Detection of Helminthosporium Leaf Blotch Disease Based on UAV Imagery
Abstract
:1. Introduction
2. Materials and Methods
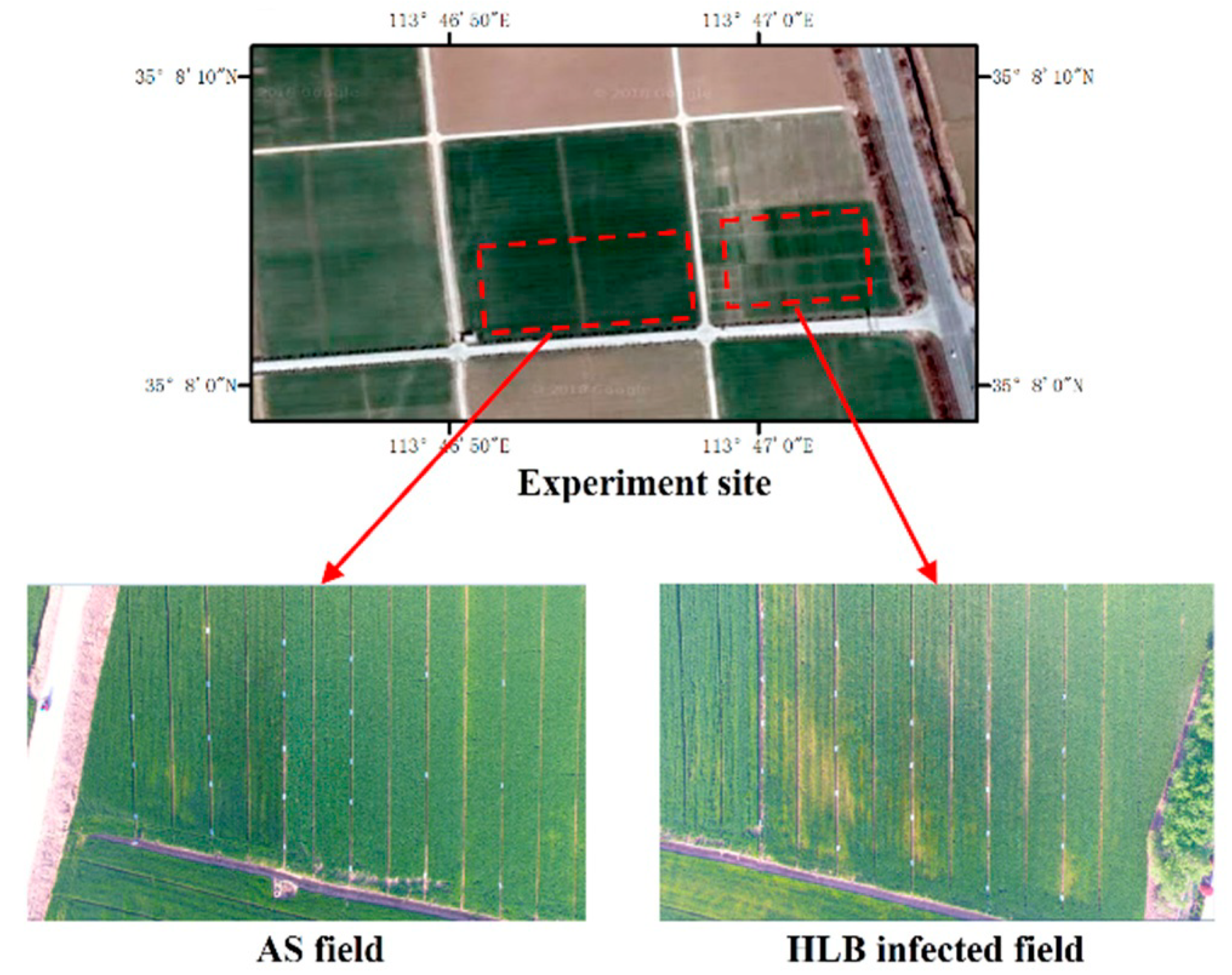
2.1. Study Site
2.2. Data Collection
2.2.1. UAV Imagery Collection
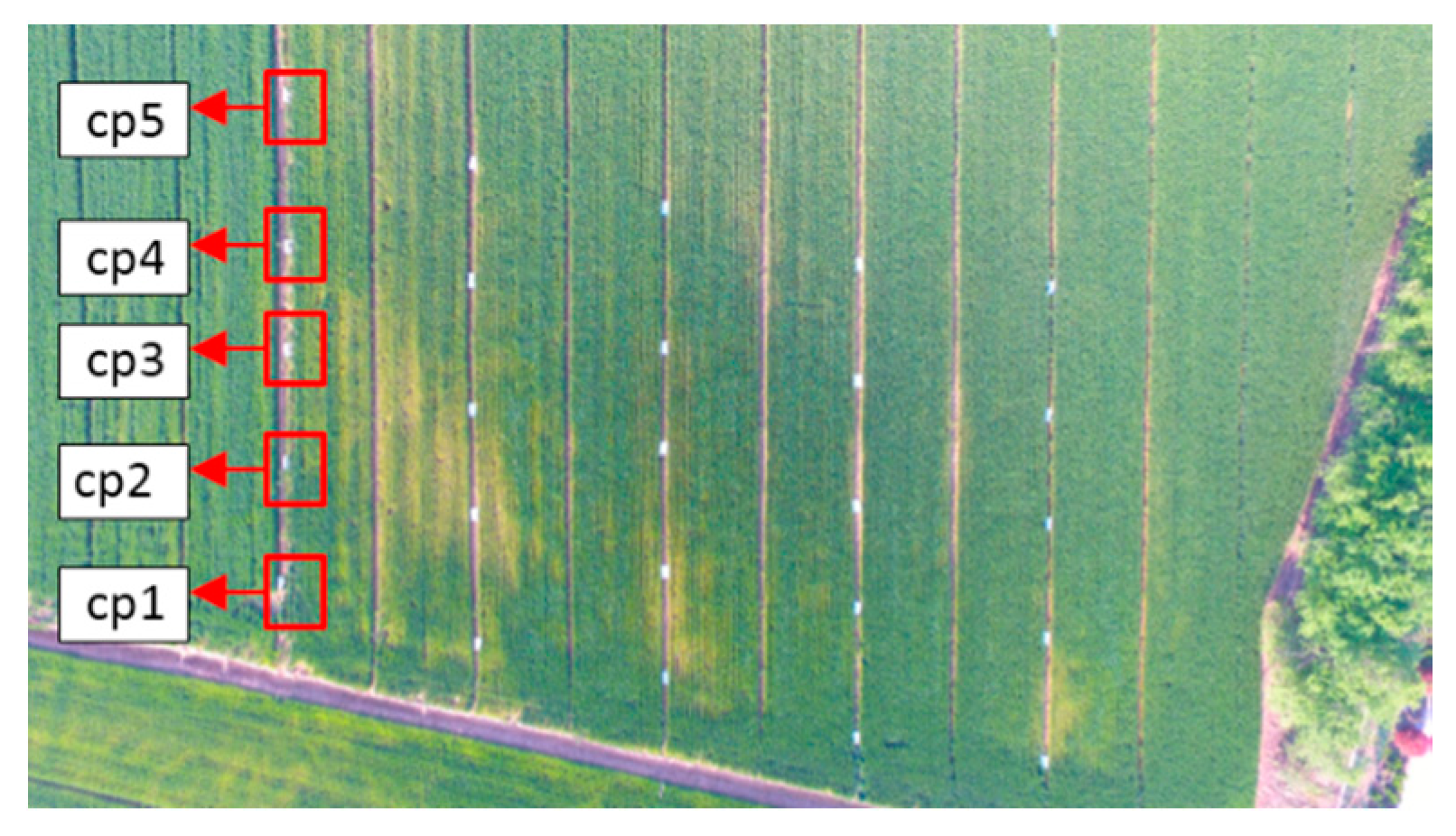
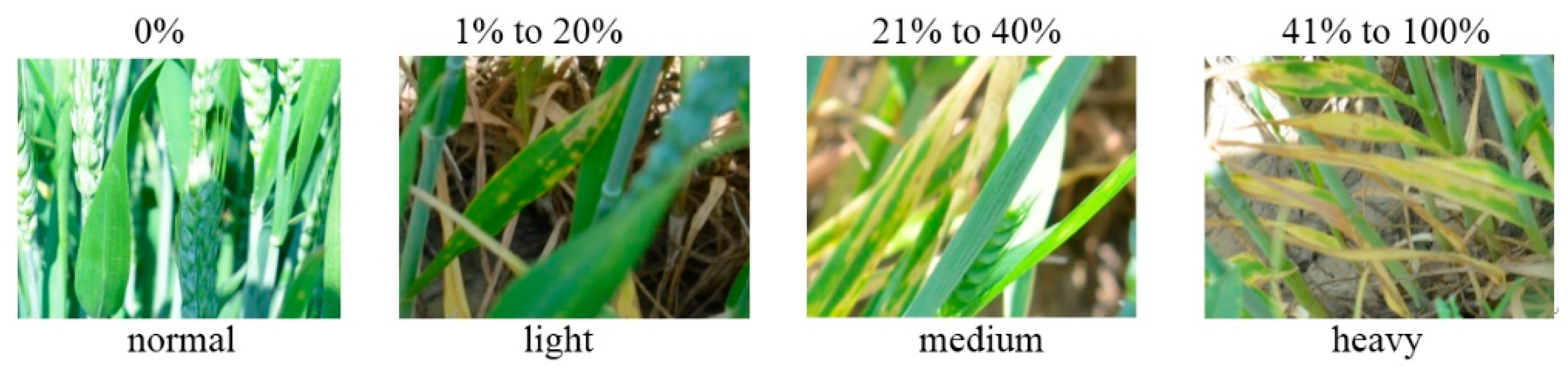
2.2.2. Ground Investigation
2.3. Data Analysis
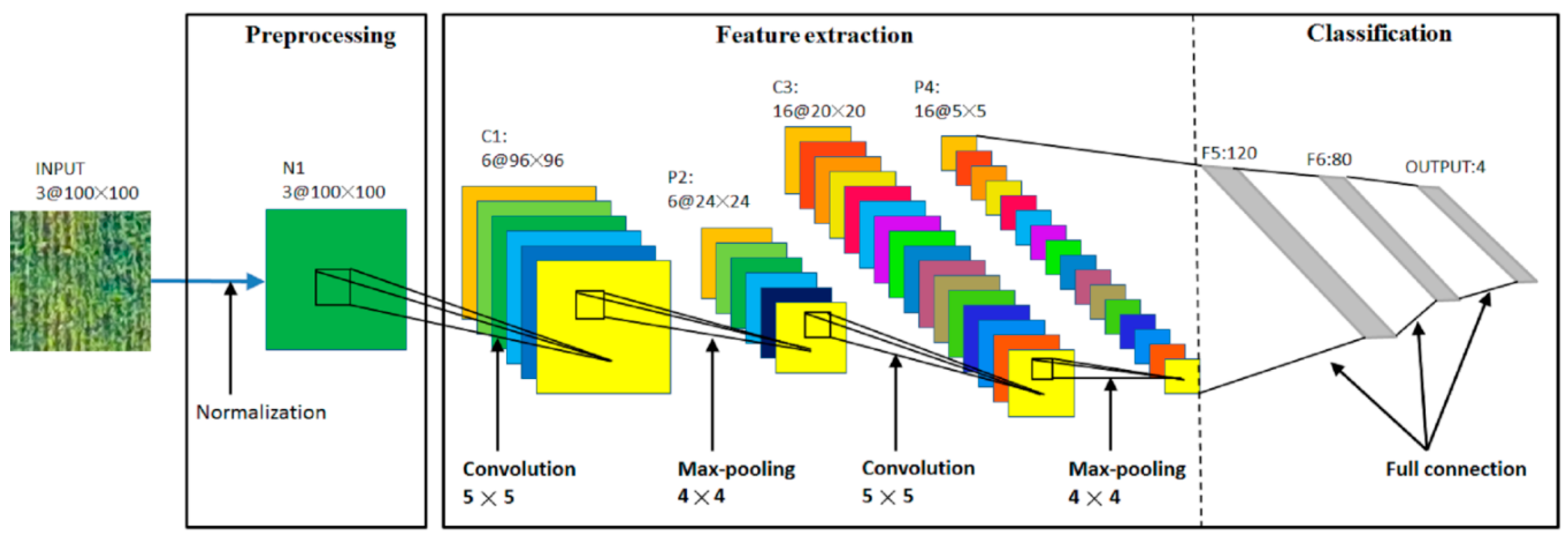
2.3.1. Preprocessing
2.3.2. Feature Extraction
2.3.3. Classification
2.4. Algorithms in Comparison
2.4.1. Color Histogram
2.4.2. Local Binary Pattern Histogram
2.4.3. Vegetation Index
2.4.4. Support Vector Machine
3. Results and Discussion
3.1. Dataset Preparation
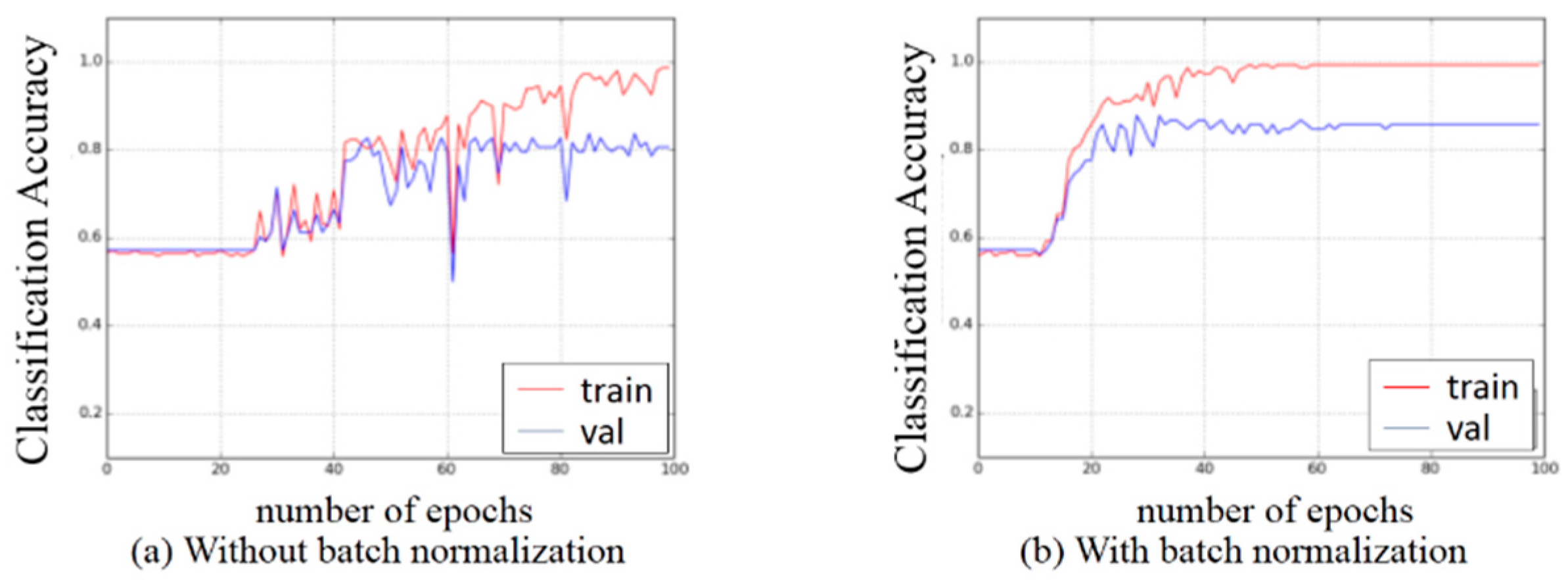
3.2. Experiments on Preprocessing
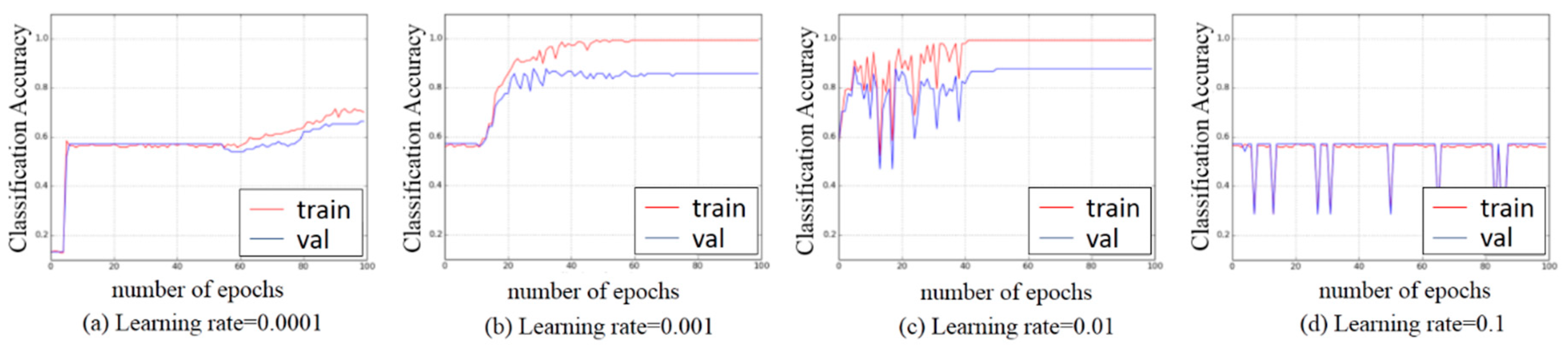
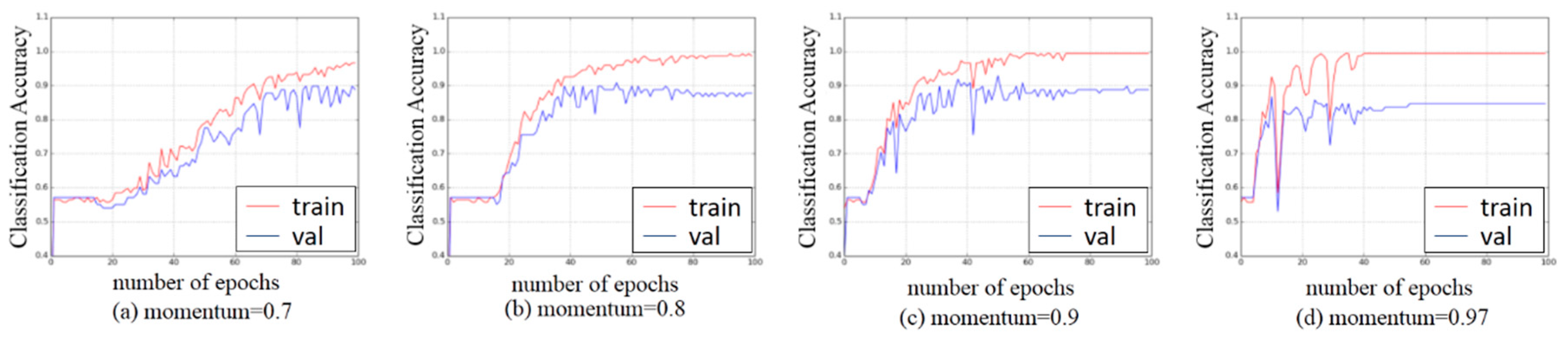
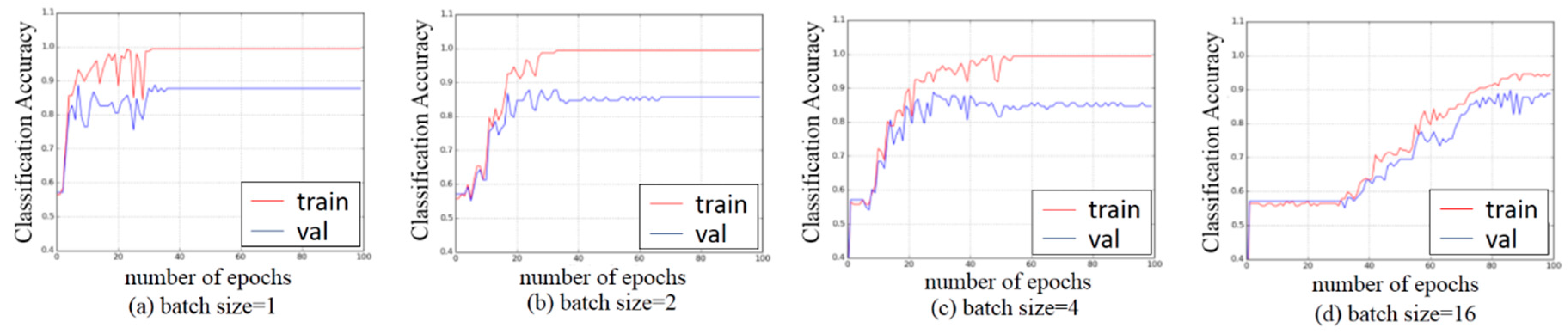
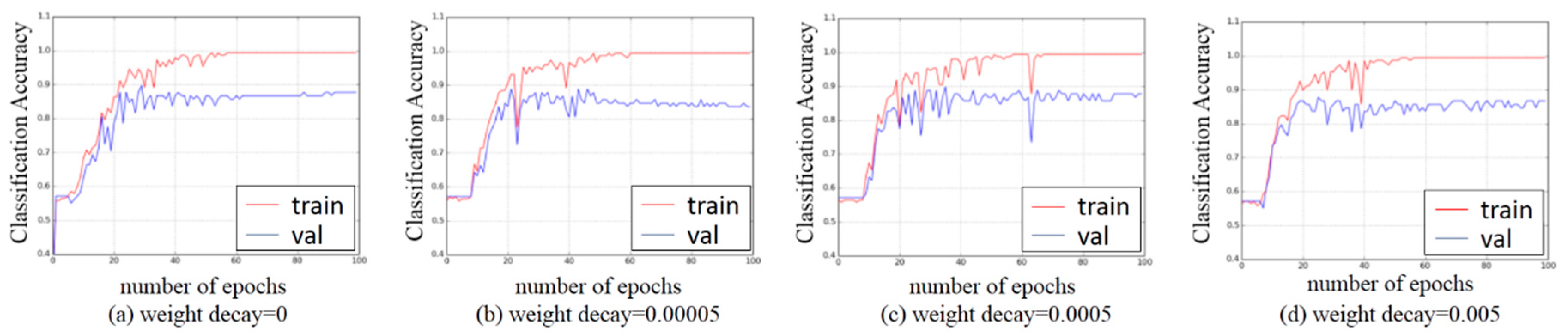
3.3. Experiments on Hyper-Parameters Tuning
3.4. Comparison with Other Methods
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhu, X.; Chang, N.; Zhou, C. Advance of research in helminthosporium leaf blotch of wheat. Agric. Sci. Technol. Equip. 2010, 08, 15–18. [Google Scholar]
- Saari, E.E. Leaf blight disease and associated soil-borne fungal pathogens of wheat in South and South East Asia. Helminthosporium Blights Wheat Spot Blotch Tan Spot 1998, 37–51. [Google Scholar]
