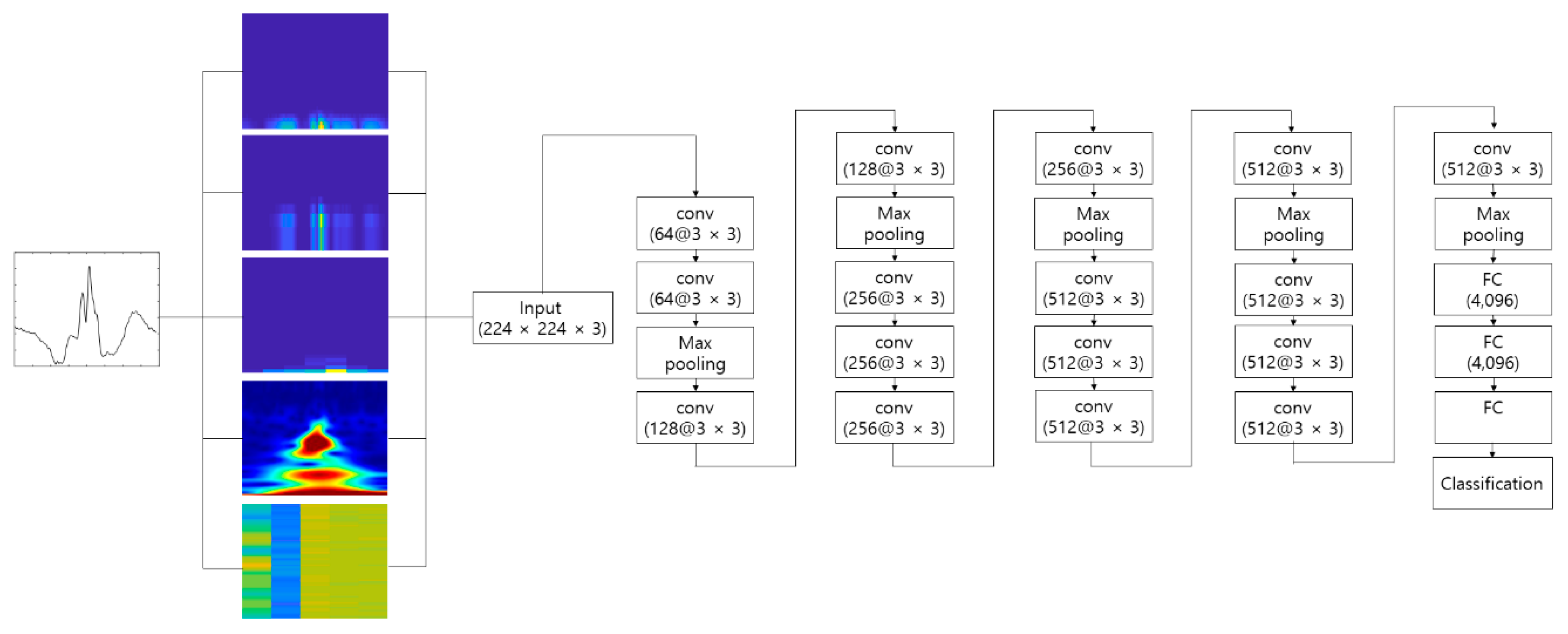
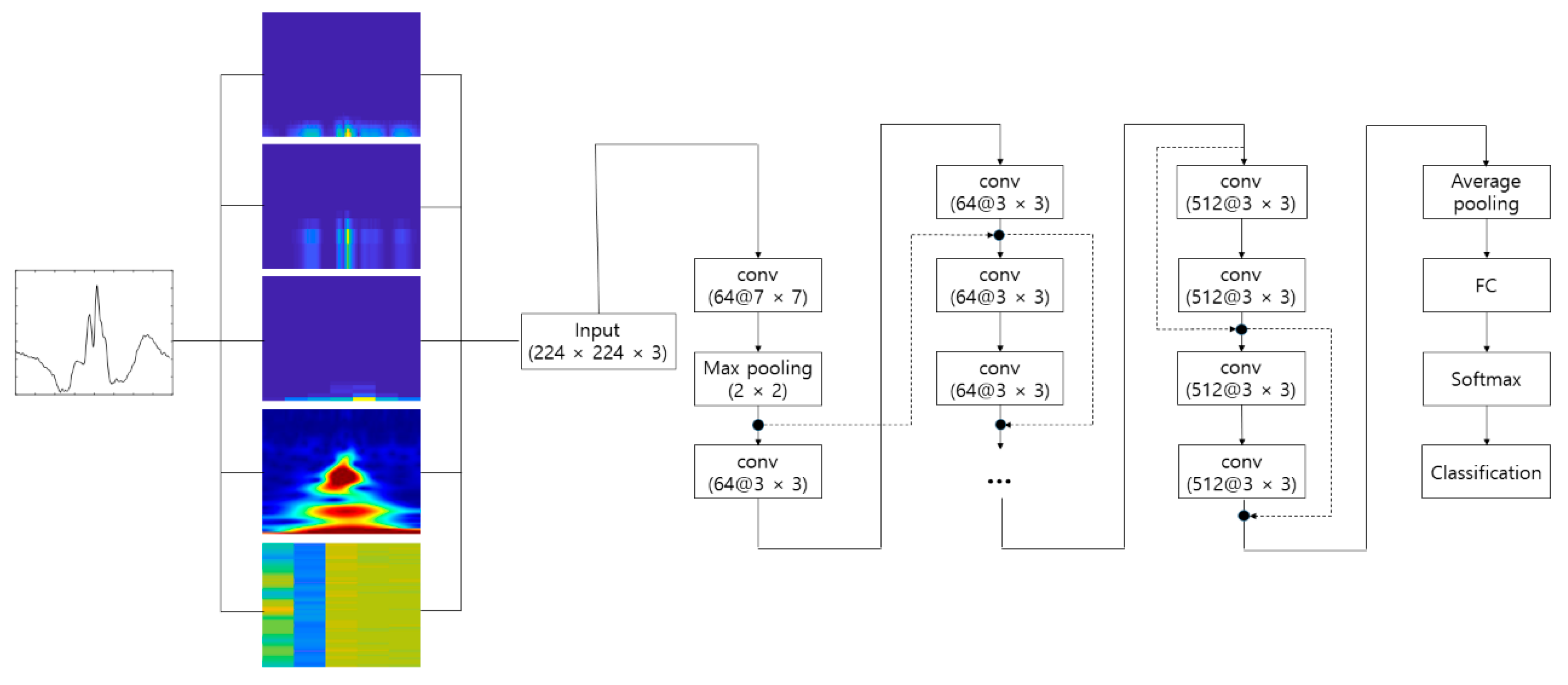
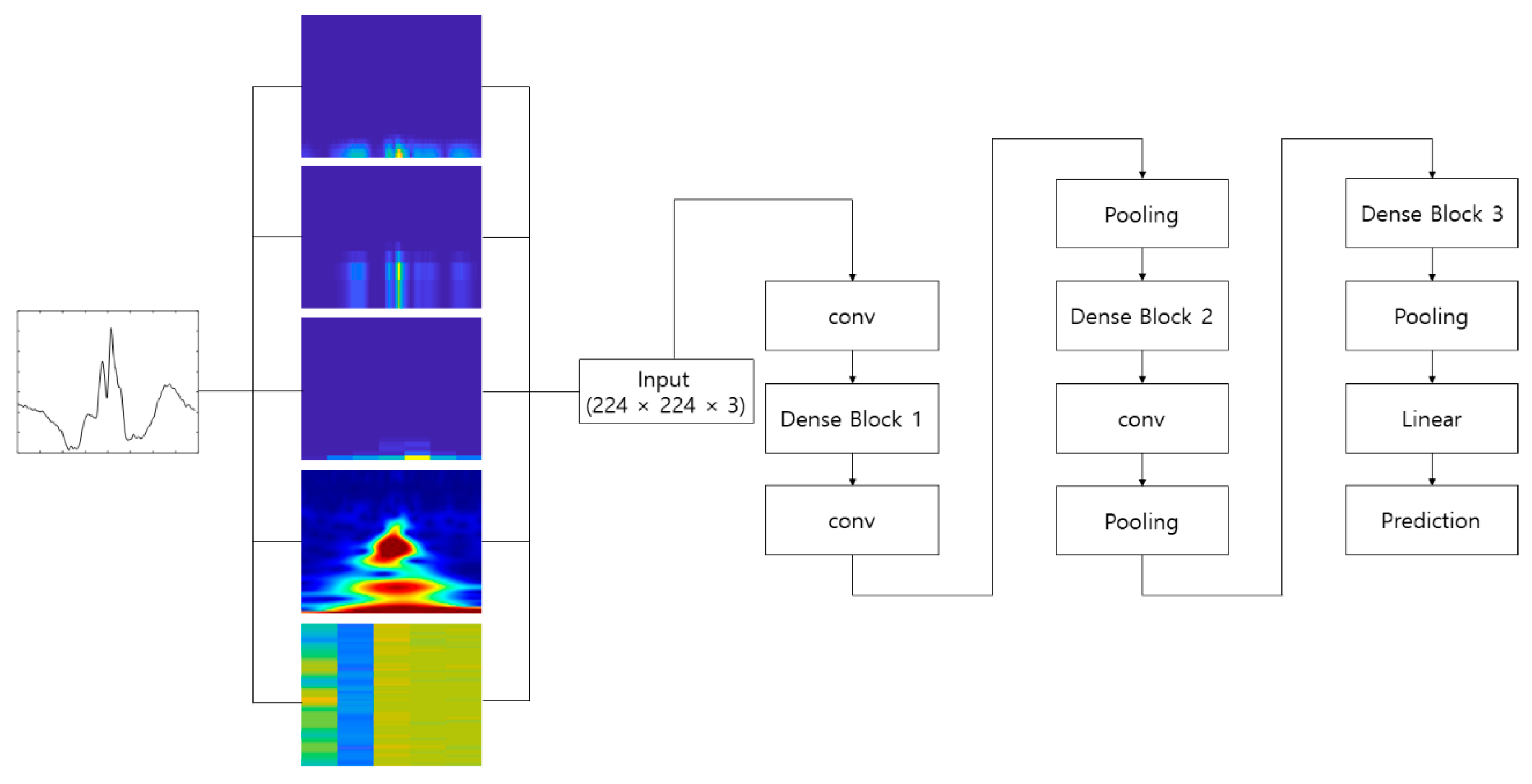
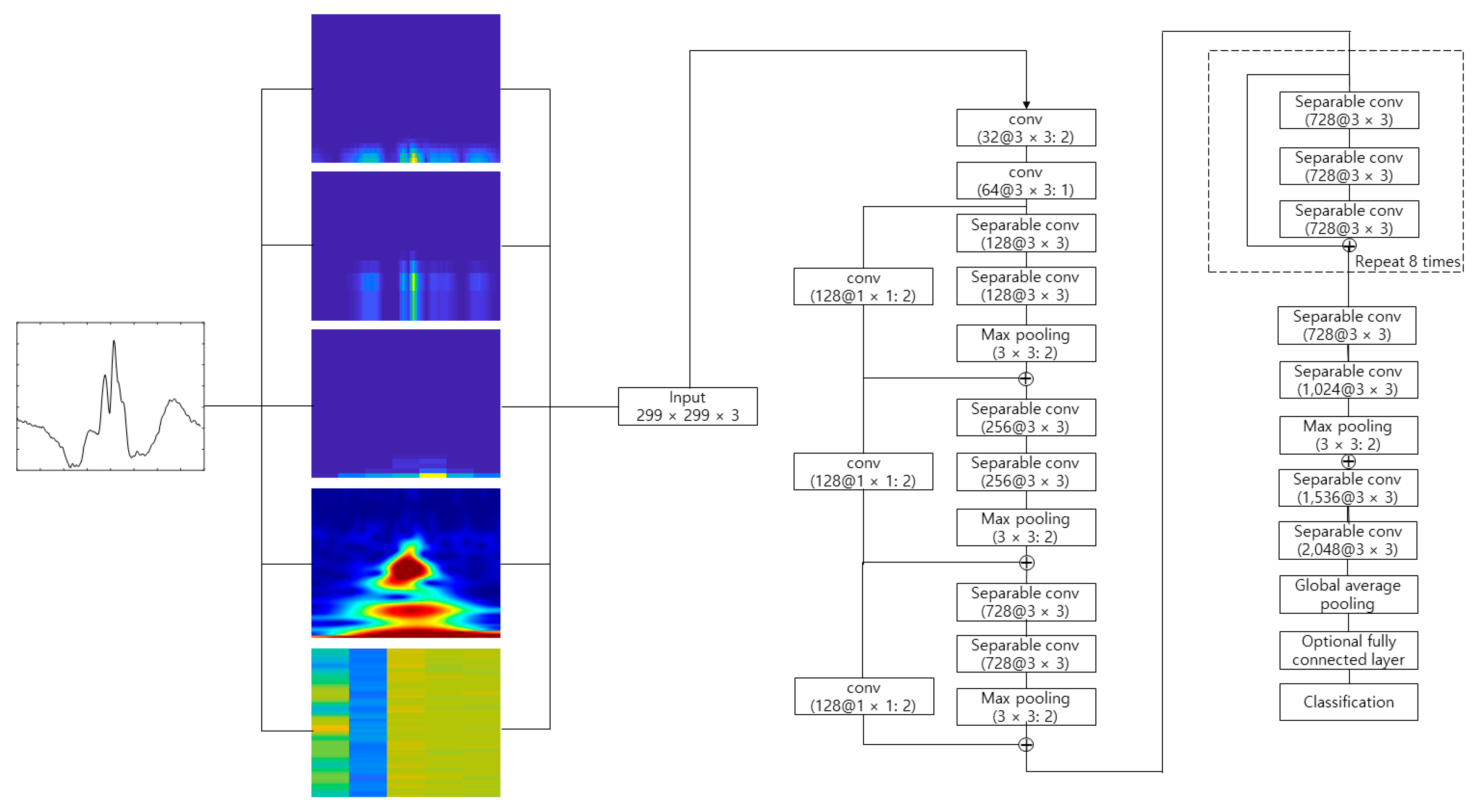
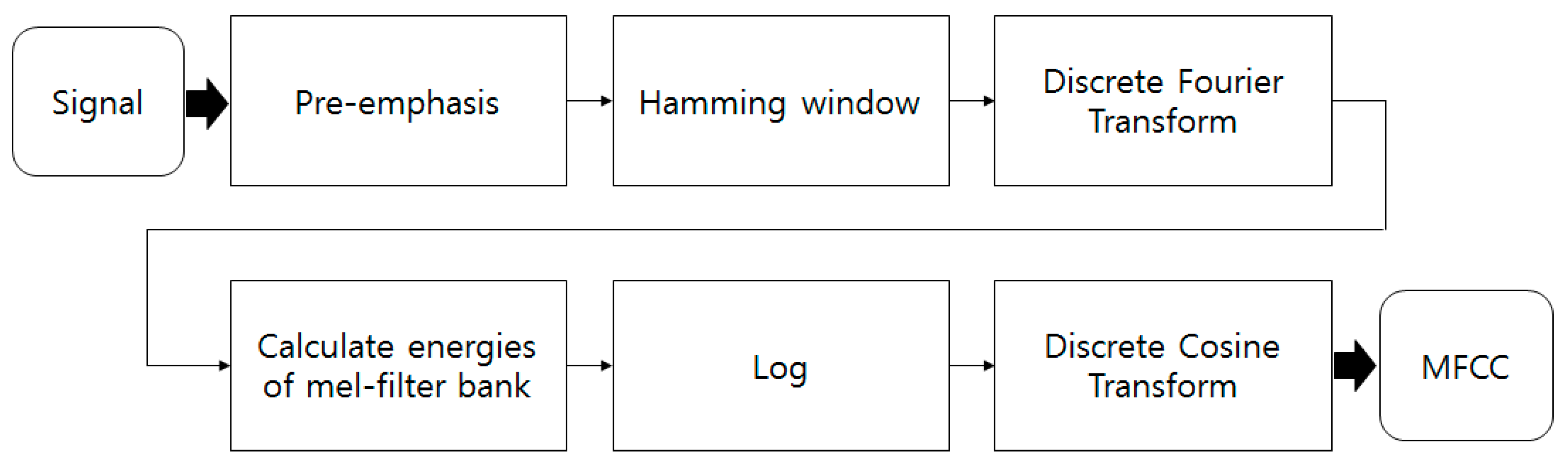
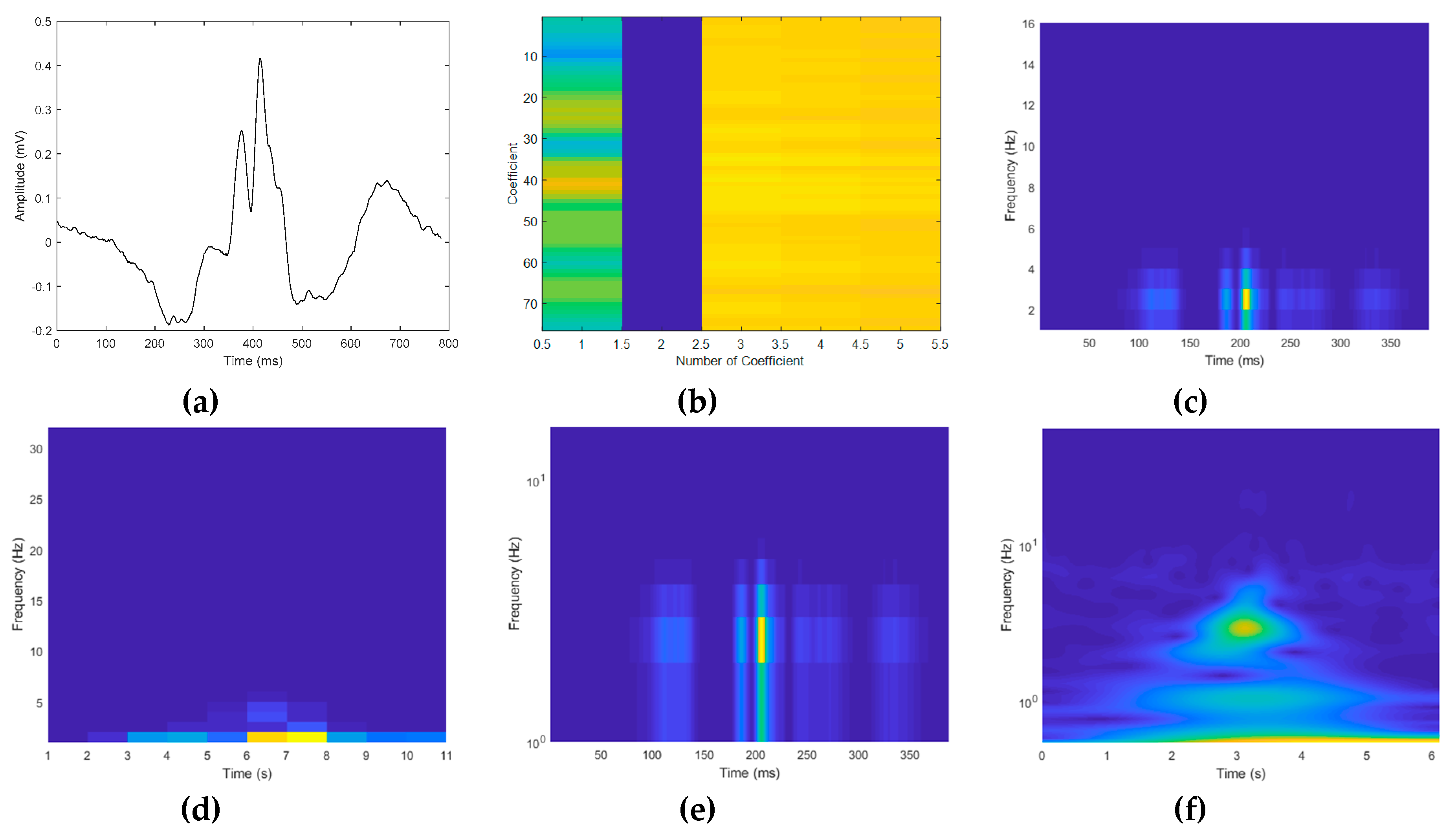
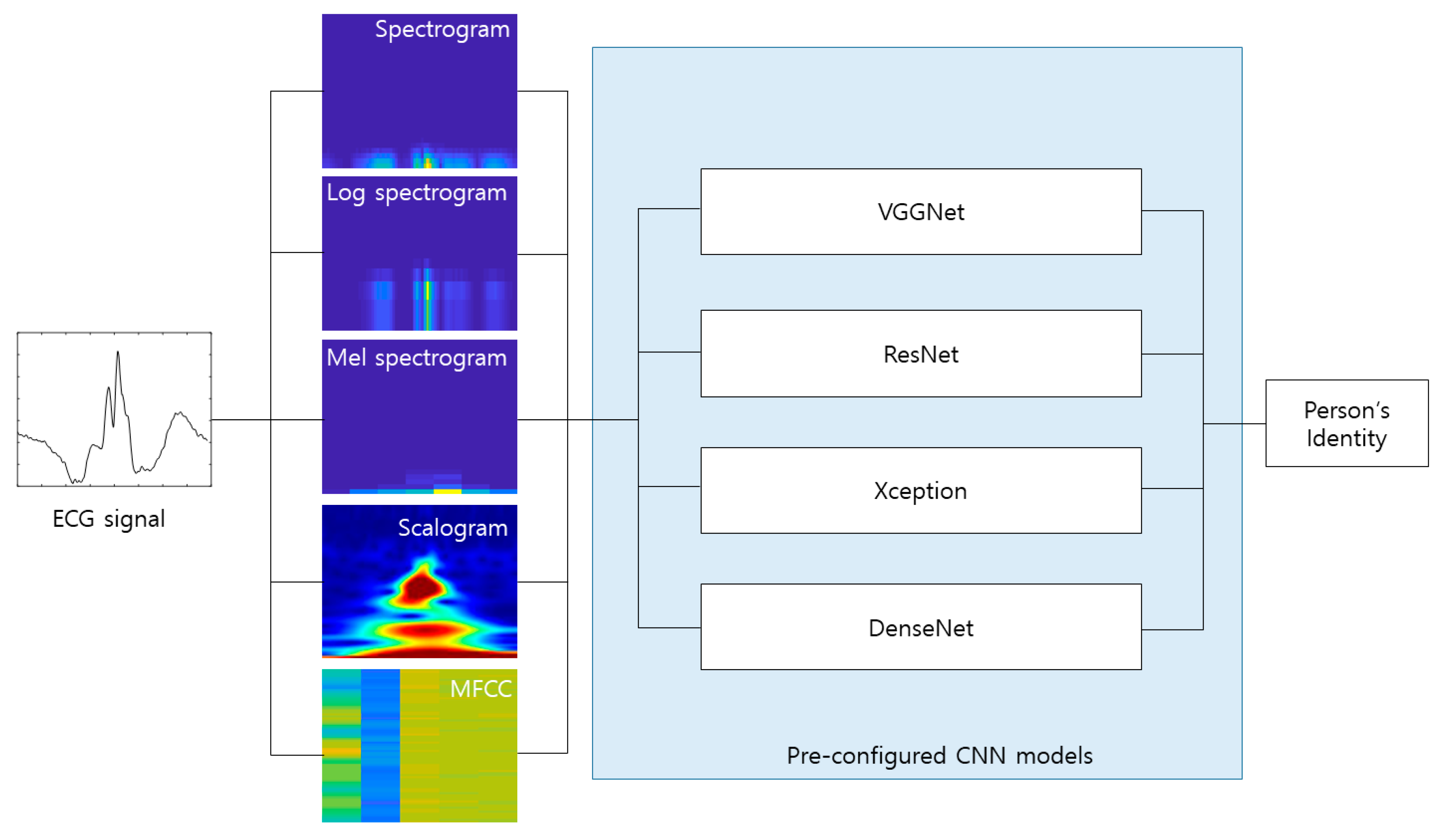
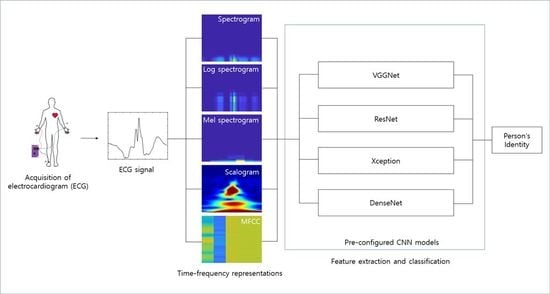
4.2. Experimental Results
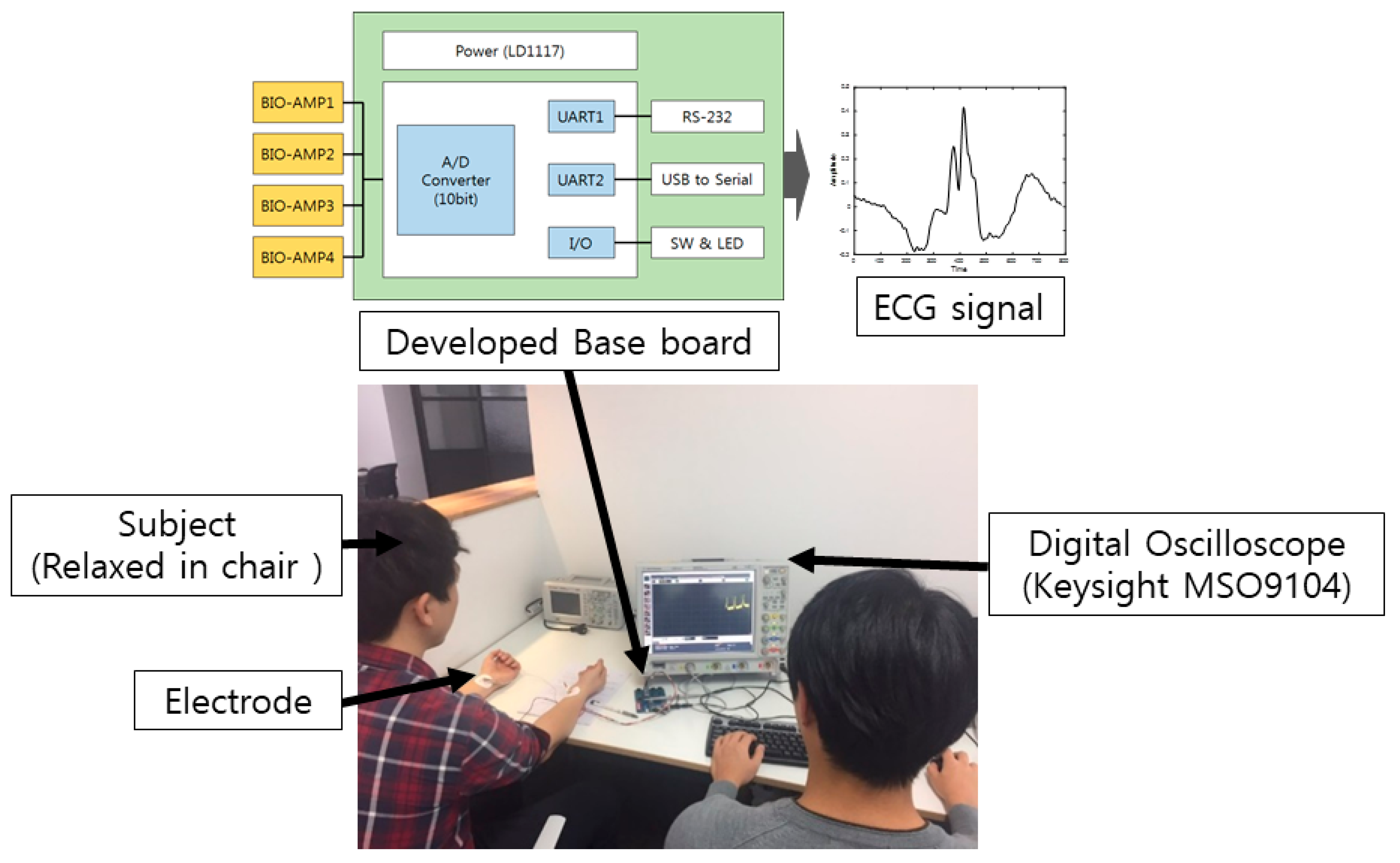
The experiment was performed using a computer with the following specifications: Nvidia GeForce GTX 1080 Ti, Intel i7-6850K central processing unit at 3.60 GHz, Windows 10 64-bit operating system, and 64 GB random-access memory. In this