Towards a Physiological Scale of Vocal Fold Agent-Based Models of Surgical Injury and Repair: Sensitivity Analysis, Calibration and Verification
Abstract
1. Introduction
2. Materials and Methods
2.1. Vocal Fold Cell Phenotyping
2.1.1. Vocal Fold Surgical Model
2.1.2. Vocal Fold Cell Isolation
2.1.3. Sample Preparation for Flow Cytometry
Experimental Samples
Control Samples
2.1.4. Flow Cytometry Data Processing and Analysis
2.1.5. Flow Cytometry Statistical Analysis
2.2. Vocal Fold Agent-based Models
2.2.1. VF-ABM Implementation
| Algorithm 1: Overview of 3D rat vocal fold agent-based models (VF-ABM) |
| Procedure VFABM Initialization of patches Initialization of chemicals Initialization of cells Initialization of ECM Initialization of damage For each tick /* Model Computation */ For each Patch Seed Cell Function ECM Function ECM Fragmentation For each Cell Cell Function For each Chemical Type Diffuse Chemical /* Model Update */ For each Patch Update ECM Update Patch Update Chemicals For each Cell Update Cell |
2.2.2. Sensitivity Analysis
| Algorithm 2: Random Forests in R |
| Library – clusterGeneration Library – mnormt Require – randomForest Library – caret Number of Trees = 600 X=Samples Y=Model Output for a time point T and cell type C Df = data.frame (Y,X) allX = paste (“X”, 1:ncol(X),sep=““) names(df) = c(“Y”, allX) fit= randomForest(factor(Y)~., data=df) VI_F = importance(fit) varImp(fit) varImpPlot(fit, type=2) |
2.2.3. Model Calibration
2.2.4. Model Verification
3. Results
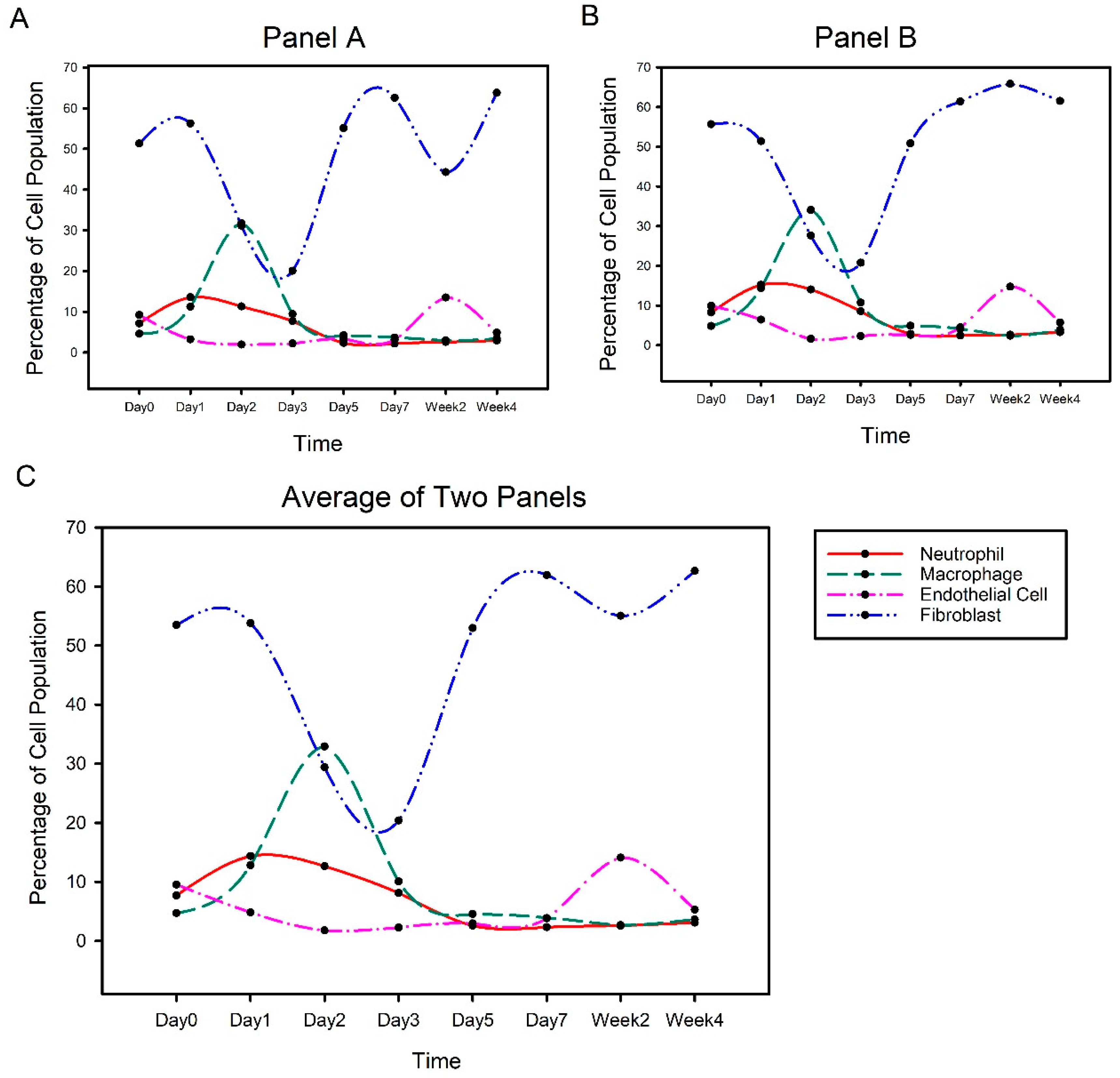
3.1. Cell Compositions in Injured Rat Vocal Folds
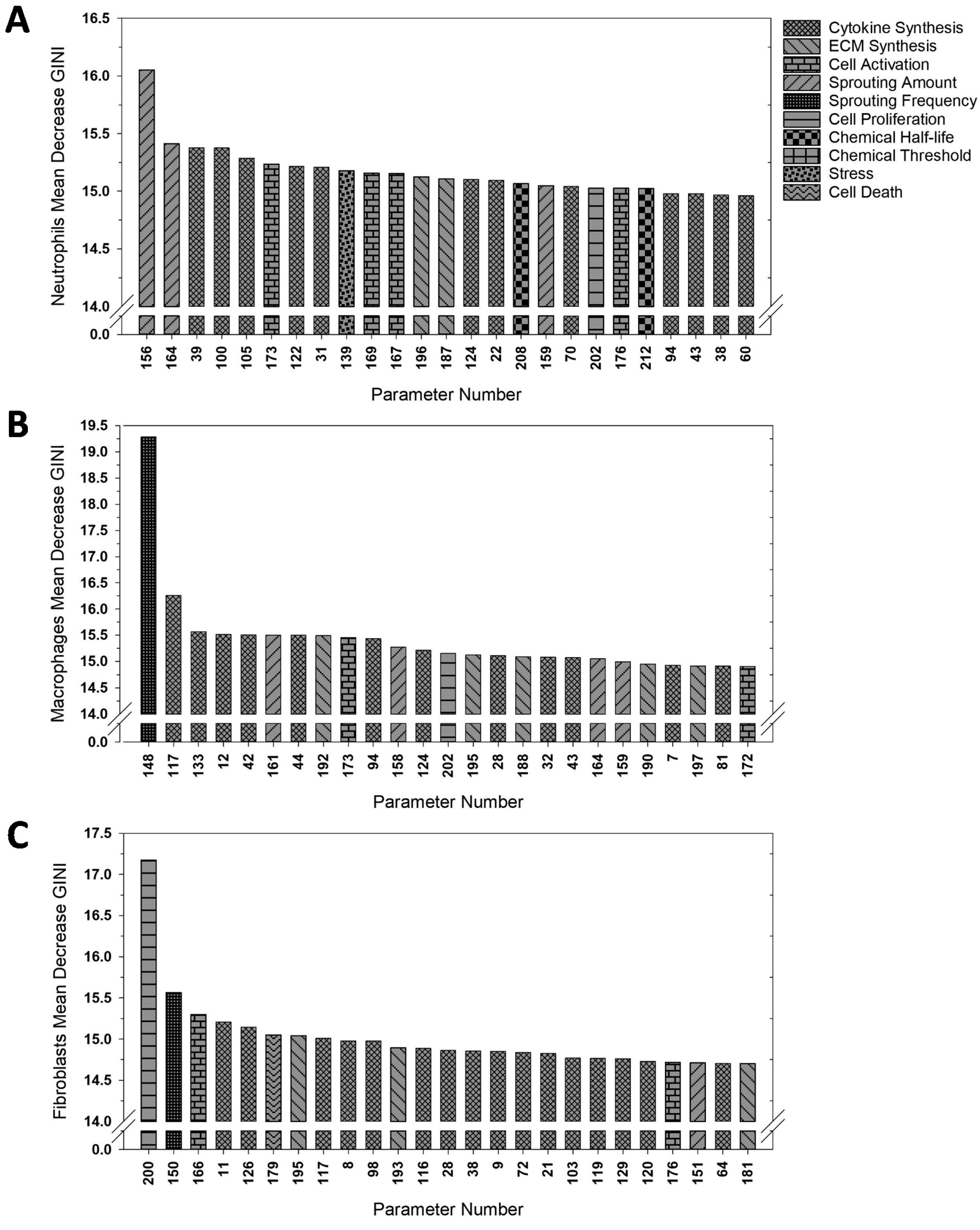
3.2. ABM Parameter Ranking
3.3. VF-ABM Calibration with Most Influential Parameters
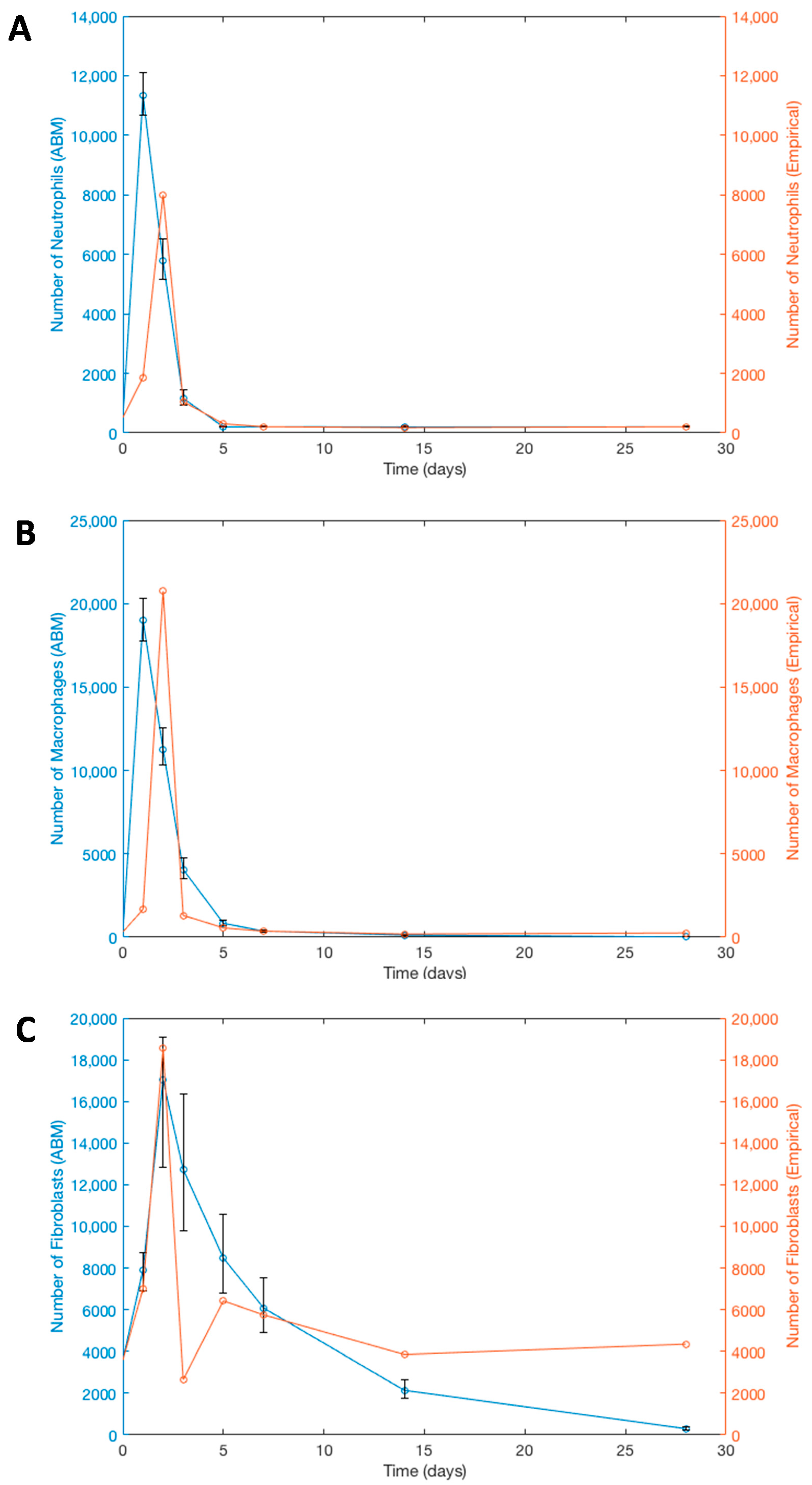
3.4. Verification of ABM-Predicted Cellular Dynamics
4. Discussion
4.1. Time-Evolution of Cell Populations in Surgical Vocal Fold Injury and Repair
4.2. Random Forests and ROPE for VF-ABM Parameter Optimization
4.3. VF-ABM Performance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedrich, G.; Dikkers, F.; Arens, C.; Remacle, M.; Hess, M.; Giovanni, A.; Duflo, S.; Hantzakos, A.; Bachy, V.; Gugatschka, M. Vocal fold scars: Current concepts and future directions. Consensus report of the phonosurgery committee of the European laryngological society. Eur. Arch. Oto Rhino Laryngol. 2013, 270, 2491–2507. [Google Scholar] [CrossRef] [PubMed]
- Hirano, S. Current treatment of vocal fold scarring. Curr. Opin. Otolaryngol. Head Neck Surg. 2005, 13, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Rosen, C.A. Vocal Fold Scar. In The Otolaryngologica Clinics of North America. Voice Disorders and Phonosurgery; Rosen, C.A., Murry, T., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2000. [Google Scholar]
- Benninger, M.S.; Alessi, D.; Archer, S.; Bastian, R.; Ford, C.; Koufman, J.; Sataloff, R.T.; Spiegel, J.R.; Woo, P. Vocal fold scarring: Current concepts and management. Otolaryngol. Neck Surg. 1996, 115, 474–482. [Google Scholar]
- Shin, Y.S.; Chang, J.W.; Yang, S.M.; Wu, H.W.; Cho, M.H.; Kim, C.H. Persistent dysphonia after laryngomicrosurgery for benign vocal fold disease. Clin. Exp. Otorhinolaryngol. 2013, 6, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.; Casper, J.; Colton, R.; Brewer, D. Diagnosis and treatment of persistent dysphonia after laryngeal surgery: A retrospective analysis of 62 patients. Laryngoscope 1994, 104, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Perouse, A.R.; Coulombeau, B. Iatrogenic scarring of the vocal folds after phonosurgery for benign lesions. A descriptive study of 108 patients. Rev. Laryngol. Otol. Rhinol. (Bord) 2014, 135, 57–61. [Google Scholar]
- Hansen, J.K.; Thibeault, S.L. Current understanding and review of the literature: Vocal fold scarring. J. Voice 2006, 20, 110–120. [Google Scholar] [CrossRef]
- An, G.; Mi, Q.; Dutta-Moscato, J.; Vodovotz, Y. Agent-based models in translational systems biology. Wiley Interdiscip. Rev. Syst. Boil. Med. 2009, 1, 159–171. [Google Scholar] [CrossRef]
- Macal, C.M.; North, M.J. Agent-based modeling and simulation: ABMS examples. In Proceedings of the 2008 Winter Simulation Conference, Miami, FL, USA, 7–10 December 2008; pp. 101–112. [Google Scholar]
- Macal, C.M.; North, M.J. Tutorial on agent-based modeling and simulation. In Proceedings of the 2018 Winter Simulation Conference (WSC), Gothenburg, Sweden, 9–12 December 2005; p. 14. [Google Scholar]