- Sharma, R.C.; Duveiller, E. Effect of helminthosporium leaf blight on performance of timely and late-seeded wheat under optimal and stressed levels of soil fertility and moisture. Field Crops Res. 2004, 89, 205–218. [Google Scholar] [CrossRef]
- Huang, H.; Lan, Y.; Deng, J.; Yang, A.; Deng, X.; Zhang, L.; Wen, S. A semantic labeling approach for accurate weed mapping of high resolution UAV imagery. Sensors 2018, 18, 2113. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Deng, J.; Lan, Y.; Yang, A.; Deng, X.; Wen, S.; Zhang, H.; Zhang, Y. Accurate weed mapping and prescription map generation based on fully convolutional networks using UAV imagery. Sensors 2018, 18, 3299. [Google Scholar] [CrossRef]
- Peña, J.; Torressánchez, J.; Serranopérez, A.; Lópezgranados, F. Weed mapping in early-season maize fields using object-based analysis of unmanned aerial vehicle (UAV) images. PLoS One 2013, 8, e77151. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Deng, J.; Lan, Y.; Yang, A.; Deng, X.; Zhang, L.; Wen, S.; Jiang, Y.; Suo, G.; Chen, P. A two-stage classification approach for the detection of spider mite- infested cotton using UAV multispectral imagery. Remote Sens. Lett. 2018, 9, 933–941. [Google Scholar] [CrossRef]
- Franke, J.; Menz, G. Multi-temporal wheat disease detection by multi-spectral remote sensing. Precis. Agric. 2007, 8, 161–172. [Google Scholar] [CrossRef]