study, ECG biometrics using pre-configured models of the CNN with various time-frequency representations were evaluated. The signals were preprocessed to reduce the noise, and the R peak points were detected to normalize the center point of the ECG data. In other words, one data point was extracted based on each detected R peak point. However, the number of R peaks detected for each recording was different, as each person had a different heart rate, and the detection rate of the R peak was different for each signal. To construct the same amount of data for each class, the total number of R peak points detected for each class was calculated. The class was excluded when the number of detected R peaks was too small. When the number of detected R peaks was large, a few detected R peaks were removed to ensure the same amount of data in each class. Considering only the data from lead I, a frame length of 784 centered on the detected R peak point was obtained. The data were then transformed to the time-frequency representations. Here, the spectrogram, log spectrogram, mel spectrogram, scalogram, and MFCC were considered as the time-frequency representations. The transformed ECG data were 2D, which could be applied to the 2D CNNs.
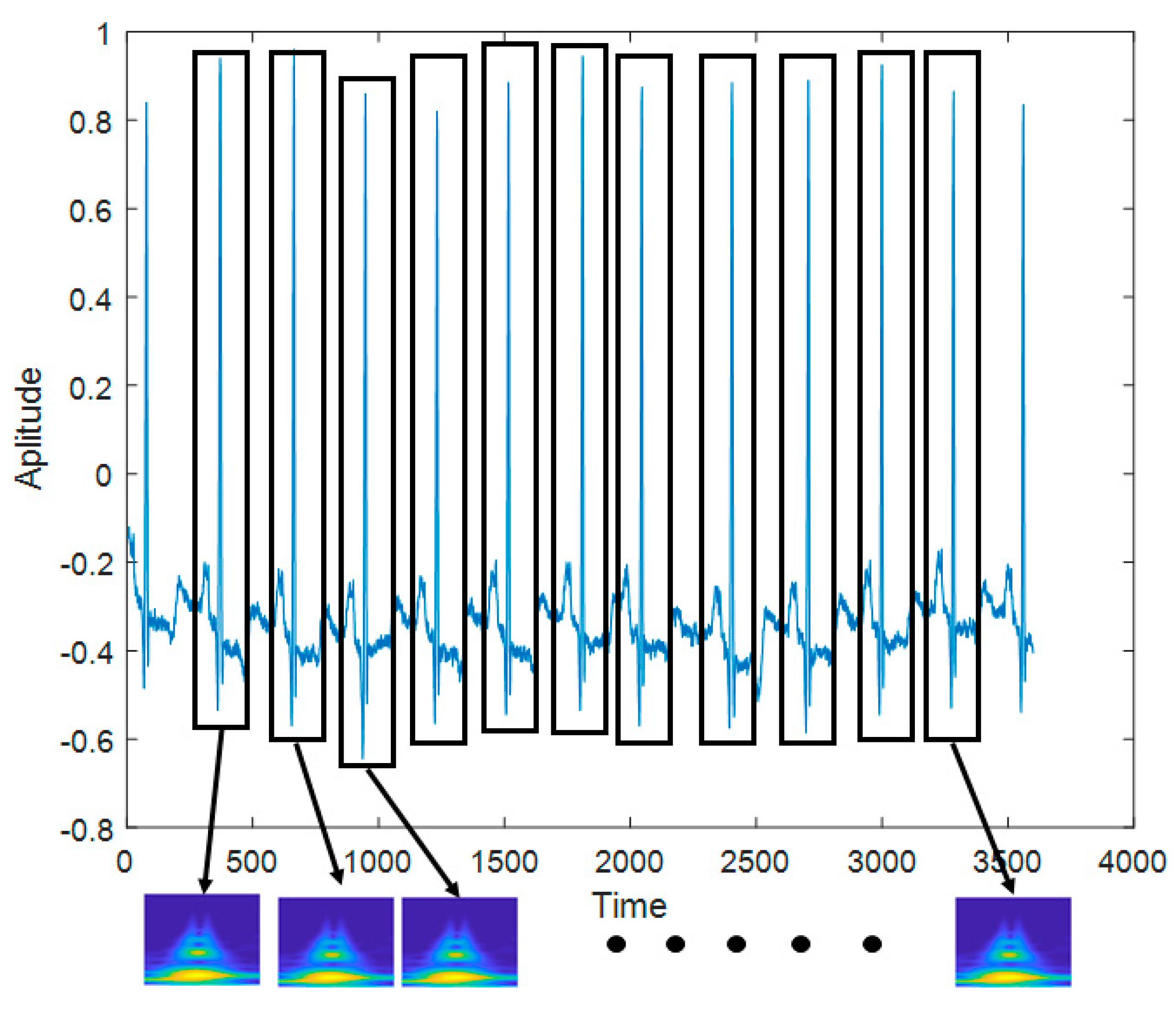
Figure 10 shows the process of making the input image from the raw ECG signal. The pre-configured models based on the 2D CNN were the VGGNet, Xception, ResNet, and DenseNet models discussed earlier. The 2D ECG image was input to the CNN, and the neural network was trained to classify its identity. The depth of the neural network in the VGGNet, ResNet, and DenseNet models was 19, 101, and 201, respectively. We did not consider a potential issue from the variability from the selection of the ECG sensor devices. A different ECG sensor device (or product) would change the shape of the ECG signals and accuracy.