- Mi, Q.; Li, N.Y.K.; Ziraldo, C.; Ghuma, A.; Mikheev, M.; Squires, R.; Okonkwo, D.; Verdolini Abbott, K.; Constantine, G.; An, G.; et al. Translational systems biology of inflammation: Applications to personalized medicine. Pers. Med. 2010, 7, 549–559. [Google Scholar] [CrossRef]
- Vodovotz, Y.; Constantine, G.; Faeder, J.; Mi, Q.; Rubin, J.; Bartels, J.; Sarkar, J.; Squires, R.; Okonkwo, D.; Gerlach, J.; et al. Translational systems approaches to the biology of inflammation and healing. Immunopharmacol. Immunotoxicol. 2010, 32, 181–195. [Google Scholar] [CrossRef][Green Version]
- Clermont, G.; Bartels, J.; Kumar, R.; Constantine, G.; Vodovotz, Y.; Chow, C. In silico design of clinical trials: A method coming of age. Crit. Care Med. 2004, 32, 2061–2070. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Clermont, G.; Vodovotz, Y.; Chow, C.C. The dynamics of acute inflammation. J. Theor. Biol. 2004, 230, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Vodovotz, Y.; Chow, C.C.; Bartels, J.; Lagoa, C.; Prince, J.M.; Levy, R.M.; Kumar, R.; Day, J.; Rubin, J.; Constantine, G.; et al. In silico models of acute inflammation in animals. Shock 2006, 26, 235–244. [Google Scholar] [CrossRef] [PubMed]
- De Castro, L.N.; Timmis, J. Artificial Immune Systems: A New Computational Intelligence Approach; Springer Science & Business Media: New York, NY, USA, 2002. [Google Scholar]
- Auffray, C.; Chen, Z.; Hood, L. Systems medicine: The future of medical genomics and healthcare. Genome Med. 2009, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Hood, L.; Flores, M. A personal view on systems medicine and the emergence of proactive P4 medicine: Predictive, preventive, personalized and participatory. New Biotechnol. 2012, 29, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Li, N.Y.K. Biosimulation of Vocal Fold Inflammation and Healing. Ph.D. Thesis, University of Pittsburgh, Pittsburgh, PA, USA, 2009. [Google Scholar]
- Li, N.Y.K.; Abbott, K.V.; Rosen, C.; An, G.; Hebda, P.A.; Vodovotz, Y. Translational systems biology and voice pathophysiology. Laryngoscope 2010, 120, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Li, N.Y.K.; Verdolini, K.; Clermont, G.; Mi, Q.; Rubinstein, E.N.; Hebda, P.A.; Vodovotz, Y. A patient-specific in silico model of inflammation and healing tested in acute vocal fold injury. PLoS ONE 2008, 3, e2789. [Google Scholar] [CrossRef]
- Li, N.Y.K.; Vodovotz, Y.; Kim, K.H.; Mi, Q.; Hebda, P.A.; Verdolini-Abbott, K. Biosimulation of acute phonotrauma: An extended model. Laryngoscope 2011, 121, 2418–2428. [Google Scholar] [CrossRef]
- Li, N.Y.K.; Vodovotz, Y.; Hebda, P.A.; Verdolini, K.A. Biosimulation of inflammation and healing in surgically injured vocal folds. Ann. Otol. Rhinol. Laryngol. 2010, 119, 412–423. [Google Scholar] [CrossRef]
- Seekhao, N.; Shung, C.; JaJa, J.; Mongeau, L.; Li-Jessen, N.Y.K. High-Performance Agent-based Modeling Applied to Vocal Fold Inflammation and Repair. Front. Physiol. 2018, 9, 304. [Google Scholar] [CrossRef]
- Seekhao, N.; JaJa, J.; Mongeau, L.; Li-Jessen, N.Y. In situ visualization for 3D agent-based vocal fold inflammation and repair simulation. Supercomput. Front. Innov. 2017, 4, 68. [Google Scholar] [PubMed]
- Seekhao, N.; Shung, C.; JaJa, J.; Mongeau, L.; Li-Jessen, N.Y. Real-time agent-based modeling simulation with in-situ visualization of complex biological systems: A case study on vocal fold inflammation and healing. In Proceedings of the 2016 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), Chicago, IL, USA, 23–27 May 2016; pp. 463–472. [Google Scholar]
- Cortés-Sol, A.; Lara-Garcia, M.; Alvarado, M.; Hudson, R.; Berbel, P.; Pacheco, P. Inner capillary diameter of hypothalamic paraventricular nucleus of female rat increases during lactation. BMC Neurosci. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.R.; Smith, C.W.; Walker, D.C. Unique structural features that influence neutrophil emigration into the lung. Physiol. Rev. 2003, 83, 309–336. [Google Scholar] [CrossRef] [PubMed]
- Hohsfield, L.A.; Ammann, C.G.; Humpel, C. Inflammatory status of transmigrating primary rat monocytes in a novel perfusion model simulating blood flow. J. Neuroimmunol. 2013, 258, 17–26. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ling, C.; Yamashita, M.; Waselchuk, E.A.; Raasch, J.L.; Bless, D.M.; Welham, N.V. Alteration in cellular morphology, density and distribution in rat vocal fold mucosa following injury. Wound Repair Regen. 2010, 18, 89–97. [Google Scholar] [CrossRef]