- Albetis, J.; Duthoit, S.; Guttler, F.; Jacquin, A.; Goulard, M.; Poilvé, H.; Féret, J.B.; Dedieu, G. Detection of flavescence dorée grapevine disease using unmanned aerial vehicle (UAV) multispectral imagery. Remote Sens. 2017, 9, 308. [Google Scholar] [CrossRef]
- Mirik, M.; Jones, D.C.; Price, J.A.; Workneh, F.; Ansley, R.J.; Rush, C.M. Satellite remote sensing of wheat infected by Wheat streak mosaic virus. Plant Dis. 2011, 95, 4–12. [Google Scholar] [CrossRef]
- Huang, W.; Lamb, D.W.; Niu, Z.; Zhang, Y.; Liu, L.; Wang, J. Identification of yellow rust in wheat using in-situ spectral reflectance measurements and airborne hyperspectral imaging. Precis. Agric. 2007, 8, 187–197. [Google Scholar] [CrossRef]
- Lan, Y.B.; Chen, S.D.; Fritz, B.K. Current status and future trends of precision agricultural aviation technologies. Int. J. Agric. Biol. Eng. 2017, 10, 1–17. [Google Scholar]
- Leng, W.F.; Wang, H.G.; Xu, Y.; Ma, Z.H. Preliminary study on monitoring wheat stripe rust with using UAV. Acta Phytopathol. Sin. 2012, 42, 202–205. [Google Scholar]
- Wei, L.; Cao, X.; Fan, J.R.; Wang, Z.; Yan, Z.; Yong, L.; West, J.S.; Xu, X.; Zhou, Y. Detecting wheat powdery mildew and predicting grain yield using unmanned aerial photography. Plant Dis. 2018, 12–17. [Google Scholar]
