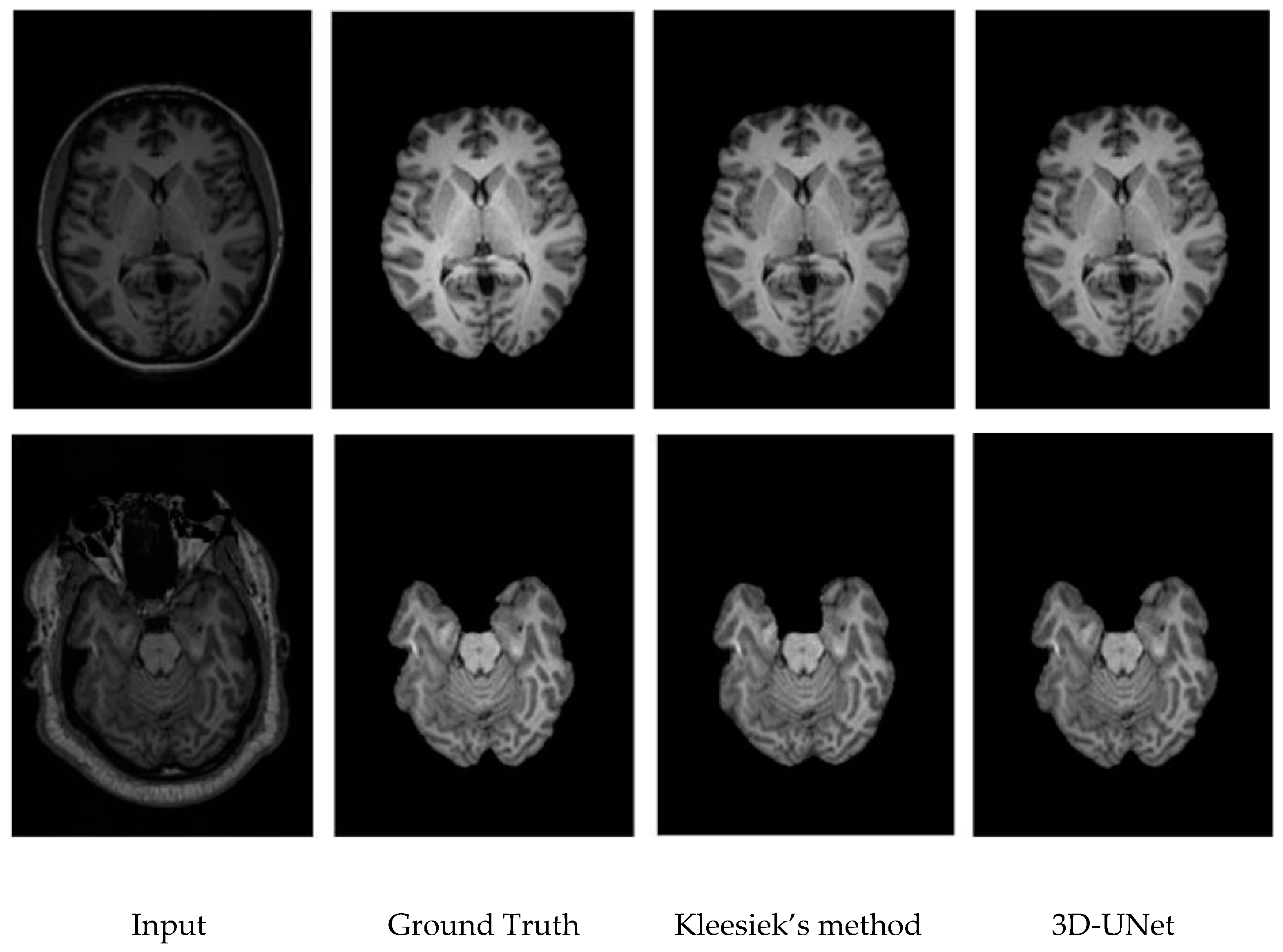
3.3. Qualitative Results.
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Uhlich, M.; Greiner, R.; Hoehn, B.; Woghiren, M.; Diaz, I.; Ivanova, T.; Murtha, A. Improved Brain Tumor Segmentation via Registration-Based Brain Extraction. Forecasting 2018, 1, 59–69. [Google Scholar] [CrossRef]
- Tosun, D.; Rettmann, M.E.; Naiman, D.Q.; Resnick, S.M.; Kraut, M.A.; Prince, J.L. Cortical reconstruction using implicit surface evolution: Accuracy and precision analysis. Neuroimage 2006, 29, 838–852. [Google Scholar] [CrossRef] [PubMed]
- Kalkers, N.F.; Ameziane, N.; Bot, J.C.J.; Minneboo, A.; Polman, C.H.; Barkhof, F. Longitudinal brain volume measurement in multiple sclerosis—Rate of brain atrophy is independent of the disease subtype. Arch. Neurol. 2002, 59, 1572–1576. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, Y.; Pan, X.; Hong, X.; Xia, D. Level set segmentation of brain magnetic resonance images based on local Gaussian distribution fitting energy. J. Neurosci. Methods 2010, 188, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ruotsalainen, U.; Hirvonen, J.; Hietala, J.; Tohka, J. Automatic cerebral and cerebellar hemisphere segmentation in 3D MRI: Adaptive disconnection algorithm. Med. Image Anal. 2010, 14, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Zhuang, Y.; Gong, H.; Zhan, J.; Grossman, M.; Wang, Z. Resting State Brain Entropy Alterations in Relapsing Remitting Multiple Sclerosis. PLoS ONE 2016, 11, e0146080. [Google Scholar] [CrossRef] [PubMed]
- Tanskanen, P.; Veijola, J.M.; Piippo, U.K.; Haapea, M.; Miettunen, J.A.; Pyhtinen, J.; Bullmore, E.T.; Jones, P.B.; Isohanni, M.K. Hippocampus and amygdala volumes in schizophrenia and other psychoses in the Northern Finland 1966 birth cohort. Schizophr. Res. 2005, 75, 283–294. [Google Scholar] [CrossRef]
- Rusinek, H.; de Leon, M.J.; George, A.E.; Stylopoulos, L.A.; Chandra, R.; Smith, G.; Rand, T.; Mourino, M.; Kowalski, H. Alzheimer disease: Measuring loss of cerebral gray matter with MR imaging. Radiology 1991, 178, 109–114. [Google Scholar] [CrossRef]
- Iglesias, J.E.; Liu, C.Y.; Thompson, P.M.; Tu, Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans. Med. Imaging 2011, 30, 1617–1634. [Google Scholar] [CrossRef]