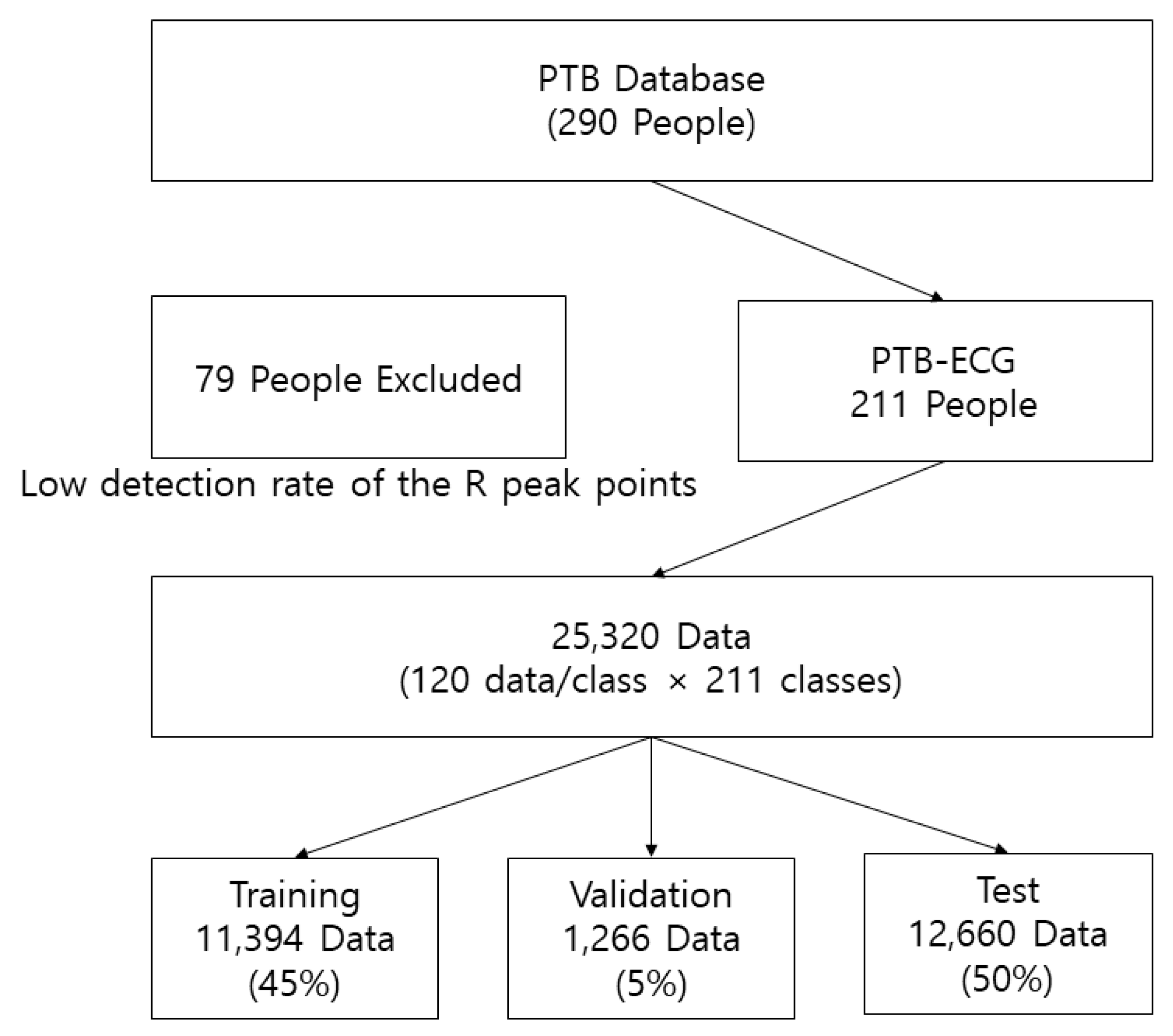
PTB-ECG is an open access database and includes ECG data from 290 people. However, the data from 211 people were used to construct the PTB-ECG database as the ECG data from the remaining 79 had only few R peak points. The R peak points from 211 people were compared and adjusted to ensure that the maximum number of data per class was 120. The database size of PTB-ECG was 784 × 25, 320 (120 data/class × 211 classes). The row refers to the dimension of the data, while the column indicates the number of data. To train the CNNs, PTB-ECG was divided into the training, validation, and test datasets with ratios of 0.45, 0.05, and 0.5, respectively. To shorten the training time, the training set was composed of 45% normally less than other literature works in the field of machine learning, and a larger ratio for the training set results in too high accuracy to compare performance. The sizes of the training, validation, and test datasets were 784 × 11,394, 784 × 1266, and 784 × 12,660, respectively.
Figure 11 shows the structure of PTB-ECG. The CNNs were trained using a mini-batch size of 30, an initial learning rate of 0.0001, the Adam (adaptive moment estimation) training optimizer, and an epoch varied according to the model. PCA (principle component analysis)-L2 was executed by entering vectors reshaped from the diminished input image and measuring Euclidean distance (L2). The PCA dimension was 20.
Table 1 shows the accuracies achieved by PCA-L2 using time-frequency representations on PTB-ECG. PCANet [
42] is a neural network that has a CNN architecture based on PCA. PCANet was executed with 4 as the patch size, 4 filters, 4 as the block size, and 0.5 as the block overlap ratio.
Table 2 shows the accuracies achieved by PCANet using time-frequency representations on PTB-ECG.
Table 3 presents the accuracies achieved by the different models based on CNN using time-frequency representations on PTB-ECG. Considering MFCC on PTB-ECG, CNN failed to train VGGNet-19. The best accuracies achieved by the time-frequency representations on PTB-ECG were 98.99% for DenseNet-201 in MFCC, 98.85% for Xception in spectrogram, 97.16% for Xception in log spectrogram, 96.92% for Xception in mel spectrogram, and 97.83% for ResNet-101 in scalogram. This paper did not consider a potential issue from the variability from the selection of the ECG sensor devices. A different ECG sensor device (or product) would change the shape of the ECG signals and accuracy. The MFCC input to the DenseNet-201 model achieved the best accuracy for the PTB-ECG dataset.