- López-Granados, F.; Torres-Sánchez, J.; Serrano-Pérez, A.; de Castro, A.I.; Mesas-Carrascosa, F.J.; Peña, J. Early season weed mapping in sunflower using UAV technology: variability of herbicide treatment maps against weed thresholds. Precs. Agric. 2016, 17, 183–199. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Ioffe, S.; Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv, 2015; arXiv:1502.03167. [Google Scholar]
- Boureau, Y.L.; Bach, F.; Lecun, Y.; Ponce, J. Learning Mid-Level Features for Recognition. In Proceedings of the 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (2010), San Francisco, CA, USA, 13 June–18 June 2010. [Google Scholar]
- Bejiga, M.; Zeggada, A.; Nouffidj, A.; Melgani, F. A convolutional neural network approach for assisting avalanche search and rescue operations with UAV imagery. Remote Sens. 2017, 9, 100. [Google Scholar] [CrossRef]
- Ammour, N.; Alhichri, H.; Bazi, Y.; Benjdira, B.; Alajlan, N.; Zuair, M. Deep learning approach for car detection in UAV imagery. Remote Sens. 2017, 9, 312. [Google Scholar] [CrossRef]
- Längkvist, M.; Kiselev, A.; Alirezaie, M.; Loutfi, A. Classification and segmentation of satellite orthoimagery using convolutional neural networks. Remote Sens. 2016, 8, 329. [Google Scholar] [CrossRef]
- Lecun, Y.; Kavukcuoglu, K.; Farabet, C.M. Convolutional networks and applications in vision. In Proceedings of the 2010 IEEE International Symposium on Circuits and Systems, Paris, France, 30 May–2 June 2010. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Proceedings of the International Conference on Neural Information Processing Systems, Lake Tahoe, Nevada, 3–8 December 2012. [Google Scholar] [CrossRef]