- Kleesiek, J.; Urban, G.; Hubert, A.; Schwarz, D.; Maier-Hein, K.; Bendszus, M.; Biller, A. Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. Neuroimage 2016, 129, 460–469. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; Volume 1, pp. 1097–1105. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, H.; van Ginneken, B.; Summers, R.M. Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans. Med. Imag. 2016, 35, 1153–1159. [Google Scholar] [CrossRef]
- Dawant, B.M.; Hartmann, S.L.; Thirion, J.P.; Maes, F.; Vandermeulen, D.; Demaerel, P. Automatic 3-D segmentation of internal structures of the head in MR images using a combination of similarity and free-form transformations: Part I, methodology and validation on normal subjects. IEEE Trans. Med. Imag. 1999, 18, 909–916. [Google Scholar] [CrossRef]
- Grau, V.; Mewes, A.U.J.; Alcaniz, M.; Kikinis, R.; Warfield, S.K. Improved watershed transform for medical image segmentation using prior information. IEEE Trans. Med. Imag. 2004, 23, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.Y.; Yue, G.H.; Liu, J.Z. Automated histogram-based brain segmentation in T1-weighted three-dimensional magnetic resonance head images. Neuroimage 2002, 17, 1587–1598. [Google Scholar] [CrossRef] [PubMed]
- Aboutanos, G.B.; Nikanne, J.; Watkins, N.; Dawant, B.M. Model creation and deformation for the automatic segmentation of the brain in MR images. IEEE Trans. Biomed. Eng. 1999, 46, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S. Two-dimensional fast magnetic resonance brain segmentation. IEEE. Eng. Med. Biol. Mag. 2001, 20, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Merisaari, H.; Parkkola, R.; Alhoniemi, E.; Teras, M.; Lehtonen, L.; Haataja, L.; Lapinleimu, H.; Nevalainen, O.S. Gaussian mixture model-based segmentation of MR images taken from premature infant brains. J. Neurosci. Methods 2009, 182, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Dale, A.M.; Fischl, B.; Sereno, M.I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999, 9, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Kobashi, S.; Fujimoto, Y.; Ogawa, M.; Ando, K.; Ishikura, R.; Kondo, K.; Hirota, S.; Hata, Y. Fuzzy-ASM Based Automated Skull Stripping Method from Infantile Brain MR Images. In Proceedings of the 2007 IEEE International Conference on Granular Computing (GRC 2007), San Jose, CA, USA, 2–4 November 2007; p. 632. [Google Scholar]
- Leung, K.K.; Barnes, J.; Modat, M.; Ridgway, G.R.; Bartlett, J.W.; Fox, N.C.; Ourselin, S.; Initia, A.D.N. Brain MAPS: An automated, accurate and robust brain extraction technique using a template library. Neuroimage 2011, 55, 1091–1108. [Google Scholar] [CrossRef] [PubMed]