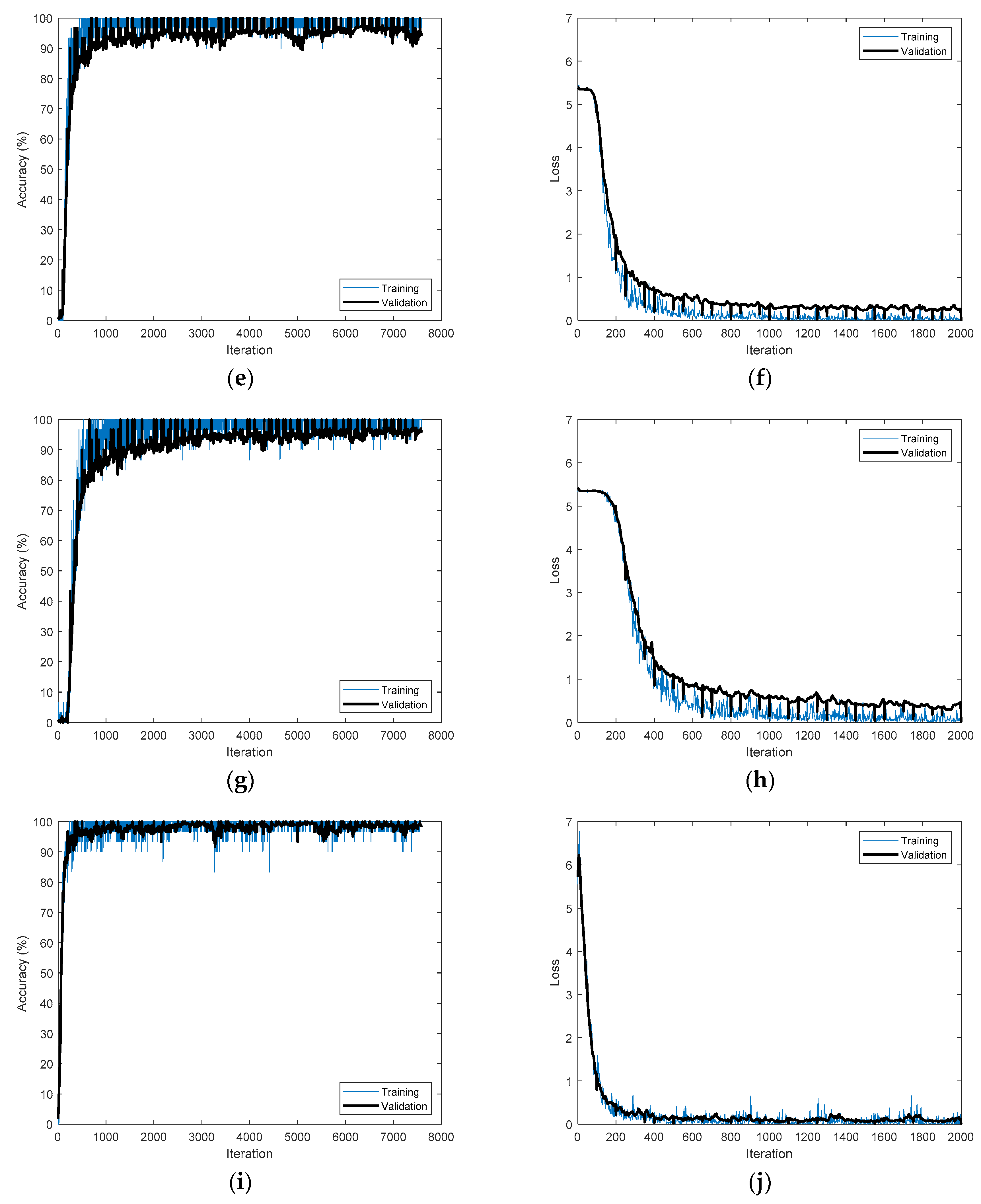
Figure 12 shows the training processes that achieved the best accuracies for the time-frequency representations applied to PTB-ECG.
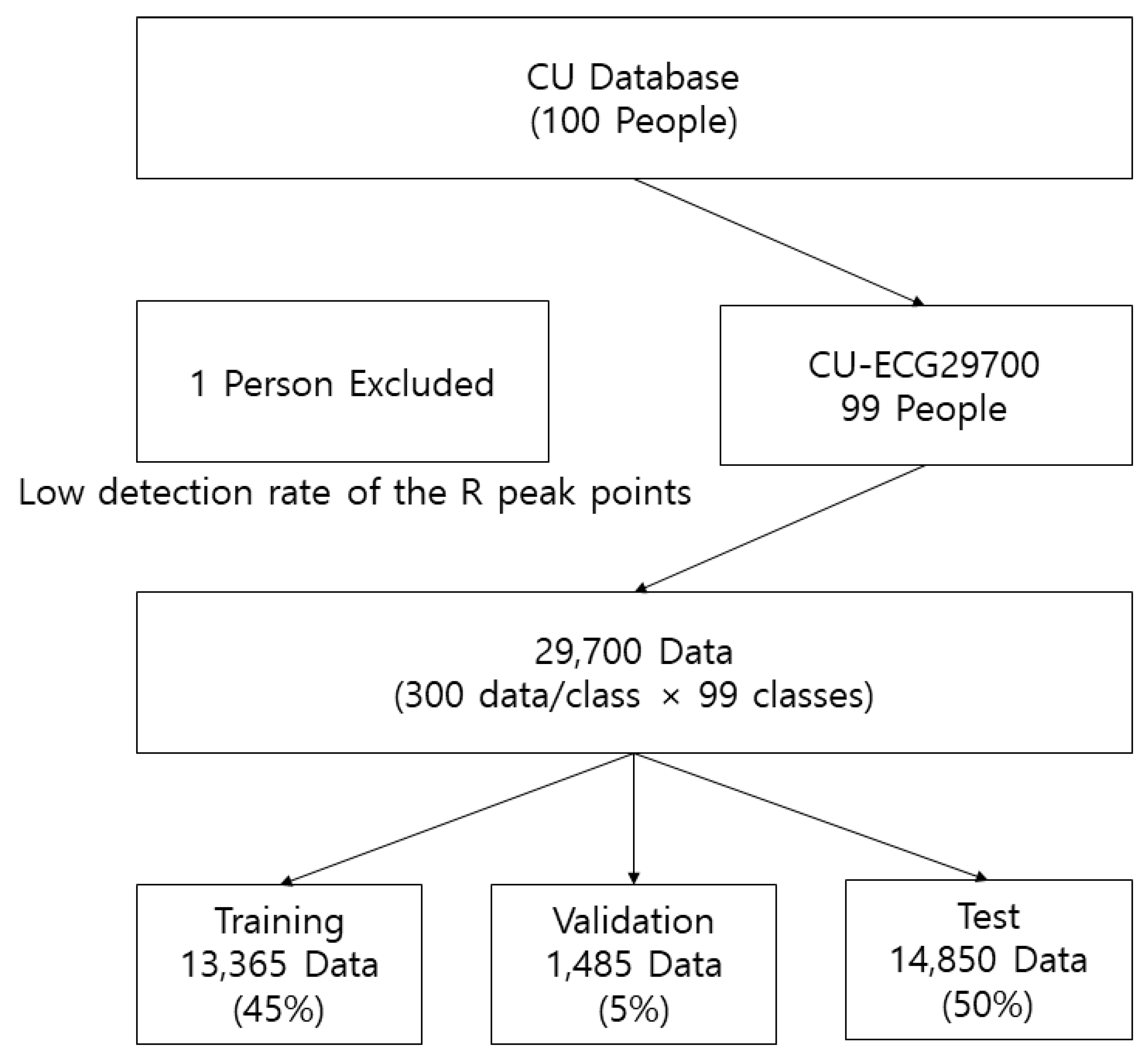
The CU database directly constructed for this study was acquired at 500 kHz. The dataset was resampled to 1 kHz because it was too large for data processing. The CU database included ECG data from 100 people. However, the data from 99 people were used to construct CU-ECG as the ECG data from one subject had very few R peak points. The R peak points from 99 people were compared and adjusted to ensure that the maximum number of data per class was 300. The size of the CU-ECG database was 784 × 29,700 (300 data/class × 99 classes). The row represents the dimension of the data, while the column indicates the number of data. To train the CNNs, CU-ECG was divided into the training, validation, and test datasets with ratios of 0.45, 0.05, and 0.5, respectively. The sizes of the training, validation, and test datasets were 784 × 13,365, 784 × 1485, and 784 × 14,850, respectively.
Figure 13 shows the structure of CU-ECG. The CNNs were trained using a mini-batch size of 30, an initial learning rate of 0.0001, the Adam training optimizer, and an epoch varied according to the model. PCA-L2 was executed by entering vectors reshaped from the diminished input image and measuring Euclidean distance. The PCA dimension was 20.