- Swain, M.J.; Ballard, D.H. Indexing via color histograms. In Proceedings of the 2010 IEEE International Symposium on Circuits and Systems, Osaka, Japan, 4–7 December 1990. [Google Scholar]
- Ojala, T.; Harwood, I. A comparative study of texture measures with classification based on feature distributions. Pattern Recognit. 1996, 29, 51–59. [Google Scholar] [CrossRef]
- Ahonen, T.; Hadid, A.; Pietikainen, M. Face description with local binary patterns: Application to face recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 12, 2037–2041. [Google Scholar]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Hunt, E.R.; Cavigelli, M.; Daughtry, C.S.T.; Mcmurtrey, J.E.; Walthall, C.L. Evaluation of digital photography from model aircraft for remote sensing of crop biomass and nitrogen status. Precis. Agric. 2005, 6, 359–378. [Google Scholar] [CrossRef]
- Wang, X.; Wang, M.; Wang, S.; Wu, Y. Extraction of vegetation information from visible unmanned aerial vehicle images. Trans. Chin. Soc. Agric. Eng. 2015, 31, 152–159. [Google Scholar]
- Sun, G.; Wang, X.; Yan, T.; Xue, L.; Man, C.; Shi, Y.; Chen, J.; University, N.A. Inversion method of flora growth parameters based on machine vision. Trans. Chin. Soc. Agric. Eng. 2014, 30, 187–195. [Google Scholar]
