1. Introduction
Studies on Electronic Health Records (EHRs) [
1] play an important role in modern society [
2]. According to the International Organization for Standardization (ISO) definition [
3], EHRs mean a repository of patient data in digital form, which includes diagnostic records, electronic medical images, patient history, allergies and laboratory test results, etc. Healthcare workers rely on EHRs to bill patients and evaluate the physical conditions of patients [
4]. In addition, more and more studies focus on using machine learning to deal with EHRs for the large amount of data. In this paper, we study the clinical time series, which originates from sensors in intensive care units (ICUs) or is recorded manually. Under most circumstances, these records are multivariate, including heart rate, blood pressure, and weight, etc. Generally, researchers analyze the multivariate time series of EHRs to accomplish the following tasks: in-hospital morality classification [
5], diagnosis classification [
6] and length of stay classification problems [
7]. Because it is unnecessary to record all variables at all times, resulting in massive missing values, one of the major challenges of clinical time series classification tasks is to deal with the massive missing data.
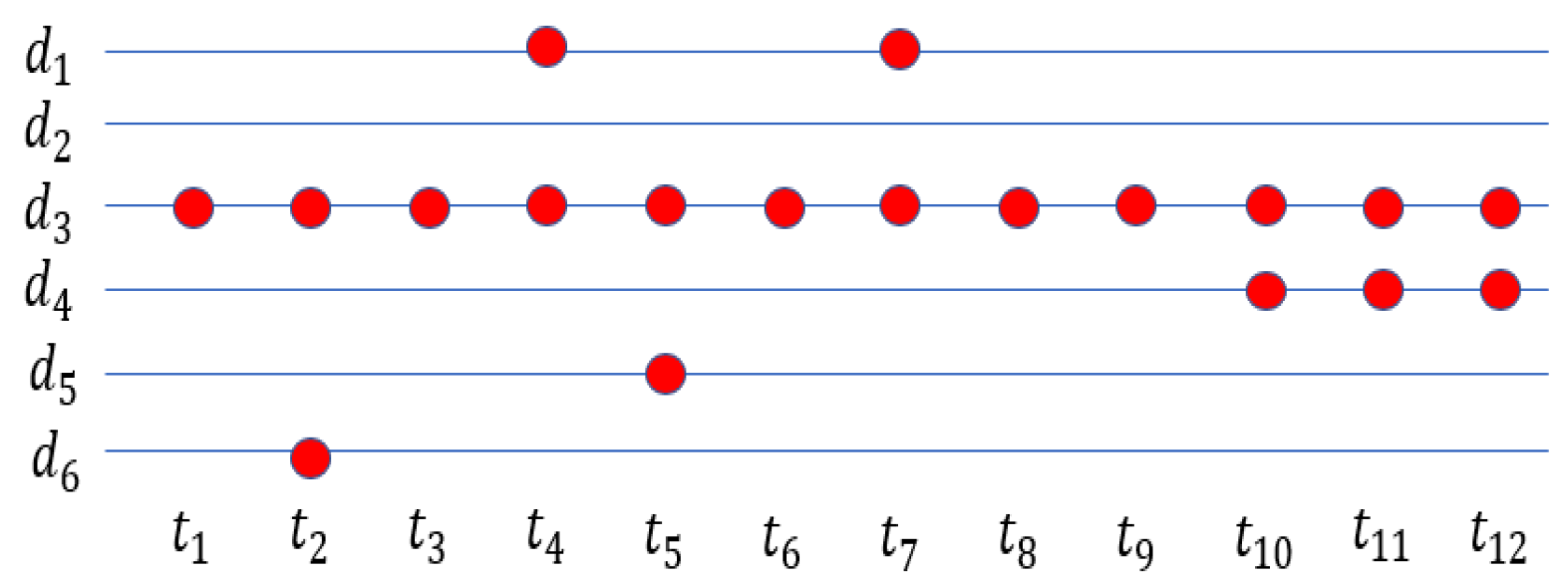
Figure 1 shows an example of a clinical time series. It should be noted that the observation frequency is usually quite different between different variables.