- Atkins, M.S.; Mackiewich, B.T. Fully automatic segmentation of the brain in MRI. IEEE Trans. Med. Imag. 1998, 17, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Rehm, K.; Schaper, K.; Anderson, J.; Woods, R.; Stoltzner, S.; Rottenberg, D. Putting our heads together: A consensus approach to brain/non-brain segmentation in T1-weighted MR volumes. Neuroimage 2004, 22, 1262–1270. [Google Scholar] [CrossRef]
- Kalavathi, P.; Prasath, V.S. Methods on skull stripping of MRI head scan images—A review. J. Digit. Imag. 2016, 29, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Brummer, M.E.; Mersereau, R.M.; Eisner, R.L.; Lewine, R.J. Automatic detection of brain contours in MRI data sets. IEEE Trans. Med. Imaging 1993, 12, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Park, J.G.; Lee, C. Skull stripping based on region growing for magnetic resonance brain images. Neuroimage 2009, 47, 1394–1407. [Google Scholar] [CrossRef]
- Somasundaram, K.; Kalaiselvi, T. Fully automatic brain extraction algorithm for axial T2-weighted magnetic resonance images. Comput. Biol. Med. 2010, 40, 811–822. [Google Scholar] [CrossRef]
- Somasundaram, K.; Kalaiselvi, T. Automatic brain extraction methods for T1 magnetic resonance images using region labeling and morphological operations. Comput. Biol. Med. 2011, 41, 716–725. [Google Scholar] [CrossRef]
- Shattuck, D.W.; Sandor-Leahy, S.R.; Schaper, K.A.; Rottenberg, D.A.; Leahy, R.M. Magnetic resonance image tissue classification using a partial volume model. Neuroimage 2001, 13, 856–876. [Google Scholar] [CrossRef]
- Smith, S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002, 17, 143–155. [Google Scholar] [CrossRef]
- Zhuang, A.H.; Valentino, D.J.; Toga, A.W. Skull-stripping magnetic resonance brain images using a model-based level set. Neuroimage 2006, 32, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Eskildsen, S.F.; Coupe, P.; Fonov, V.; Manjon, J.V.; Leung, K.K.; Guizard, N.; Wassef, S.N.; Ostergaard, L.R.; Collins, D.L.; Alzheimer’s Disease Neuroimaging, I. BEaST: Brain extraction based on nonlocal segmentation technique. Neuroimage 2012, 59, 2362–2373. [Google Scholar] [CrossRef] [PubMed]
- Heckemann, R.A.; Ledig, C.; Gray, K.R.; Aljabar, P.; Rueckert, D.; Hajnal, J.V.; Hammers, A. Brain Extraction Using Label Propagation and Group Agreement: Pincram. PLoS ONE 2015, 10, e0129211. [Google Scholar] [CrossRef]