- Louhaichi, M.; Borman, M.; Johnson, D. Spatially located platform and aerial photography for documentation of grazing impacts on wheat. Geocarto Int. 2001, 16, 65–70. [Google Scholar] [CrossRef]
- Gamon, J.A.; Surfus, J.S. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 2010, 143, 105–117. [Google Scholar] [CrossRef]
- Suykens, J.A.K. Support vector machines: A nonlinear modelling and control perspective. Eur. J. Control. 2001, 7, 311–327. [Google Scholar] [CrossRef]
- Chi, M.; Feng, R.; Bruzzone, L. Classification of hyperspectral remote-sensing data with primal SVM for small-sized training dataset problem. Adv. Space Res. 2008, 41, 1793–1799. [Google Scholar] [CrossRef]
- Domhan, T.; Springenberg, J.T.; Hutter, F. Speeding up automatic hyperparameter optimization of deep neural networks by extrapolation of learning curves. In Proceedings of the 24th International Conference on Artificial Intelligence, Buenos Aires, Argentina, 25–31 July 2015. [Google Scholar]
- Guggenmoos-Holzmann, I. The meaning of kappa: probabilistic concepts of reliability and validity revisited. J. Clin. Epidemiol. 1996, 49, 775–782. [Google Scholar] [CrossRef]


| Phantom 4 | Quad-Rotor UAV |
|---|---|
| Cross weight | 1380 g |
| Diagonal Size | 350 mm |
| Max Speed | 20 m/s |
| Max Flight Time | 28 minutes |