Traditionally, in order to do the classification task on clinical time series, we will impute the missing values in the time series first. After imputing the missing data or simply omitting it [
8], we feed the processed data to the auto-regressive integrated moving average (ARIMA) model [
9] and the Kalman filter [
10], treating it as a normal time series classification problem [
11,
12,
13]. Most people impute missing values with the mean value in the training set (mean imputation) or the last observation (forward imputation) for effectiveness and efficiency [
14]. We can apply not only the simple methods mentioned above but also various advanced methods, such as matrix factorization [
15], kernel methods [
16], and the EM algorithm [
17], to perform the imputation. However, missing data imputation only serves as an auxiliary function to improve classification accuracy, and some advanced methods may cause time-consuming and expensive computational problems without classification performance improvement. Moreover, many imputation methods may fail in dealing with such massive missing values [
18]. Thus, the solutions of time series with no or few missing values and time series with massive missing values in classification tasks should be different.
When most of the multivariate time series is missing, we need to find more features besides the time series itself. The missing values in clinical time series are caused by many reasons. The most important one is that it costs much money and time to ask healthcare workers to record every possible variable of the patients [
19]. Usually, they only choose to record the variables related to the medical condition of the patients. For example, heart rate may be recorded more frequently when the patients have heart disease. In addition, when the condition of the patients get worse, the recording frequency becomes higher. Finally, automated monitoring devices might fail to record the variables sometimes, but it is rarer than the previous cases we discuss above. We define the information contained in whether to record the variable (missing mark) and how often to record it (missing rate) as a missing pattern of a variable in the multivariate clinical time series. Take the patients with heart disease as examples: we believe the missing pattern of variables related to heart conditions behaves differently from others. Moreover, we should process the missing patterns of different variables separately to fully exploit the information from the multivariate time series.