- Tateya, I.; Tateya, T.; Bless, D.M. Homeostasis of hyaluronic acid in scarred rat vocal folds. In Proceedings of the International Conference on Voice Physiology and Biomechanics, Marseille, France, 18–20 August 2004. [Google Scholar]
- Tateya, I.; Tateya, T.; Sohn, J.-H.; Bless, D.M. Histological effect of basic fibroblast growth factor on chronic vocal fold scarring in a rat model. Clin. Exp. Otorhinolaryngol. 2016, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Tateya, T.; Tateya, I.; Munoz-del-Rio, A.; Bless, D.M. Postnatal development of rat vocal folds. Ann. Otol. Rhinol. Laryngol. 2006, 115, 215–224. [Google Scholar] [CrossRef]
- Welham, N.V.; Montequin, D.W.; Tateya, I.; Tateya, T.; Choi, S.H.; Bless, D.M. A rat excised larynx model of vocal fold scar. J. Speech Lang. Hear. Res. 2009, 52, 1008–1020. [Google Scholar] [CrossRef]
- Tateya, I.; Tateya, T.; Lim, X.; Sohn, J.H.; Bless, D.M. Cell production in injured vocal folds: A rat study. Ann. Otol. Rhinol. Laryngol. 2006, 115, 135–143. [Google Scholar] [CrossRef]
- Tateya, T.; Tateya, I.; Sohn, J.H.; Bless, D.M. Histological study of acute vocal fold injury in a rat model. Ann. Otol. Rhinol. Laryngol. 2006, 115, 285–292. [Google Scholar] [CrossRef]
- Welham, N.V.; Lim, X.; Tateya, I.; Bless, D.M. Inflammatory factor profiles one hour following vocal fold injury. Ann. Otol. Rhinol. Laryngol. 2008, 117, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Lim, X.; Bless, D.M.; Munoz-Del-Rio, A.; Welham, N.V. Changes in cytokine signaling and extracellular matrix production induced by inflammatory factors in cultured vocal fold fibroblasts. Ann. Otol. Rhinol. Laryngol. 2008, 117, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Tateya, T.; Tateya, I.; Sohn, J.H.; Bless, D.M. Histologic characterization of rat vocal fold scarring. Ann. Otol. Rhinol. Laryngol. 2005, 114, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Folcik, V.A.; An, G.C.; Orosz, C.G. The Basic Immune Simulator: An agent-based model to study the interactions between innate and adaptive immunity. Theor. Biol. Med. Model. 2007, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Grimm, V.; Revilla, E.; Berger, U.; Jeltsch, F.; Mooij, W.M.; Railsback, S.F.; Thulke, H.-H.; Weiner, J.; Wiegand, T.; DeAngelis, D.L. Pattern-oriented modeling of agent-based complex systems: Lessons from ecology. Science 2005, 310, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, J.; Hawkins-Daarud, A.; Massey, S.C.; Swanson, K.; Anderson, A. Hybrid approach for parameter estimation in agent-based models. bioRxiv 2017, 175661. [Google Scholar]
- Hussain, F.; Langmead, C.J.; Mi, Q.; Dutta-Moscato, J.; Vodovotz, Y.; Jha, S.K. Automated parameter estimation for biological models using Bayesian statistical model checking. BMC Bioinform. 2015, 16, S8. [Google Scholar] [CrossRef]
- Li, T.; Cheng, Z.; Zhang, L. Developing a Novel Parameter Estimation Method for Agent-Based Model in Immune System Simulation under the Framework of History Matching: A Case Study on Influenza A Virus Infection. Int. J. Mol. Sci. 2017, 18, 2592. [Google Scholar] [CrossRef]
- Moedomo, R.L.; Pancoro, A.; Ibrahim, J.; Ahmad, A.S.; Mardiyanto, M.S.; Belatiff, M.B.; Tasman, H. Simulation of influenza pandemic based on genetic algorithm and agent-based modeling: A multi-objective optimization problem solving. J. Matemat. Dan Sains 2010, 15, 47–59. [Google Scholar]
- Tong, X.; Chen, J.; Miao, H.; Li, T.; Zhang, L. Development of an agent-based model (ABM) to simulate the immune system and integration of a regression method to estimate the key ABM parameters by fitting the experimental data. PloS ONE 2015, 10, e0141295. [Google Scholar] [CrossRef]
- Wise, S.M.; Lowengrub, J.S.; Frieboes, H.B.; Cristini, V. Three-dimensional multispecies nonlinear tumor growth—I: Model and numerical method. J. Theor. Biol. 2008, 253, 524–543. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T.; Jeltsch, F.; Hanski, I.; Grimm, V. Using pattern-oriented modeling for revealing hidden information: A key for reconciling ecological theory and application. Oikos 2003, 100, 209–222. [Google Scholar] [CrossRef]