| Size | 4000 × 300 pixels |
| Lens | FOV 94°20 mm |
| Typical spatial resolution (at 80 m altitude) | 3.4 cm/pixel |
| Index | Formula | References |
|---|---|---|
| Normalized Green–Red Difference Index | [27] | |
| Normalized Green–Blue Difference Index | [28] | |
| Excess Green | ExG = 2 × Green-Red-Blue | [29] |
| Excess Red | ExR = 1.4 × Red-Green | [30] |
| Excess Green - Excess Red | ExGR = 3×Green-2.4 × Red-Blue | [30] |
| Green Leaf Index | [31] | |
| Red–Green Rate Index | RGRI = Red/Green | [32] |
| Blue–Green Rate Index | BGRI = Blue/Green | [32] |
| Categories | Training | Validation |
|---|---|---|
| normal | 84 | 56 |
| light | 44 | 29 |
| medium | 20 | 13 |
| heavy | 0 | 0 |
| Total. | 148 | 98 |
| Hyper-parameter | Learning Rate | Momentum | Batch Size | Weight Decay |
|---|---|---|---|---|
| Value | 0.001 | 0.9 | 4 | 0 |
| Method | OA (%) | SE (%) |
|---|---|---|
| Color Histogram + SVM | 85.92 | 1.31 |
| LBPH + SVM | 65.10 | 2.86 |
| VI + SVM | 87.65 | 1.17 |
| Color Histogram + LBPH + VI + SVM | 90.00 | 0.96 |
| CNN | 91.43 | 0.83 |
| Method | GT/Predicted Class | Normal (%) | Light (%) | Medium (%) |
|---|---|---|---|---|
| Color Histogram + SVM | normal | 89.11 | 10.89 | 0.00 |
| light | 13.79 | 84.48 | 1.72 | |
| medium | 2.31 | 22.31 | 75.38 | |
| LBPH + SVM | normal | 98.57 | 1.43 | 0.00 |
| light | 72.07 | 27.59 | 0.34 | |
| medium | 48.46 | 46.92 | 4.62 | |