- Wang, Y.; Nie, J.; Yap, P.T.; Shi, F.; Guo, L.; Shen, D. Robust deformable-surface-based skull-stripping for large-scale studies. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Toronto, ON, Canada, 18–22 September 2011; Volume 14, pp. 635–642. [Google Scholar]
- Hariharan, B.; Arbeláez, P.; Girshick, R.; Malik, J. Hypercolumns for object segmentation and fine-grained localization. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 447–456. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical image computing and computer-assisted intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Seyedhosseini, M.; Sajjadi, M.; Tasdizen, T. Image Segmentation with Cascaded Hierarchical Models and Logistic Disjunctive Normal Networks. In Proceedings of the 2013 IEEE International Conference on Computer Vision, Sydney, NSW, Australia, 1–8 December 2013; pp. 2168–2175. [Google Scholar]
- Milletari, F.; Ahmadi, S.A.; Kroll, C.; Plate, A.; Rozanski, V.; Maiostre, J.; Levin, J.; Dietrich, O.; Ertl-Wagner, B.; Botzel, K.; et al. Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput. Vis. Image Underst. 2017, 164, 92–102. [Google Scholar] [CrossRef]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Auto-Context Convolutional Neural Network (Auto-Net) for Brain Extraction in Magnetic Resonance Imaging. IEEE Trans. Med. Imag. 2017, 36, 2319–2330. [Google Scholar] [CrossRef] [PubMed]
- Duy, N.H.M.; Duy, N.M.; Truong, M.T.N.; Bao, P.T.; Binh, N.T. Accurate brain extraction using Active Shape Model and Convolutional Neural Networks. arXiv, 2018; arXiv:1802.01268. [Google Scholar]
- Dey, R.; Hong, Y. CompNet: Complementary Segmentation Network for Brain MRI Extraction. arXiv, 2018; arXiv:1804.00521. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Puccio, B.; Pooley, J.P.; Pellman, J.S.; Taverna, E.C.; Craddock, R.C. The preprocessed connectomes project repository of manually corrected skull-stripped T1-weighted anatomical MRI data. Gigascience 2016, 5, 45. [Google Scholar] [CrossRef]
- Fischl, B. FreeSurfer. Neuroimage 2012, 62, 774–781. [Google Scholar] [CrossRef]
- Fedorov, A.; Johnson, J.; Damaraju, E.; Ozerin, A.; Calhoun, V.; Plis, S. End-to-end learning of brain tissue segmentation from imperfect labeling. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 3785–3792. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv, 2015; preprint. arXiv:1502.03167. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv, 2014; preprint. arXiv:1412.6980. [Google Scholar]