Table 4 shows the accuracies achieved by PCA-L2 using time-frequency representations on CU-ECG. PCANet was executed with 4 as the patch size, 4 filters, 4 as the block size, and 0.5 as the block overlap ratio.
Table 5 shows the accuracies achieved by PCANet using time-frequency representations on CU-ECG.
Table 6 presents the accuracies achieved by the different models based on the CNN using time-frequency representations on CU-ECG. The best accuracies achieved by the time-frequency representations on CU-ECG were 93.82% for DenseNet-201 in MFCC, 94.03% for Xception in spectrogram, 90.65% for Xception in log spectrogram, 88.84% for Xception in mel spectrogram, and 93.49% for Xception in scalogram. The spectrogram input to the Xception model achieved the best accuracy for the CU-ECG dataset.
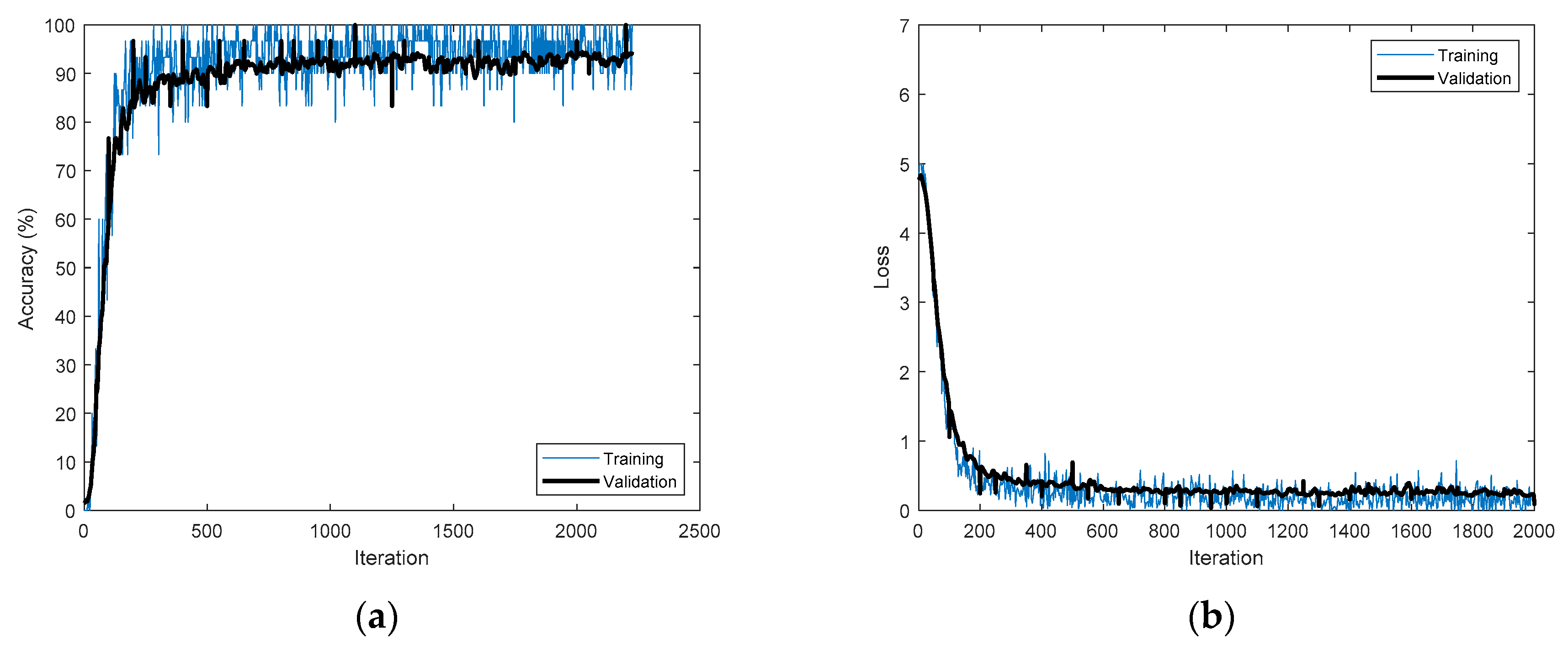
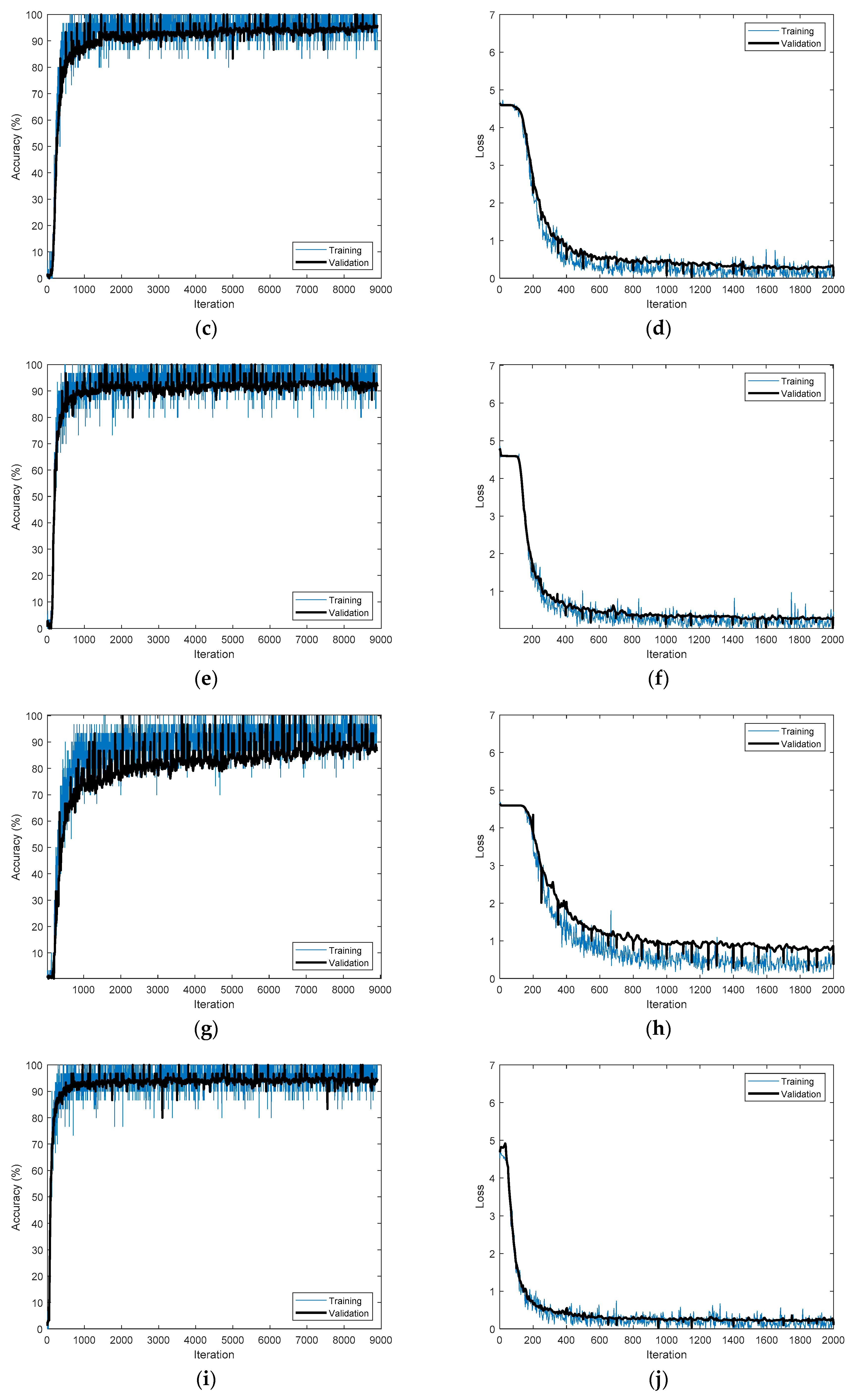
Figure 14 shows the training processes that achieved the best accuracies for the different time-frequency representations applied to CU-ECG.