With the development of deep learning, recurrent neural networks (RNN) have become one of the most widely used models to solve time series problems for the reason that they can directly handle time series of varying length [
20]. Long short-term memory (LSTM) and gated recurrent units (GRU) are the most popular RNN models for capturing long-term and short-term impacts in time series [
21,
22]. Even though few studies focus on handling multivariate time series with massive missing data, there are still some outstanding works that have been proposed recently. GRU-D proposed by Che et al. [
23] focuses on the missing pattern of multivariate time series. However, GRU-D treats multivariate time series as a whole and it does not consider the missing rate differences. Harutyunyan et al. separate the multivariate time series into univariate time series and use an independent LSTM network for every variable to fully extract the single variable feature and its missing pattern at the cost of time consumption [
24].
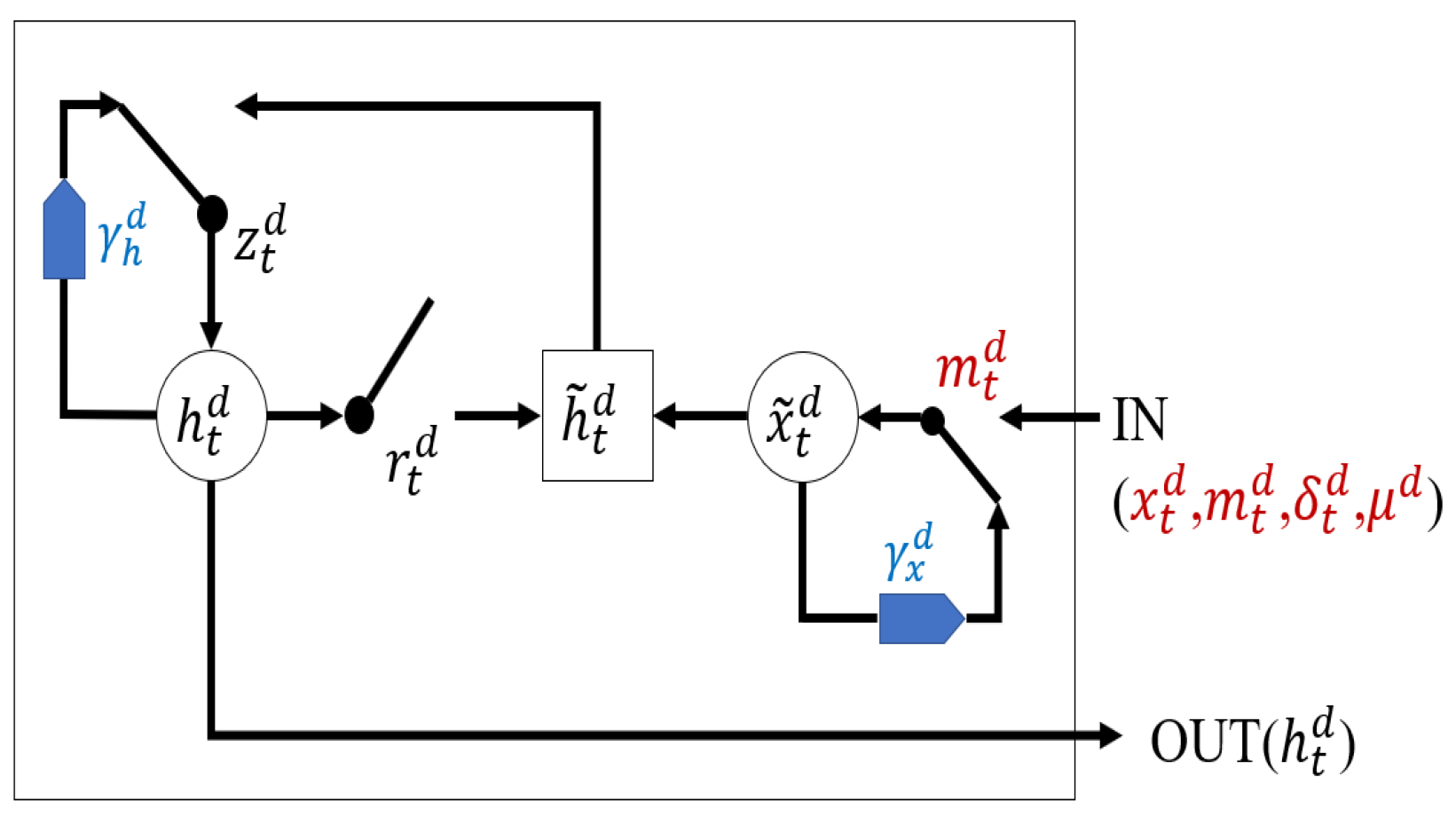
In this paper, we propose our method called variable sensitive GRU (VS-GRU), which has the following contributions:
In clinical time series, we believe the missing rate of variable is related to its characteristic. In addition to the missing mark, VS-GRU considers the missing rate of different variables, seeing it as another input of GRU.
VS-GRU processes variables separately at the same time in a simple structure. Variables can maintain its characteristics before being mixed with others, which increases robustness when dealing with time series with some variables that are almost completely missing.
VS-GRU considers the classification result at every time step, which decreases the probability of learning error from the whole time series.
In
Section 2, we present the related recent research in time series with missing values’ classification problems and clinical time series analyses using deep learning. In