- Grimm, V.; Railsback, S.F. Pattern-oriented modelling: A ‘multi-scope’for predictive systems ecology. Phil. Trans. R. Soc. B 2012, 367, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Du, W.; Nian, F. An adaptive particle swarm optimization algorithm based on directed weighted complex network. Math. Probl. Eng. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Katare, S.; Bhan, A.; Caruthers, J.M.; Delgass, W.N.; Venkatasubramanian, V.; Katare, S.; Bhan, A.; Caruthers, J.M.; Delgass, W.N.; Venkatasubramanian, V. A hybrid genetic algorithm for efficient parameter estimation of large kinetic models. Comput. Chem. Eng. 2004, 28, 2569–2581. [Google Scholar] [CrossRef]
- Crooks, A.; Castle, C.; Batty, M. Key challenges in agent-based modelling for geo-spatial simulation. Comput. Environ. Urban Syst. 2008, 32, 417–430. [Google Scholar] [CrossRef]
- Windrum, P.; Fagiolo, G.; Moneta, A. Empirical validation of agent-based models: Alternatives and prospects. J. Artif. Soc. Soc. Simul. 2007, 10, 8. [Google Scholar]
- Fehler, M.; Klügl, F.; Puppe, F. In Approaches for resolving the dilemma between model structure refinement and parameter calibration in agent-based simulations. In Proceedings of the Fifth International Joint Conference on Autonomous Agents and Multiagent Systems, Hakodate, Japan, 8–12 May 2006; pp. 120–122. [Google Scholar]
- Iooss, B.; Lemaître, P. A review on global sensitivity analysis methods. In Uncertainty Management in Simulation-Optimization of Complex Systems; Springer: Boston, MA, USA, 2015; pp. 101–122. [Google Scholar]
- Marino, S.; Hogue, I.B.; Ray, C.J.; Kirschner, D.E. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 2008, 254, 178–196. [Google Scholar] [CrossRef]
- Ten Broeke, G.; Van Voorn, G.; Ligtenberg, A. Which sensitivity analysis method should I use for my agent-based model? J. Artif. Soc. Soc. Simul. 2016, 19, 1–5. [Google Scholar] [CrossRef]
- Saltelli, A.; Tarantola, S.; Chan, K.-S. A quantitative model-independent method for global sensitivity analysis of model output. Technometrics 1999, 41, 39–56. [Google Scholar] [CrossRef]
- McRae, G.J.; Tilden, J.W.; Seinfeld, J.H. Global sensitivity analysis—A computational implementation of the Fourier amplitude sensitivity test (FAST). Comput. Chem. Eng. 1982, 6, 15–25. [Google Scholar] [CrossRef]
- Henkel, T.; Wilson, H.; Krug, W. In Global sensitivity analysis of nonlinear mathematical models—An implementation of two complementing variance-based algorithms. In Proceedings of the 2012 Winter Simulation Conference, Berlin, Germany, 9–12 December 2012; p. 154. [Google Scholar]
- Strobl, C.; Boulesteix, A.-L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef] [PubMed]
- Criminisi, A.; Shotton, J.; Konukoglu, E. Decision forests: A unified framework for classification, regression, density estimation, manifold learning and semi-supervised learning. Found. Trends® Comput. Gr. Vis. 2012, 7, 81–227. [Google Scholar] [CrossRef]
- Strobl, C.; Boulesteix, A.-L.; Zeileis, A.; Hothorn, T. Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinform. 2007, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L.; Cutler, A. Random Forests-Classification Description. Available online: https://www.stat.berkeley.edu/~breiman/RandomForests/cc_home.htm (accessed on 20 May 2019).
- Ling, C.; Yamashita, M.; Zhang, J.; Bless, D.M.; Welham, N.V. Reactive response of fibrocytes to vocal fold mucosal injury in rat. Wound Repair Regen. 2010, 18, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Li, N.Y.; Lee, B.J.; Thibeault, S.L. Temporal and spatial expression of high-mobility group box 1 in surgically injured rat vocal folds. Laryngoscope 2012, 122, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Pilling, D.; Fan, T.; Huang, D.; Kaul, B.; Gomer, R.H. Identification of markers that distinguish monocyte-derived fibrocytes from monocytes, macrophages, and fibroblasts. PLoS ONE 2009, 4, e7475. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Li, Z. Role of CD11a and CD11b in corneal wound healing and inflammatory process. Investig. Ophthalmol. Vis. Sci. 2004, 45, 125. [Google Scholar]
- Mizgerd, J.P.; Kubo, H.; Kutkoski, G.J.; Bhagwan, S.D.; Scharffetter-Kochanek, K.; Beaudet, A.L.; Doerschuk, C.M. Neutrophil emigration in the skin, lungs, and peritoneum: Different requirements for CD11/CD18 revealed by CD18-deficient mice. J. Exp. Med. 1997, 186, 1357–1364. [Google Scholar] [CrossRef]