| VI + SVM | normal | 88.39 | 11.61 | 0.00 |
| light | 8.97 | 88.62 | 2.41 | |
| medium | 0.77 | 16.92 | 82.31 | |
| Color Histogram + LBPH + VI + SVM | normal | 94.33 | 5.67 | 0.00 |
| light | 9.16 | 89.28 | 1.56 | |
| medium | 1.80 | 21.48 | 76.72 | |
| CNN | normal | 93.93 | 6.07 | 0.00 |
| light | 7.93 | 88.62 | 3.45 | |
| medium | 0.00 | 13.08 | 86.92 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, H.; Deng, J.; Lan, Y.; Yang, A.; Zhang, L.; Wen, S.; Zhang, H.; Zhang, Y.; Deng, Y. Detection of Helminthosporium Leaf Blotch Disease Based on UAV Imagery. Appl. Sci. 2019, 9, 558. https://doi.org/10.3390/app9030558
Huang H, Deng J, Lan Y, Yang A, Zhang L, Wen S, Zhang H, Zhang Y, Deng Y. Detection of Helminthosporium Leaf Blotch Disease Based on UAV Imagery. Applied Sciences. 2019; 9(3):558. https://doi.org/10.3390/app9030558
Chicago/Turabian StyleHuang, Huasheng, Jizhong Deng, Yubin Lan, Aqing Yang, Lei Zhang, Sheng Wen, Huihui Zhang, Yali Zhang, and Yusen Deng. 2019. "Detection of Helminthosporium Leaf Blotch Disease Based on UAV Imagery" Applied Sciences 9, no. 3: 558. https://doi.org/10.3390/app9030558
APA StyleHuang, H., Deng, J., Lan, Y., Yang, A., Zhang, L., Wen, S., Zhang, H., Zhang, Y., & Deng, Y. (2019). Detection of Helminthosporium Leaf Blotch Disease Based on UAV Imagery. Applied Sciences, 9(3), 558. https://doi.org/10.3390/app9030558