Section 3, VS-GRU is presented in detail. We evaluate our method on two public clinical datasets and compare it with the state-of-the-art in
Section 4. Finally, we make our conclusions in
Section 5.
4. Experiment
4.1. Dataset
We use two clinical public datasets from the real world, PhysioNet Challenge 2012 (PhysioNet) [
42] and MIMIC-III dataset (MIMIC-III) [
43] to perform the multivariate time series classification experiments.The detail to process these two datasets can be found in [
23].
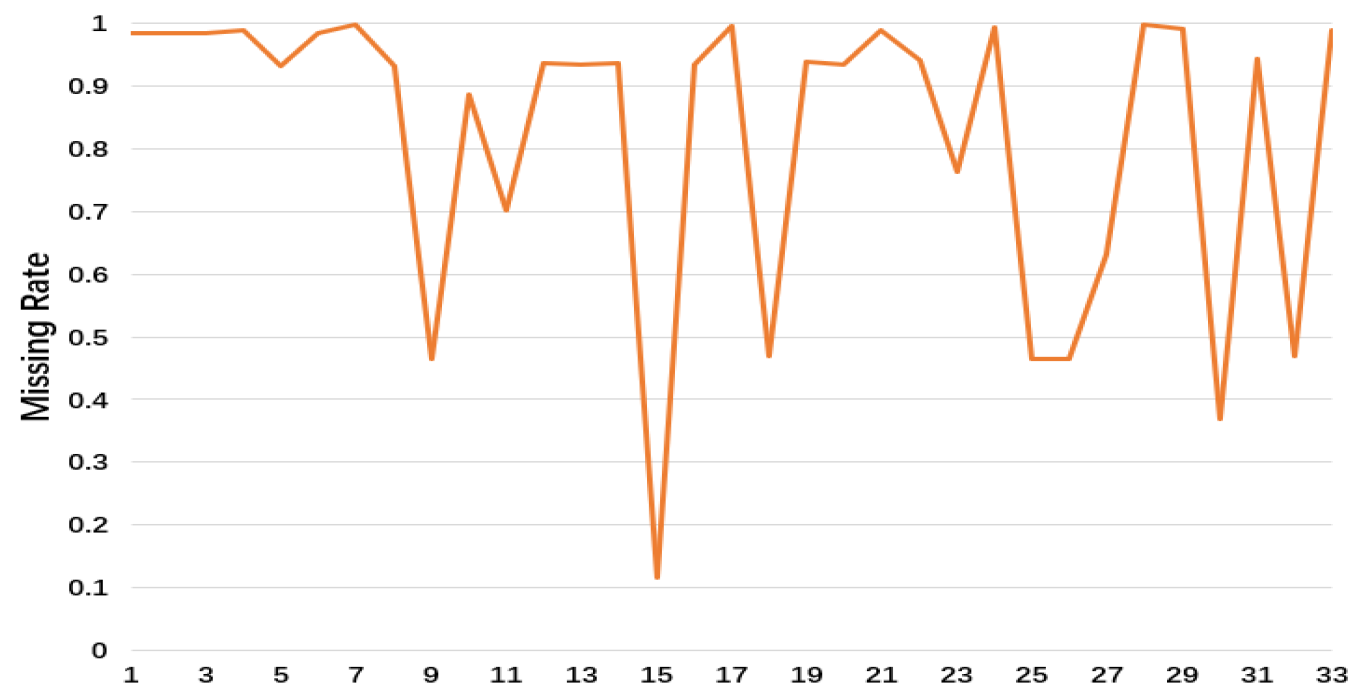
The PhysioNet dataset consists of three parts, each with 4000 patient records in the ICU, including heart rate, body temperature, and the number of red blood cells, etc. Each patient’s records are at least 48 h, with a total of 12,000 records. Here, we take the first part of the dataset, and 33 variables are extracted. We only consider the first 48 h after admission. In order to maintain the original observation, we choose the first and the last time step and time steps with more real observation than others, which is 49 time steps in total.
Figure 5 shows the missing rate of 33 variables in PhysioNet. This dataset has the following two classification tasks:
The MIMIC-III dataset includes health care records for more than 40,000 patients in the ICU of the Beth Israel Deaconess Medical Center between 2001 and 2012, with a total of 58,976 admission records. We extracted 19,671 admission records, each with 99 variables (e.g., the number of white blood cells, the number of red blood cells, and the pH of the blood). Similarly, each time series is taken only within the first 48 h after admission, and a maximum 49 time steps are chosen, the same as PhysioNet.
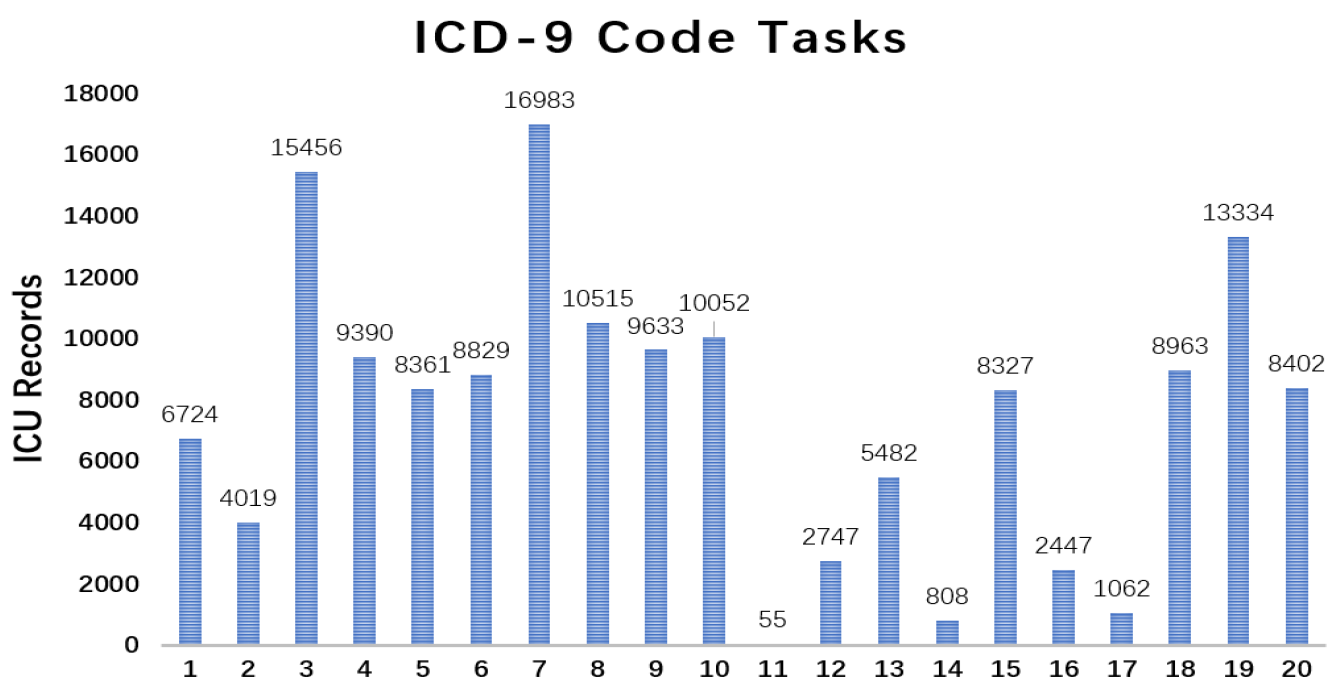
Figure 7 shows the missing rate of 99 variables in MIMIC-III. This dataset has the following two classification tasks:
4.2. Baseline
In nonRNN algorithms, we choose logistic regression and random forests. Because they cannot handle time series with different lengths, we will impute the time series less than the maximum time step with zero. In the deep learning algorithms, we choose LSTM and GRU.