- Biosciences, B. CD Marker Handbook. BD Biosciences, 2014. Available online: https://www.bdbiosciences.com/documents/cd_marker_handbook.pdf (accessed on 10 July 2019).
- Ahmed, N.; Vogel, B.; Rohde, E.; Strunk, D.; Grifka, J.; Schulz, M.B.; Grassel, S. CD45-positive cells of haematopoietic origin enhance chondrogenic marker gene expression in rat marrow stromal cells. Int. J. Mol. Med. 2006, 18, 233. [Google Scholar] [CrossRef][Green Version]
- Kundrotas, G. Surface markers distinguishing mesenchymal stem cells from fibroblasts. Acta Medica Litu. 2012, 19. [Google Scholar] [CrossRef]
- Inoue, T.; Plieth, D.; Venkov, C.D.; Xu, C.; Neilson, E.G. Antibodies against macrophages that overlap in specificity with fibroblasts. Kidney Int. 2005, 67, 2488–2493. [Google Scholar] [CrossRef] [PubMed]
- Van Landuyt, K.B.; Jones, E.A.; McGonagle, D.; Luyten, F.P.; Lories, R.J. Flow cytometric characterization of freshly isolated and culture expanded human synovial cell populations in patients with chronic arthritis. Arthritis Res. Ther. 2010, 12, R15. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, C.; Döpp, E.; Joling, P.; Kraal, G. The heterogeneity of mononuclear phagocytes in lymphoid organs: Distinct macrophage subpopulations in rat recognized by monoclonal antibodies ED1, ED2 and ED3. In Microenvironments in the Lymphoid System; Springer: London, UK, 1985; pp. 409–419. [Google Scholar]
- Fecho, K.; Lysle, D.T. Morphine-induced enhancement in the granulocyte response to thioglycollate administration in the rat. Inflammation 2002, 26, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Thomas, A.; Bunyan, K.; Tiidus, P.M. Progesterone and estrogen influence postexercise leukocyte infiltration in overiectomized female rats. Appl. Physiol. Nutr. Metab. 2008, 33, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2001, 21, A.3B.1–A.3B.2. [Google Scholar]
- Roederer, M. Spectral compensation for flow cytometry: Visualization artifacts, limitations, and caveats. Cytom. Part A 2001, 45, 194–205. [Google Scholar] [CrossRef]
- Roederer, M. Compensation in flow cytometry. Curr. Protoc. Cytom. 2002, 22, 1.14.1–1.14.20. [Google Scholar]
- Perfetto, S.P.; Chattopadhyay, P.K.; Roederer, M. Seventeen-colour flow cytometry: Unravelling the immune system. Nat. Rev. Immunol. 2004, 4, 648–655. [Google Scholar] [CrossRef]
- Arnoulet, C.; Béné, M.C.; Durrieu, F.; Feuillard, J.; Fossat, C.; Husson, B.; Jouault, H.; Maynadié, M.; Lacombe, F. Four-and five-color flow cytometry analysis of leukocyte differentiation pathways in normal bone marrow: A reference document based on a systematic approach by the GTLLF and GEIL. Cytom. Part B Clin. Cytom. 2010, 78, 4–10. [Google Scholar] [CrossRef]
- Lugli, E.; Roederer, M.; Cossarizza, A. Data analysis in flow cytometry: The future just started. Cytom. Part A 2010, 77, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Autissier, P.; Soulas, C.; Burdo, T.H.; Williams, K.C. Evaluation of a 12-color flow cytometry panel to study lymphocyte, monocyte, and dendritic cell subsets in humans. Cytom. Part A 2010, 77, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Pyne, S.; Hu, X.; Wang, K.; Rossin, E.; Lin, T.-I.; Maier, L.M.; Baecher-Allan, C.; McLachlan, G.J.; Tamayo, P.; Hafler, D.A. Automated high-dimensional flow cytometric data analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 8519–8524. [Google Scholar] [CrossRef] [PubMed]
- Lugli, E.; Pinti, M.; Nasi, M.; Troiano, L.; Ferraresi, R.; Mussi, C.; Salvioli, G.; Patsekin, V.; Robinson, J.P.; Durante, C. Subject classification obtained by cluster analysis and principal component analysis applied to flow cytometric data. Cytom. Part A 2007, 71, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Steinbrich-Zöllner, M.; Grün, J.R.; Kaiser, T.; Biesen, R.; Raba, K.; Wu, P.; Thiel, A.; Rudwaleit, M.; Sieper, J.; Burmester, G.R. From transcriptome to cytome: Integrating cytometric profiling, multivariate cluster, and prediction analyses for a phenotypical classification of inflammatory diseases. Cytom. Part A 2008, 73, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, Y.; Watanabe, H.; Inoue, J.; Takeda, N.; Sakata, J.; Mishima, Y.; Hitomi, J.; Yamamoto, T.; Utsuyama, M.; Niwa, O. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat. Immunol. 2003, 4, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, H.M. Practical Flow Cytometry; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Houtz, B.; Trotter, J.; Sasaki, D. BD FACService™ TECHNOTES. Custom. Focus. Solut. 2004, 9, 1–9. [Google Scholar]