- Sled, J.G.; Zijdenbos, A.P.; Evans, A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imag. 1998, 17, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Shattuck, D.W.; Leahy, R.M. BrainSuite: An automated cortical surface identification tool. Med. Image Anal. 2002, 6, 129–142. [Google Scholar] [CrossRef]
- Iglesias, J.E. ROBEX 1.2. Available online: https://www.nitrc.org/projects/robex (accessed on 10 June 2018).
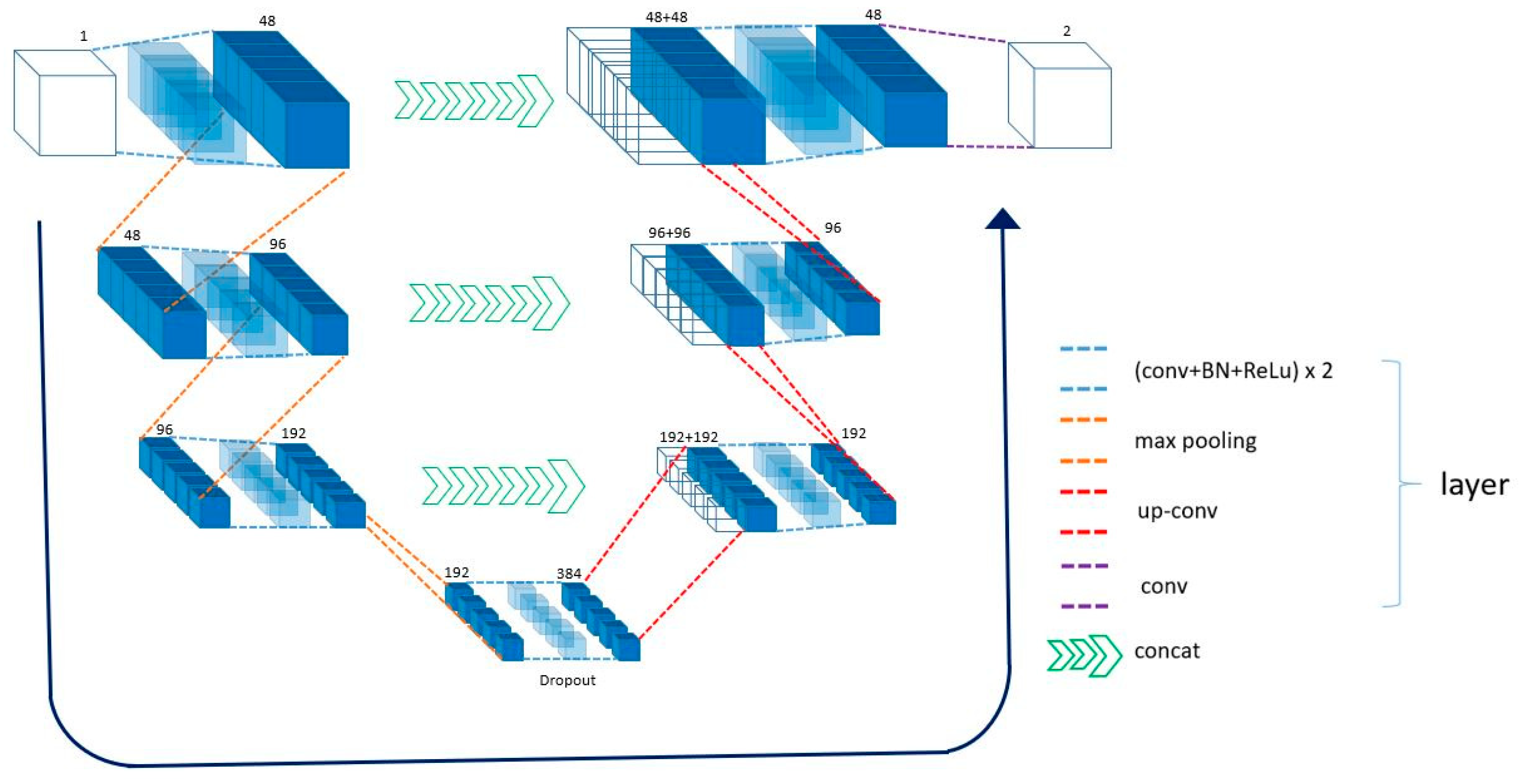

| Block | Kernel Size | Stride | MaxPool | Activation Function | Batch Norm. | Repeat | Input | Output | Number of Parameters |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 33 | 1 | No | ReLu | Yes | 2 | 643 × 1 | 643 × 48 | 63,504 |
| 2 | 33 | 2 | 33 | - | No | 1 | 643 × 48 | 323 × 48 | - |
| 3 | 33 | 1 | No | ReLu | Yes | 2 | 323 × 48 | 323 × 96 | 373,248 |
| 4 | 33 | 2 | 33 | - | No | 1 | 323 × 96 | 163 × 96 | - |
| 5 | 33 | 1 | No | ReLu | Yes | 2 | 163 × 96 | 163 × 192 | 1,492,992 |
| 6 | 33 | 2 | 33 | - | No | 1 | 163 × 192 | 83 × 192 | - |
| 7 | 33 | 1 | No | ReLu | Yes | 2 | 83 × 192 | 83 × 384 | 5,971,968 |
| 8 | 23 | 2 | No | ReLu | Yes | 1 | 83 × 384 | 163 × (192 + 192) | 1,179,648 |
| 9 | 33 | 1 | No | ReLu | Yes | 2 | 163 × (192 + 192) | 163 × 192 | 2,985,984 |
| 10 | 23 | 2 | No | ReLu | Yes | 1 | 163 × 192 | 323 × (96 + 96) | 294,912 |
| 11 | 33 | 1 | No | ReLu | Yes | 2 | 323 × (96 + 96) | 323 × 96 | 746,496 |
| 12 | 23 | 2 | No | ReLu | Yes | 1 | 323 × 96 | 643 × (48 + 48) | 73,728 |
| 13 | 33 | 1 | No | ReLu | Yes | 2 | 643 × (48 + 48) | 643 × 48 | 186,624 |
| 14 | 13 | 1 | No | SoftMax | No | 1 | 643 × 48 | 643 × 2 | 96 |
| Total number of parameters | 13,369,200 | ||||||||
| Subject I.D. | Dice Coefficient | Sensitivity | Specificity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BSE | ROBEX | 3D UNet | BSE | ROBEX | 3D UNet | BSE | ROBEX | 3D UNet | |
| A00061276 | 0.9435 | 0.9568 | 0.9908 | 0.8883 | 0.9413 | 0.9853 | 0.9929 | 0.9710 | 0.9962 |
| A00061387 | 0.9547 | 0.9431 | 0.9911 | 0.9186 | 0.9530 | 0.9865 | 0.9876 | 0.9342 | 0.9956 |
| A00061709 | 0.9583 | 0.9528 | 0.9888 | 0.9306 | 0.9512 | 0.9890 | 0.9838 | 0.9543 | 0.9885 |
| A00061711 | 0.9663 | 0.9467 | 0.9896 | 0.9435 | 0.9395 | 0.9856 | 0.9876 | 0.9531 | 0.9935 |
| A00061806 | 0.9637 | 0.8793 | 0.9876 | 0.9313 | 0.8511 | 0.9797 | 0.9939 | 0.9015 | 0.9953 |
| A00062210 | 0.964 | 0.9508 | 0.9913 | 0.9336 | 0.9216 | 0.9891 | 0.9923 | 0.9774 | 0.9934 |
| A00062248 | 0.9585 | 0.9579 | 0.9914 | 0.9208 | 0.9434 | 0.9858 | 0.9933 | 0.9713 | 0.9969 |