Three imputation methods of missing values will be used in the four algorithms mentioned above, which are mean imputation (Equation (
21)), forward imputation (Equation (
22)) and zero imputation (Equation (
23)). We refer these approaches as RF-forward, RF-mean, RF-zero, LR-forward, LR-mean, LR-zero, LSTM-forward, LSTM-mean, LSTM-zero, GRU-forward, GRU-mean and GRU-zero. In addition to the imputed time series, we input the binary mask indicator into the four models we choose by concatenating them together. The purpose is to know whether the models are able to capture missing pattern information in the mask indicator and improve classification performances. We refer these approaches as RF-mask-forward, RF-mask-mean, LR-mask-forward, LR-mask-mean, LSTM-mask-forward, LSTM-mask-mean, GRU-mask-forward and GRU-mask-mean. Moreover, in such sparse time series (missing rate more than 80%), feature engineering is one of the effective ways to ease from the lack of data. We use logistic regression and random forests to perform the experiments using feature engineering. Besides the mask indicator, the maximum, minimum, mean values and missing rate of every variables are input into the models along with time series. We refer these approaches as RF-mask-forward-m and LR-mask-forward-m. In the interest of testing if the missing rate improves the classification performances, we do contrast experiments by simply removing it from the baselines. We refer these approaches as RF-mask-forward-m w/o
and LR-mask-forward-m w/o
. We do the feature extraction and imputation before time step sampling, so that only when the variables are completely missing can we not find its last observation or the maximum, minimum, mean values:
indicates the mean value of variable d in the training set. indicates the last observation of variable d in time series x if there is an observation before time step t; otherwise, it is zero.
We compare our method with the state-of-the-art GRU-D. GRU-D does not need to choose the imputation method because of the dynamic imputation mechanism.
Because the filling of missing data will directly affect the experimental results and we are not sure whether the dynamic imputation mechanism is effective, we separate it from VS-GRU and VS-GRU-i, which are VS-GRU w/o di and VS-GRU-i w/o di, using forward imputation instead.
4.3. Setting
All deep learning models are implemented with PyTorch 1.0. Logistic regression and random forests are implemented with Scikit-learn in Python. The learning rate of all RNN models is 0.005. We use the Adam optimization method to train all RNN models. The hyper-parameter in deep supervision is set to 0.5.
All experimental data are normalized to have 0 mean and 1 standard deviation. All experiments were performed using a 5-fold cross-validation method. We calculated the area under the ROC curve (AUC) of each method to evaluate the classification performance.
4.4. Result
Table 1 and
Table 2 present the experimental results of 25 baselines and four versions of our models in the four tasks on PhysioNet and MIMIC-III. The model we propose and the best results we obtained are highlighted. Moreover, we mark the top five results in the four tasks.
4.4.1. Compare VS-GRU with Baselines
It is apparent that forward imputation outperforms mean imputation and zero imputation in all models, which suggests that maintaining the time series feature in itself is important. Because zero imputation can carry information from missing patterns like mask indicators, zero imputation beats mean imputation in many cases.
With the mask indicator, RNN models can achieve better performances except that LSTM shows almost the same performances in the ICD-9 Code tasks on MIMIC-III. On the other hand, sometimes nonRNN models fail with the mask indicator. We can see that the missing pattern is beneficial for helping the RNN model to get better results. On the other hand, results on the two datasets show that applying simple feature engineering on the time series can lead to better performances. If we remove the missing rate from the input, all experimental results decrease except logistic regression in dealing with the multi-task problem on PhysioNet, which is almost the same. It is safe to say that the missing rate plays an effective role in the problem of multivariate time series with massive missing values.