- López-Sánchez, N.; Frade, J.M. Genetic evidence for p75NTR-dependent tetraploidy in cortical projection neurons from adult mice. J. Neurosci. 2013, 33, 7488–7500. [Google Scholar] [CrossRef] [PubMed]
- Herzenberg, L.A.; Tung, J.; Moore, W.A.; Herzenberg, L.A.; Parks, D.R. Interpreting flow cytometry data: A guide for the perplexed. Nat. Immunol. 2006, 7, 681–685. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Menze, B.H.; Kelm, B.M.; Masuch, R.; Himmelreich, U.; Bachert, P.; Petrich, W.; Hamprecht, F.A. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinform. 2009, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Galiano, V.; Chica-Olmo, M.; Abarca-Hernandez, F.; Atkinson, P.M.; Jeganathan, C. Random Forest classification of Mediterranean land cover using multi-seasonal imagery and multi-seasonal texture. Remote Sens. Environ. 2012, 121, 93–107. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Houska, T.; Kraft, P.; Chamorro-Chavez, A.; Breuer, L. SPOTting model parameters using a ready-made python package. PLoS ONE 2015, 10, e0145180. [Google Scholar] [CrossRef] [PubMed]
- Krauße, T.; Cullmann, J. Towards a more representative parametrisation of hydrologic models via synthesizing the strengths of Particle Swarm Optimisation and Robust Parameter Estimation. Hydrol. Earth Syst. Sci. 2012, 16, 603–629. [Google Scholar] [CrossRef]
- Bárdossy, A.; Singh, S. Robust estimation of hydrological model parameters. Hydrol. Earth Syst. Sci. 2008, 12, 1273–1283. [Google Scholar] [CrossRef]
- Kirsner, R.S.; Eaglstein, W. The wound healing process. Dermatol. Clin. 1993, 11, 629–640. [Google Scholar] [CrossRef]
- Martin, P. Wound healing--aiming for perfect skin regeneration. Science 1997, 276, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Witte, M.B.; Barbul, A. General principles of wound healing. Surg. Clin. N. Am. 1997, 77, 509–528. [Google Scholar] [CrossRef]
- Robson, M.C.; Steed, D.L.; Franz, M.G. Wound healing: Biologic features and approaches to maximize healing trajectories. Curr. Probl. Surg. 2001, 38, A1–A140. [Google Scholar] [CrossRef] [PubMed]
- Broughton, G., 2nd; Janis, J.E.; Attinger, C.E. The basic science of wound healing. Plast. Reconstr. Surg. 2006, 117, 12S–34S. [Google Scholar] [CrossRef] [PubMed]
- Enoch, S.; Leaper, D.J. Basic science of wound healing. Surgery (Oxford) 2008, 26, 31–37. [Google Scholar] [CrossRef]
- Pohlman, T.H.; Stanness, K.A.; Beatty, P.G.; Ochs, H.D.; Harlan, J.M. An endothelial cell surface factor (s) induced in vitro by lipopolysaccharide, interleukin 1, and tumor necrosis factor-alpha increases neutrophil adherence by a CDw18-dependent mechanism. J. Immunol. 1986, 136, 4548–4553. [Google Scholar] [PubMed]
- Branski, R.C.; Verdolini, K.; Rosen, C.A.; Hebda, P.A. Acute vocal fold wound healing in a rabbit model. Ann. Otol. Rhinol. Laryngol. 2005, 114, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Li-Jessen, N.Y.; Powell, M.; Choi, A.J.; Lee, B.J.; Thibeault, S.L. Cellular source and proinflammatory roles of high-mobility group box 1 in surgically injured rat vocal folds. Laryngoscope 2016, 127, E193–E200. [Google Scholar] [CrossRef]
- King, S.N.; Chen, F.; Jetté, M.E.; Thibeault, S.L. Vocal fold fibroblasts immunoregulate activated macrophage phenotype. Cytokine 2013, 61, 228–236. [Google Scholar] [CrossRef]
- Hanson, S.E.; Kim, J.; Johnson, B.H.Q.; Bradley, B.; Breunig, M.J.; Hematti, P.; Thibeault, S.L. Characterization of mesenchymal stem cells from human vocal fold fibroblasts. Laryngoscope 2010, 120, 546–551. [Google Scholar] [CrossRef]
- Jiang, P.; Wu, H.; Wang, W.; Ma, W.; Sun, X.; Lu, Z. MiPred: Classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007, 35, W339–W344. [Google Scholar] [CrossRef]
- Virag, J.I.; Murry, C.E. Myofibroblast and endothelial cell proliferation during murine myocardial infarct repair. Am. J. Pathol. 2003, 163, 2433–2440. [Google Scholar] [CrossRef]
- Butterfield, T.A.; Best, T.M.; Merrick, M.A. The dual roles of neutrophils and macrophages in inflammation: A critical balance between tissue damage and repair. J. Athl. Train. 2006, 41, 457. [Google Scholar]