| A00062266 | 0.9482 | 0.9546 | 0.9915 | 0.8919 | 0.9521 | 0.9863 | 0.9989 | 0.9569 | 0.9966 |
| A00062282 | 0.9587 | 0.9466 | 0.9897 | 0.9314 | 0.9519 | 0.9826 | 0.9838 | 0.9417 | 0.9967 |
| A00062288 | 0.9674 | 0.9473 | 0.9905 | 0.9438 | 0.9397 | 0.9842 | 0.9895 | 0.9542 | 0.9968 |
| A00062351 | 0.9554 | 0.9488 | 0.9909 | 0.9235 | 0.9328 | 0.9907 | 0.9846 | 0.9633 | 0.9912 |
| A00062917 | 0.9557 | 0.9549 | 0.9897 | 0.9278 | 0.9338 | 0.9839 | 0.9813 | 0.9741 | 0.9954 |
| A00062934 | 0.9197 | 0.9451 | 0.9921 | 0.8329 | 0.9409 | 0.9887 | 0.9935 | 0.9487 | 0.9954 |
| A00062942 | 0.9608 | 0.9507 | 0.9922 | 0.9225 | 0.9164 | 0.9877 | 0.9962 | 0.9817 | 0.9966 |
| A00063008 | 0.9705 | 0.9588 | 0.9915 | 0.9437 | 0.9334 | 0.9883 | 0.9957 | 0.9823 | 0.9947 |
| A00063103 | 0.9544 | 0.9602 | 0.9884 | 0.9116 | 0.9434 | 0.9794 | 0.9935 | 0.9757 | 0.9972 |
| A00063326 | 0.9436 | 0.9503 | 0.9896 | 0.8956 | 0.9372 | 0.9811 | 0.9864 | 0.9621 | 0.9979 |
| A00063368 | 0.953 | 0.9478 | 0.9861 | 0.9030 | 0.9370 | 0.9751 | 0.9985 | 0.9575 | 0.9967 |
| A00063589 | 0.9607 | 0.9512 | 0.9916 | 0.9349 | 0.9440 | 0.9877 | 0.9846 | 0.9577 | 0.9956 |
| A00064081 | 0.9647 | 0.9511 | 0.9923 | 0.9296 | 0.9209 | 0.9888 | 0.9974 | 0.9785 | 0.9958 |
| Mean | 0.9561 | 0.9477 | 0.9903 | 0.9179 | 0.9342 | 0.9853 | 0.9908 | 0.9599 | 0.9953 |
| Standard deviation | 0.0113 | 0.0168 | 0.0016 | 0.0260 | 0.0221 | 0.0040 | 0.0055 | 0.0192 | 0.0022 |
| Mean & Standard Deviation | Dice Coefficient | Sensitivity | Specificity | |||
|---|---|---|---|---|---|---|
| Method | J Kleesiek | 3D UNet | J Kleesiek | 3D UNet | J Kleesiek | 3D UNet |
| Mean | 0.9950 | 0.9903 | 0.9745 | 0.9853 | 0.9973 | 0.9953 |
| Std. | 0.0008 | 0.0016 | 0.0063 | 0.0040 | 0.0007 | 0.0022 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, H.; Rehman, H.Z.U.; Lee, S. 3D U-Net for Skull Stripping in Brain MRI. Appl. Sci. 2019, 9, 569. https://doi.org/10.3390/app9030569
Hwang H, Rehman HZU, Lee S. 3D U-Net for Skull Stripping in Brain MRI. Applied Sciences. 2019; 9(3):569. https://doi.org/10.3390/app9030569
Chicago/Turabian StyleHwang, Hyunho, Hafiz Zia Ur Rehman, and Sungon Lee. 2019. "3D U-Net for Skull Stripping in Brain MRI" Applied Sciences 9, no. 3: 569. https://doi.org/10.3390/app9030569
APA StyleHwang, H., Rehman, H. Z. U., & Lee, S. (2019). 3D U-Net for Skull Stripping in Brain MRI. Applied Sciences, 9(3), 569. https://doi.org/10.3390/app9030569