Next, we will discuss the experimental results of four versions of our model. Except for all four tasks problems in PhysioNet, all four versions of our model rank within the top five. In the single-label classification problem of the two datasets, there is a significant improvement between VS-GRU and the other baselines. Interestingly, with or without a dynamic imputation mechanism, VS-GRU-i does not improve the performance of the model. Because VS-GRU-i processes multivariate time series separately at first and then applies a second GRU layer to integrate all variables, it should strike a balance between these two procedures. However, it fails when dealing with such a simple problem. We indicate that a simple framework such as VS-GRU is effective enough to solve the single-label classification problem. A fully connected layer can integrate the variable well. Comparing the results between VS-GRU and VS-GRU w/o di in two datasets, the dynamic imputation mechanism is effective in PhysioNet but fails in MIMIC-III. Because the average missing rate is 0.9559 in MIMIC-III, it is very common that time series in MIMIC-III has completely missing variables. When dealing with completely missing variables, a dynamic imputation method is equal to mean imputation. When we replace dynamic imputation method with forward imputation, it is equal to zero imputation when dealing with completely missing variables. As the experimental results shown from
Table 1 and
Table 2, zero imputation outperforms mean imputation. Thus, we should notice the limit of dynamic imputation mechanism and try to use it carefully.
On the other hand, situations are quite the opposite in multi-label classification problems. VS-GRU-i performs the best in both datasets. The dynamic imputation mechanism fails in the multi-task classification problems. We suggest that using only a few parameters of each variable to impute the missing values cannot correctly map the multi-label into feature space. It may be effective when dealing with a single label, but it cannot capture all the information from multi-labels at the same time. We indicate that it is also the same reason why VS-GRU fails in multi-task classification problems due to its lack of parameters.
To verify our suggestion, we solve the multi-label classification problem as single-label classification, which means we run every binary label independently and average the results. Here, we compare the best model VS-GRU in the binary classification problem with the best model VS-GRU-i w/o di in the multi-label classification problem.
The results in
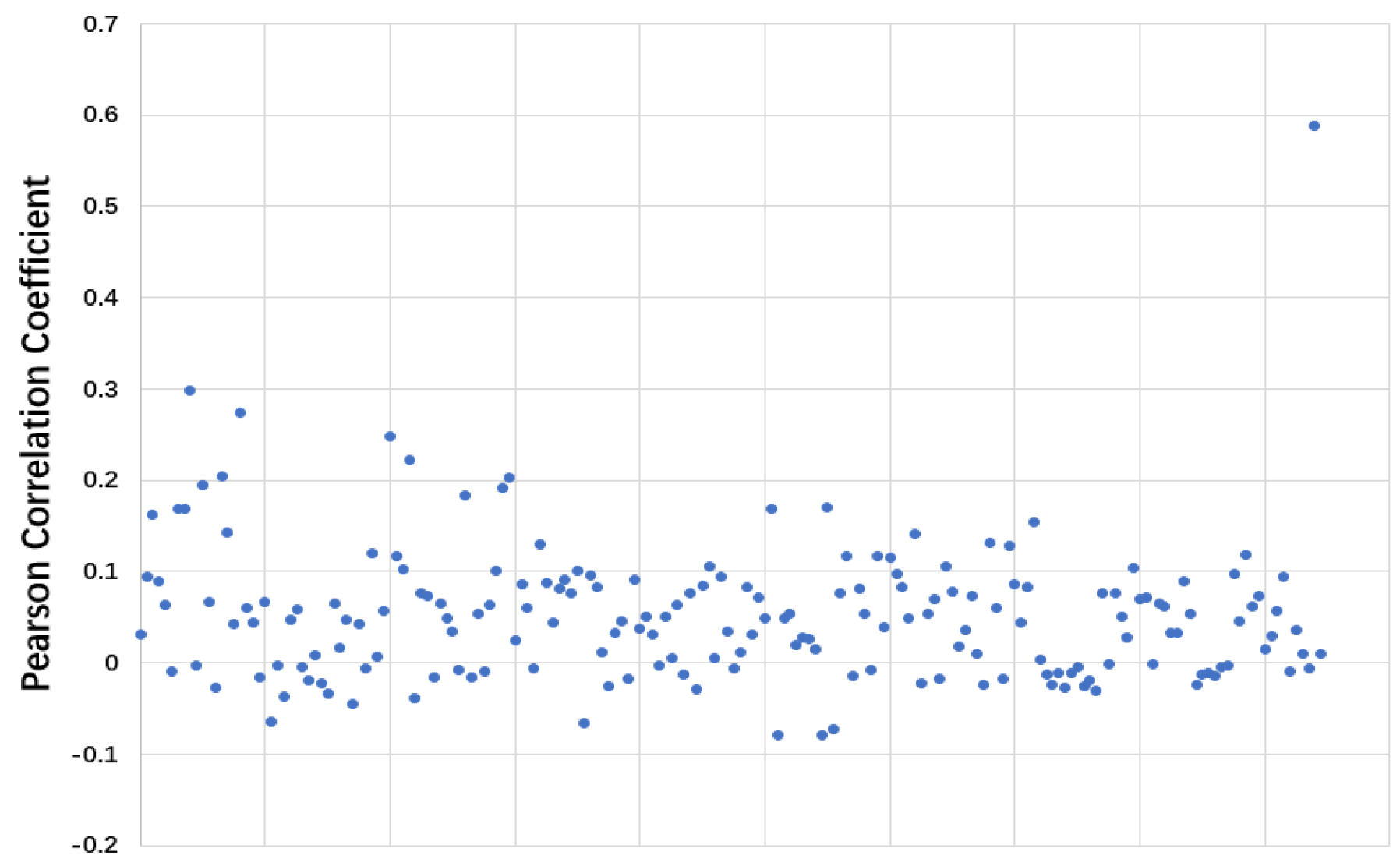