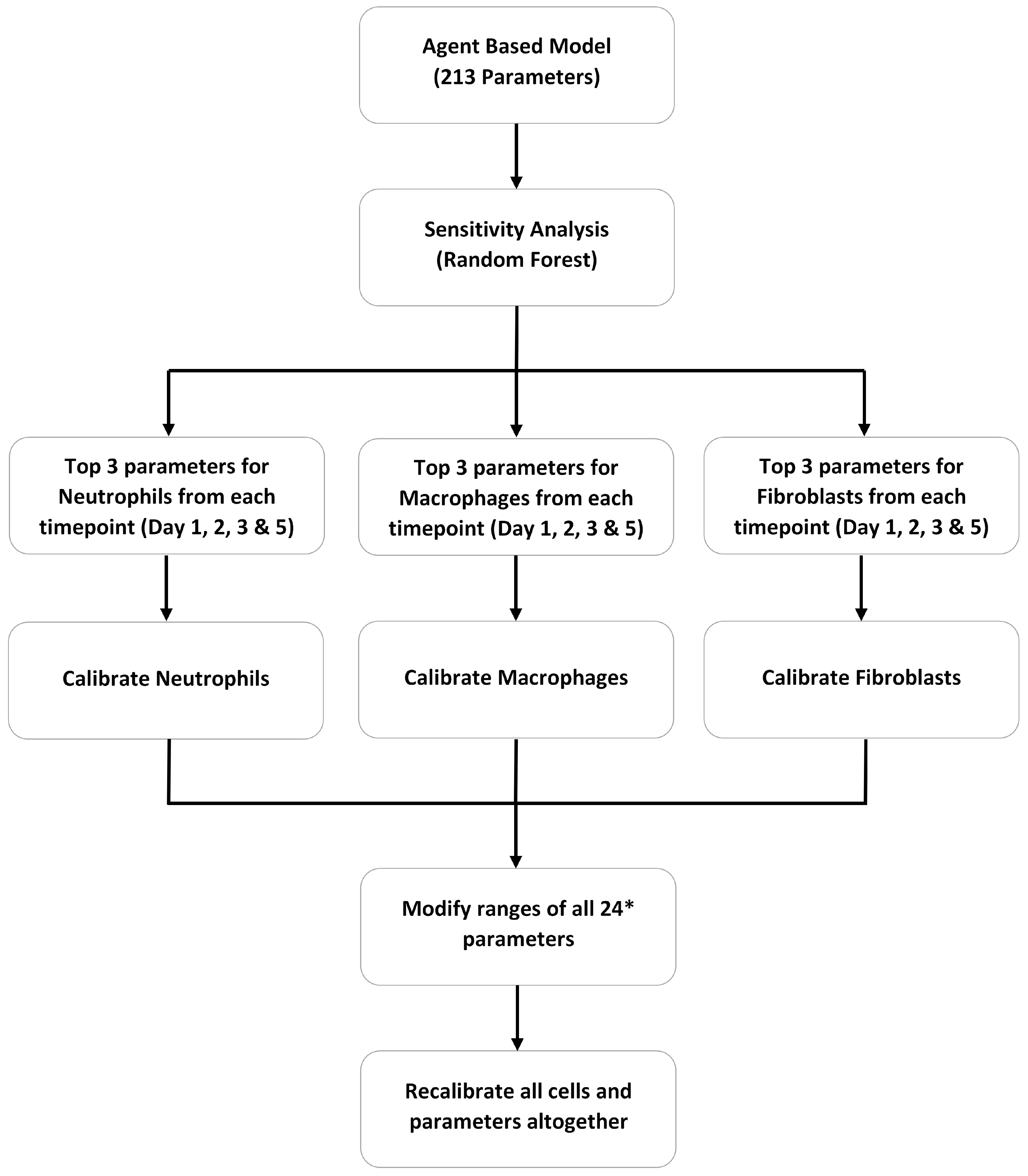
| Study | Calibration Method | Number of Unknown Parameters | Field of ABM Application |
|---|---|---|---|
| Folcik et al., 2007 [41] | Parameter Sweeping | 87 | Basic Immune Simulator |
| Grimm et al., 2005 [42] | Pattern-Oriented Approach | unreported | Ecology |
| Gallaher et al., 2017 [43] | Genetic algorithm with random weighted sampling | 16 | Glioblastoma multiforme model |
| Hussain et al., 2015 [44] | Bayesian Approach | 24 | Dynamics of acute inflammation |
| Li et al., 2017 [45] | Particle swarm optimization (PSO) | 50 | Immune System |
| Moedomo et al., 2010 [46] | Genetic Algorithm | 6 | Avian Influenza (H5N1) viruses mutation |
| Tong et al., 2015 [47] | Greedy algorithm and Regression | 4 | Immune System |
| Wise et al., 2008 [48] | Nonlinear multigrid/finite difference method | 20 | Three-dimensional multispecies nonlinear tumor growth |
| Marker | Fluorochrome | Neutrophil | Macrophage | Endothelial Cell | Fibroblast | References |
|---|---|---|---|---|---|---|
| CD11b/c | FITC | + | + | - | - | [68,69,70,71] |
| CD29 | PE-Cy7 | + | + | + | + | [68,71,72,73] |
| CD44H | APC-Cy7 | - | + | + | + | [68,71,73] |
| CD45 | PerCP-Cy5.5 | + | + | - | - | [68,71,72,73,74,75] |
| CD68 | PE-Texas Red | + | + | - | + | [68,71,74,76] |
| CD105 | PE | - | + | + | + | [68,71,73,75] |
| CD106 | Brilliant Violet 421 | - | - | + | - | [68,73] |
| His48 | APC | + | - | - | - | [77,78] |
| Cell Viability Dye | AmCyan | + | + | + | + | [79] |
| Marker | Fluorochrome | Neutrophil | Macrophage | Endothelial Cell | Fibroblast | References |
|---|---|---|---|---|---|---|
| CD31 | APC | + | + | + | - | [68,71,73] |
| CD45 | PerCP-Cy5.5 | + | + | - | - | [68,71,72,73,74,75] |
| CD90 | FITC | - | - | + | + | [68,71,73,75] |
| CD163 | PE-Cy7 | - | + | - | - | [68,71,75] |
| His48 | PE | + | - | - | - | [77,78] |
| Cell Viability Dye | AmCyan | + | + | + | + | [79] |
| Item | Unit | Size | Reference |
|---|---|---|---|
| Vocal Fold Width | mm | 1 | [35] |
| patches | 142 | ||
| Vocal Fold Height | mm | 1 | [35] |
| patches | 142 | ||
| Vocal Fold Thickness | mm | 0.2 | [35] |
| patches | 28 | ||
| Vocal Fold Epithelium Thickness | mm | 0.01 | [31] |
| patches | 1 | ||
| Capillary Diameter | µm | 7 | [28] |
| patches | 1 | ||
| Capillary Gap | µm | 12.89 | [28] |
| patches | 1 | ||
| Patch size | µm | 7 × 7 × 7 | |
| Total number of patches | patches | 564,592 | - |
| Total number of non-epi patches | patches | 544,428 | - |
| Total number of capillary patches | patches | 138,450 | - |
| Total number of tissue patches | patches | 405,978 | - |
| Simulated time-step | Minutes | 30 | - |
| Neutrophils | µm | 7 | [29] |
| Cells | 517 | Flow data | |
| Macrophages | µm | 6 | [30] |
| Cells | 316 | Flow data | |
| Fibroblasts | µm | 6 | [29] |
| Cells | 3594 | Flow data |
| Time Point | Cell | Parameter | Biological Significance |
|---|---|---|---|
| Day 1 | Neutrophils | 156--WhSproutAmount5 | Determine the effect of damage on the rate of neutrophil extravasation |
| 164--WhSproutAmount13 | Determine the baseline rate of fibroblast sprouting | ||
| 39--MacCytSynth14 | Determine the macrophage’s baseline synthesis rate of TNF-alpha | ||
| Macrophages | 148--WhSproutFreq3 | Frequency macrophages enter vocal fold capillaries when damage is present | |
| 117--FibCytSynth10 | Determine a fibroblast’s baseline synthesis rate of FGF | ||
| 133--FibCytSynth26 | Determine a fibroblast’s baseline synthesis rate of IL-8 | ||
| Fibroblasts | 200--FibProlif0 | Frequency of inactivated fibroblast proliferation in hours | |
| 150--WhSproutFreq5 | Frequency of fibroblasts sprouting in tissue in the presence of damage | ||
| 166--FibActivat1 | Probability (in percent) of fibroblast activation in the absence of TGF-beta | ||
| Day 2 | Neutrophils | 156--WhSproutAmount5 | Determine the effect of damage on the rate of neutrophil extravasation |
| 181--FibECMsynth0 | Determine the fibroblast’s baseline synthesis rate of collagen | ||
| 103--MacCytSynth78 | Determine the macrophage’s baseline synthesis rate of IL-10 | ||
| Macrophages | 159--WhSproutAmount8 | Determine the baseline rate of macrophage extravasation in the presence of damage | |
| 158--WhSproutAmount7 | Determine the effect of damage on the rate of macrophage extravasation | ||