The MIT(Massachusetts Institute of Technology)-BIH(Beth Israel Hospital) arrhythmia database consists of ECG signals from 48 subjects. The data had a sample rate of 360 Hz and a 10 s length. Most data had the MLII (Modified Limb lead II) signal except two subjects. Therefore, MLII data from 46 subjects were used for biometrics. Noise was eliminated from the signal, and the R peaks were detected by the Pan–Tompkins algorithm. The number of detected R peaks was small because the length of MIT-BIH arrhythmia had a short length signal. The R peak points from 46 people were compared and adjusted to ensure that the maximum number of data per class was five. Data were captured with 289 samples putting the R peak as the center. The size of the MIT-BIH-arrhythmia-ECG database was 289 × 230 (5 data/class × 46 classes). The row represents the dimension of the data, while the column indicates the number of data. To train the CNNs, MIT-BIH-arrhythmia-ECG was divided into the training and test datasets with ratios of 0.6 and 0.4, respectively. The sizes of the training and test datasets were 289 × 138 and 289 × 92, respectively. The CNNs were trained using a mini-batch size of 10, an initial learning rate of 0.0001, a max epoch of 5, and the Adam training optimizer. PCA-L2 was executed by entering vectors reshaped from the diminished input image and measuring Euclidean distance. The PCA dimension was 20.
Table 7 shows the accuracies achieved by PCA-L2 using time-frequency representations on MIT-BIH-arrhythmia-ECG. PCANet was executed with 4 as the patch size, 4 filters, 4 as the block size, and 0.5 as the block overlap ratio.
Table 8 shows the accuracies achieved by PCANet using time-frequency representations on MIT-BIH-arrhythmia-ECG.
Table 9 presents the accuracies achieved by the different models based on CNN using time-frequency representations on MIT-BIH-arrhythmia-ECG. The best accuracies achieved by the time-frequency representations on MIT-BIH-arrhythmia-ECG were 48.91% for DenseNet-201 in MFCC, 63.04% for DenseNet-201 in spectrogram, 84.78% for DenseNet-201 in log spectrogram, 38.04% for DenseNet-201 in mel spectrogram, and 75.00% for DenseNet-201 in scalogram. The log spectrogram input to the DenseNet-201 model achieved the best accuracy for the MIT-BIH-arrhythmia-ECG dataset. There were some cases of learning failure because the size of the dataset was too small.
The mean accuracies of the different time-frequency representations applied to PTB-ECG were 98.56% for MFCC, 97.84% for spectrogram, 95.83% for log spectrogram, 94.72% for mel spectrogram, and 97.34% for scalogram. The mean accuracies of the different time-frequency representations applied to the CU-ECG were 91.53% for MFCC, 91.34% for spectrogram, 89.06% for log spectrogram, 87.58% for mel spectrogram, and 92.25% for scalogram. The mean accuracies of PTB-ECG and CU-ECG corresponding to the time-frequency representations were 95.04% for MFCC, 94.59% for spectrogram, 92.45% for log spectrogram, 91.15% for mel spectrogram, and 94.79% for scalogram. The MFCC accuracies were 0.45%, 2.60%, 3.90%, and 0.25% higher than the spectrogram, log spectrogram, mel spectrogram, and scalogram accuracies, respectively.
Figure 15 shows a comparison between the accuracies of the different time-frequency representations.
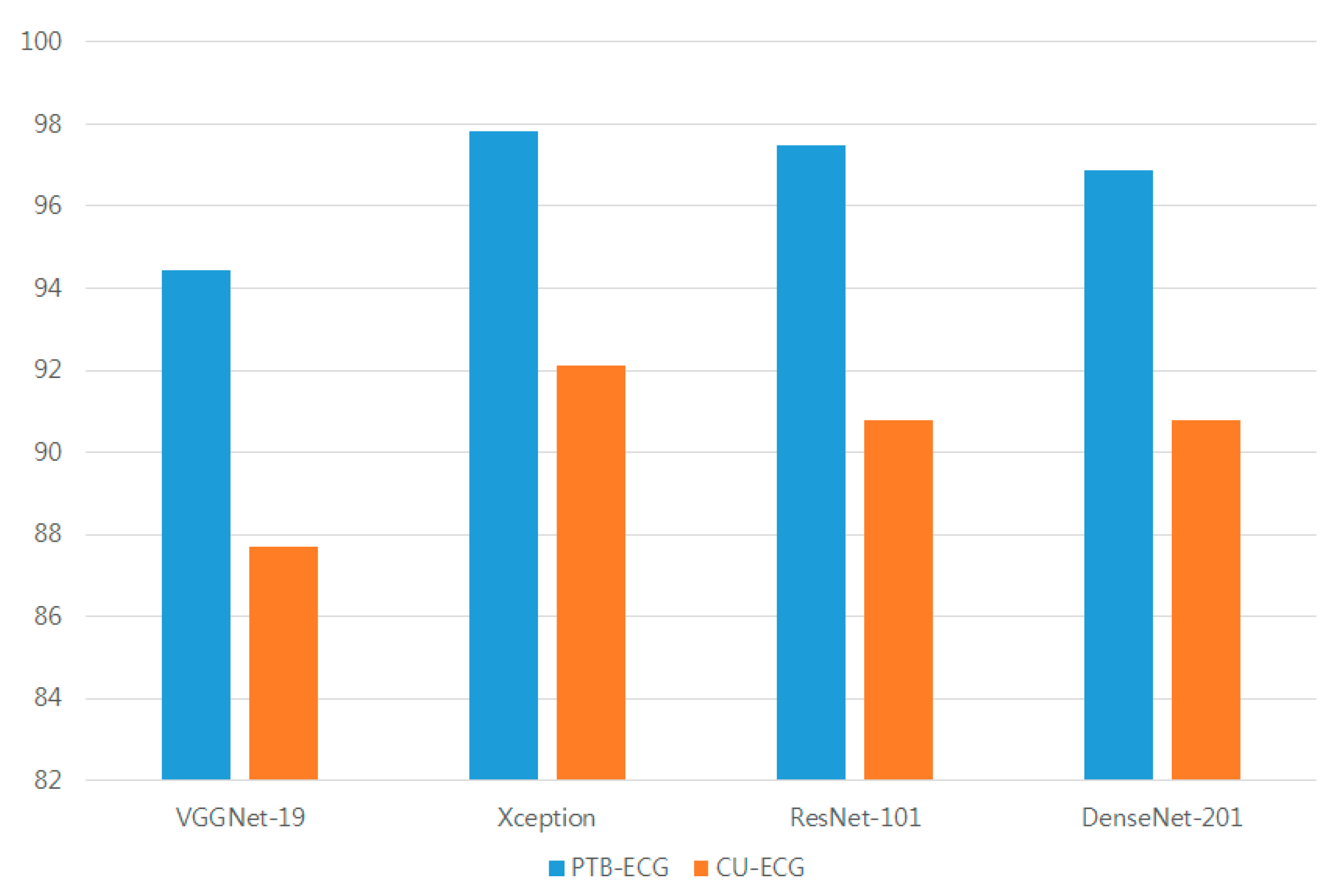
The mean accuracies of the different models in PTB-ECG were 94.43% for VGGNet-19, 97.82% for Xception, 97.48% for ResNet-101, and 96.87% for DenseNet-201. The mean accuracies of the different models in CU-ECG were 87.70% for VGGNet-19, 92.13% for Xception, 90.78% for ResNet-101, and 90.80% for DenseNet-201. The mean accuracies of PTB-ECG and CU-ECG for the different models were 91.06% for VGGNet-19, 94.97% for Xception, 94.13% for ResNet-201, and 93.84% for DenseNet-201. The Xception accuracies were 3.91%, 0.84%, and 1.14% higher than the VGGNet-19, ResNet-101, and DenseNet-201 accuracies, respectively.
Figure 16 shows a comparison between the accuracies of various models based on the CNN. The mean accuracies of PCA-L2 and PCANet were 97.81 and 98.54 in PTB-ECG, 88.75 and 90.42 in CU-ECG, and 2.4 and 84.78 in MIT-BIH-arrhythmia-ECG, respectively. The Xception in PTB-ECG was 0.1% higher than PCA-L2 and 0.72% lower than PCANet. The Xception in CU-ECG was 3.38% and 1.71% higher than PCA-L2 and PCANet, respectively.