Table 3 confirm our suggestion. The dynamic imputation mechanism also fails on MIMIC-III tasks. We can make the conclusion that the weak performance of VS-GRU in solving the multi-task classification problem is because of its lack of parameters to deal with multi-label simultaneously, which is approached in VS-GRU-i. However, if we solve the multi-label classification problem separately, VS-GRU even outperforms VS-GRU-i at the price of time consumption. These results may be against the common sense that people use multi-tasks to solve multi-label problems to improve the performances because relevant labels may help each other to be learned better. We take ICD-9 Code tasks on MIMIC-III as an example and calculate the Pearson correlation coefficient between labels. As shown in
Figure 9, among 190 label pairs in 20 labels, there are 158 label pairs under 0.1, which means the correlation is weak between those labels. Thus, in such a sparse situation, exploiting the feature within a single variable rather than exploiting the relation between labels is more important. VS-GRU can do a better job in keeping the characteristic from a single variable.
4.4.2. Missing Factor in VS-GRU
In order to further discuss the missing factor impact on classification problems, we implement two experiments: replacing the missing factor with missing rate (VS-GRU-
and VS-GRU-i-
) and removing the missing factor from the update functions (VS-GRU w/o
and VS-GRU-i w/o
). To be specific, we replace the missing factor
with missing rate
in the first layer of VS-GRU-i-
and replace the missing factor
with
in penalized mechanism in the second layer. Thus, the variables with a higher missing rate get more punishment. The experimental results are shown in
Table 4 and
Table 5. We can see from the results that missing factor is beneficial to model performances. In single-task problems, VS-GRU-
outperforms VS-GRU w/o
. However, the situation is quite the opposite in multi-task problems. We suggest that the reason is that the missing factor is not only used in the update functions in the first layer but also used as penalized mechanism in the second layer of VS-GRU-i. Given the massive missing values in the datasets, the missing rate of many variables is close to 1. The missing factor can learn the slight differences between missing rates and provide more learnable features to the models. In addition, instead of calculating the missing rate using all the records, we calculate the missing rate within the first and last 24 h, which are called VS-GRU-update and VS-GRU-i-update. Then we test the models if we can update the missing rate every 24 h without significant changes in model performance. We execute the two tasks experiments on MIMIC-III using the update models and original models we propose without the dynamic imputation mechanism. We can see from
Table 6 that the performances of VS-GRU and VS-GRU-i stay stable in the case of updating the missing rate.