| 177--NeuActivat2 | Probability (in percent) of neutrophil activation in the presence of high TNF-alpha concentration | ||
| Fibroblasts | 14--NeuCytSynth10 | Effect of local TGF-beta concentration on neutrophil MMP-8 synthesis | |
| 72--MacCytSynth47 | Effect of local IL-1beta concentration on macrophage IL-6 synthesis | ||
| 11--NeuCytSynth7 | Determine the neutrophil’s baseline synthesis rate of MMP-8 | ||
| Day 3 | Neutrophils | 156--WhSproutAmount5 | Determine the effect of damage on the rate of neutrophil extravasation |
| 200--FibProlif0 | Frequency of inactivated fibroblast proliferation in hours | ||
| 148--WhSproutFreq3 | Frequency macrophages enter vocal fold capillaries when damage is present | ||
| Macrophages | 149--WhSproutFreq4 | Frequency macrophages extravasate in the presence of damage | |
| 160--WhSproutAmount9 | Determine the effect of damage on the rate of macrophage extravasation | ||
| 103--MacCytSynth78 | Determine the macrophage’s baseline synthesis rate of IL-10 | ||
| Fibroblasts | 110--FibCytSynth3 | Determine the fibroblast’s baseline synthesis rate of TNF-alpha | |
| 51--MacCytSynth26 | Effect of local TNF-alpha concentration on macrophage IL1-beta synthesis | ||
| 117--FibCytSynth10 | Determine a fibroblast’s baseline synthesis rate of FGF | ||
| Day 5 | Neutrophils | 156--WhSproutAmount5 | Determine the effect of damage on the rate of neutrophil extravasation |
| 155--WhSproutAmount4 | Determine the baseline rate of neutrophil extravasation in the presence of damage | ||
| 153--WhSproutAmount2 | Determine the rate of neutrophil extravasation in the absence of damage | ||
| Macrophages | 154--WhSproutAmount3 | Determine the rate of macrophage extravasation in the absence of damage | |
| 159--WhSproutAmount8 | Determine the baseline rate of macrophage extravasation in the presence of damage | ||
| 149--WhSproutFreq4 | Frequency macrophages extravasate in the presence of damage | ||
| Fibroblasts | 200--FibProlif0 | Frequency of inactivated fibroblast proliferation in hours | |
| 51--MacCytSynth26 | Effect of local TNF-alpha concentration on macrophage IL1-beta synthesis | ||
| 110--FibCytSynth3 | Determine the fibroblast’s baseline synthesis rate of TNF-alpha |
| Time-Point | Cell Types | Empirical Data of Cell Quantities | 95% Confidence Interval (and Mean) of ABM-Simulated Cell Quantities |
|---|---|---|---|
| Day 7 | Neutrophils | 214 | 198.5–228 * (212.29) |
| Macrophages | 359 | 310–406 * (359.15) | |
| Fibroblasts | 5751 | 4920.5–7541.5 * (6083.45) | |
| Empirical Data within 95% CI | 100% | ||
| Week 2 | Neutrophils | 180 | 188.5–222.5 (204.87) |
| Macrophages | 186 | 103.5–155.5 (126.8) | |
| Fibroblasts | 3854 | 1759–2640 (2142) | |
| Empirical Data within 95% CI | 0% | ||
| Week 4 | Neutrophils | 214 | 192.5–227 * (210.16) |
| Macrophages | 252 | 10–27 (17.07) | |
| Fibroblasts | 4347 | 254–378 (304.45) | |
| Empirical Data within 95% CI | 33% | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garg, A.; Yuen, S.; Seekhao, N.; Yu, G.; Karwowski, J.A.C.; Powell, M.; Sakata, J.T.; Mongeau, L.; JaJa, J.; Li-Jessen, N.Y.K. Towards a Physiological Scale of Vocal Fold Agent-Based Models of Surgical Injury and Repair: Sensitivity Analysis, Calibration and Verification. Appl. Sci. 2019, 9, 2974. https://doi.org/10.3390/app9152974
Garg A, Yuen S, Seekhao N, Yu G, Karwowski JAC, Powell M, Sakata JT, Mongeau L, JaJa J, Li-Jessen NYK. Towards a Physiological Scale of Vocal Fold Agent-Based Models of Surgical Injury and Repair: Sensitivity Analysis, Calibration and Verification. Applied Sciences. 2019; 9(15):2974. https://doi.org/10.3390/app9152974
Chicago/Turabian StyleGarg, Aman, Samson Yuen, Nuttiiya Seekhao, Grace Yu, Jeannie A. C. Karwowski, Michael Powell, Jon T. Sakata, Luc Mongeau, Joseph JaJa, and Nicole Y. K. Li-Jessen. 2019. "Towards a Physiological Scale of Vocal Fold Agent-Based Models of Surgical Injury and Repair: Sensitivity Analysis, Calibration and Verification" Applied Sciences 9, no. 15: 2974. https://doi.org/10.3390/app9152974
APA StyleGarg, A., Yuen, S., Seekhao, N., Yu, G., Karwowski, J. A. C., Powell, M., Sakata, J. T., Mongeau, L., JaJa, J., & Li-Jessen, N. Y. K. (2019). Towards a Physiological Scale of Vocal Fold Agent-Based Models of Surgical Injury and Repair: Sensitivity Analysis, Calibration and Verification. Applied Sciences, 9(15), 2974. https://doi.org/10.3390/app9152974