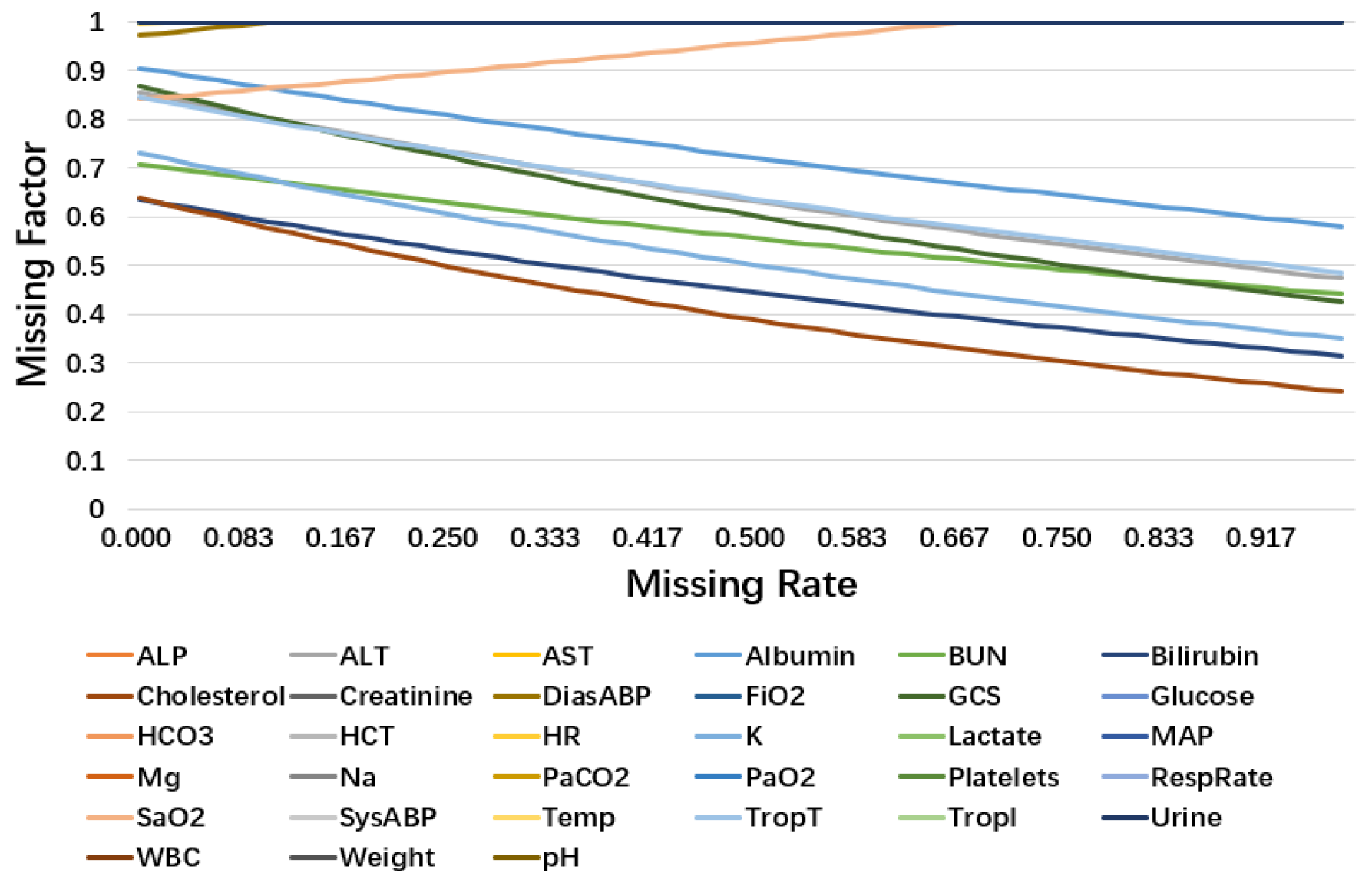
Figure 10 plots the learned missing factor of 33 variables in PhysioNet. More than half of the variables have an unchanged missing factor during the changing missing rate, which means that the missing rate of these variables may have little impact on this classification task. In other situations, the missing factor decreases as the missing rate increases, which fit the common sense that we should pay more attention to the variables with a low missing rate. This indicates that the characteristics of these variables are related to their observation frequency. However, two variables behave in the opposite way: TropT (missing rate 0.9983) and Urine (missing rate 0.9917). We suggest that variables with relatively high missing rates lack training examples for missing factors to learn the pattern at a low missing rate, resulting in different trends from other variables. In addition, in the test set, the examples of these variables in the low missing rate are rare. It is safe to say that it causes little impact on classification performance.
4.4.3. Deep Supervision in VS-GRU
Deep supervision is designed to improve the training procedure of the time series classification problem, not aiming to address its missing values. To further discuss the modeling ability of multivariate time series with a high missing rate, we separate deep supervision from the model and only use the last time step as supervision. We evaluate it on the four tasks again, comparing it with the strong baseline GRU-D. The best results are highlighted in the following tables.
As
Table 7 and
Table 8 show, even without deep supervision, our models still achieve the best results in all four tasks.