Toward Automatic Cardiomyocyte Clustering and Counting through Hesitant Fuzzy Sets
Abstract
:Featured Application
Abstract
1. Introduction
2. Related Work
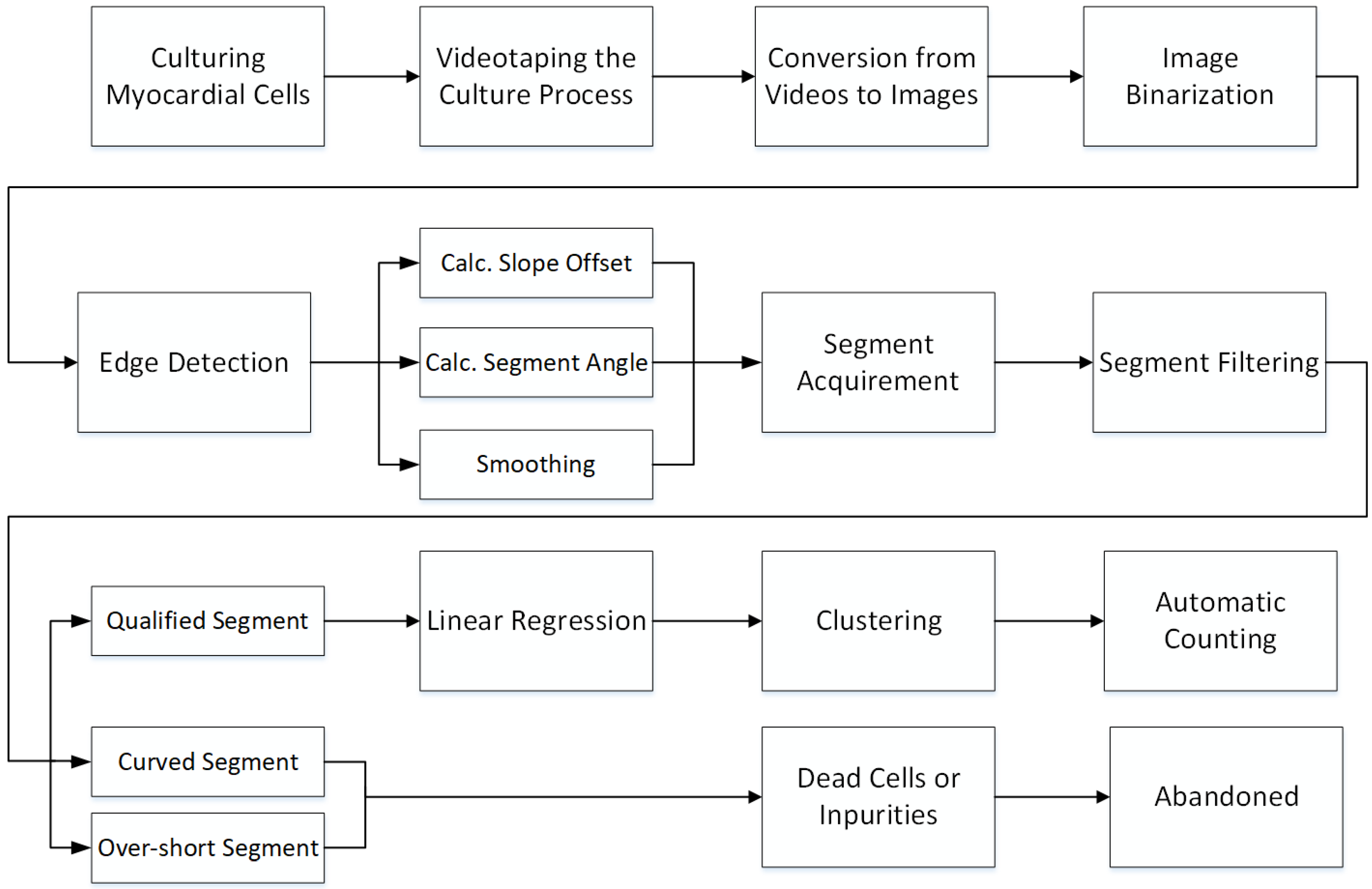
3. Methods and Algorithms
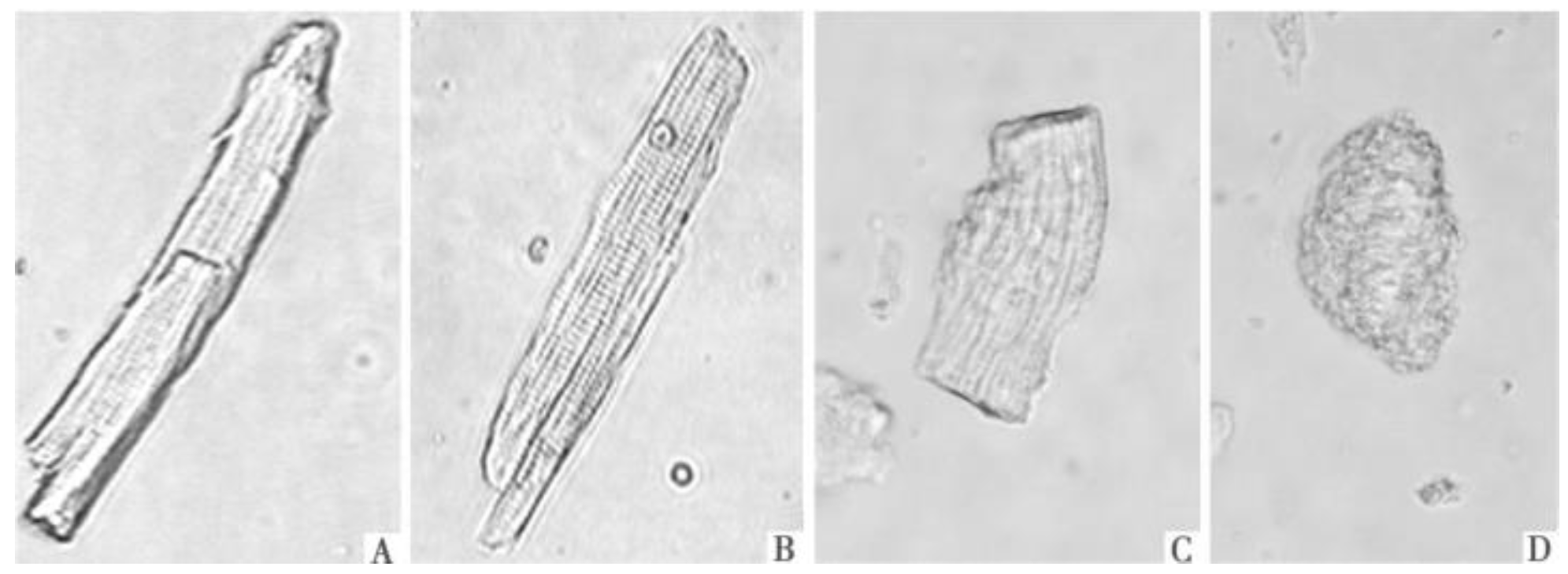
3.1. Isolation and Culture of Adult Rat Cardiomyocytes
3.2. Videotaping the Procedure
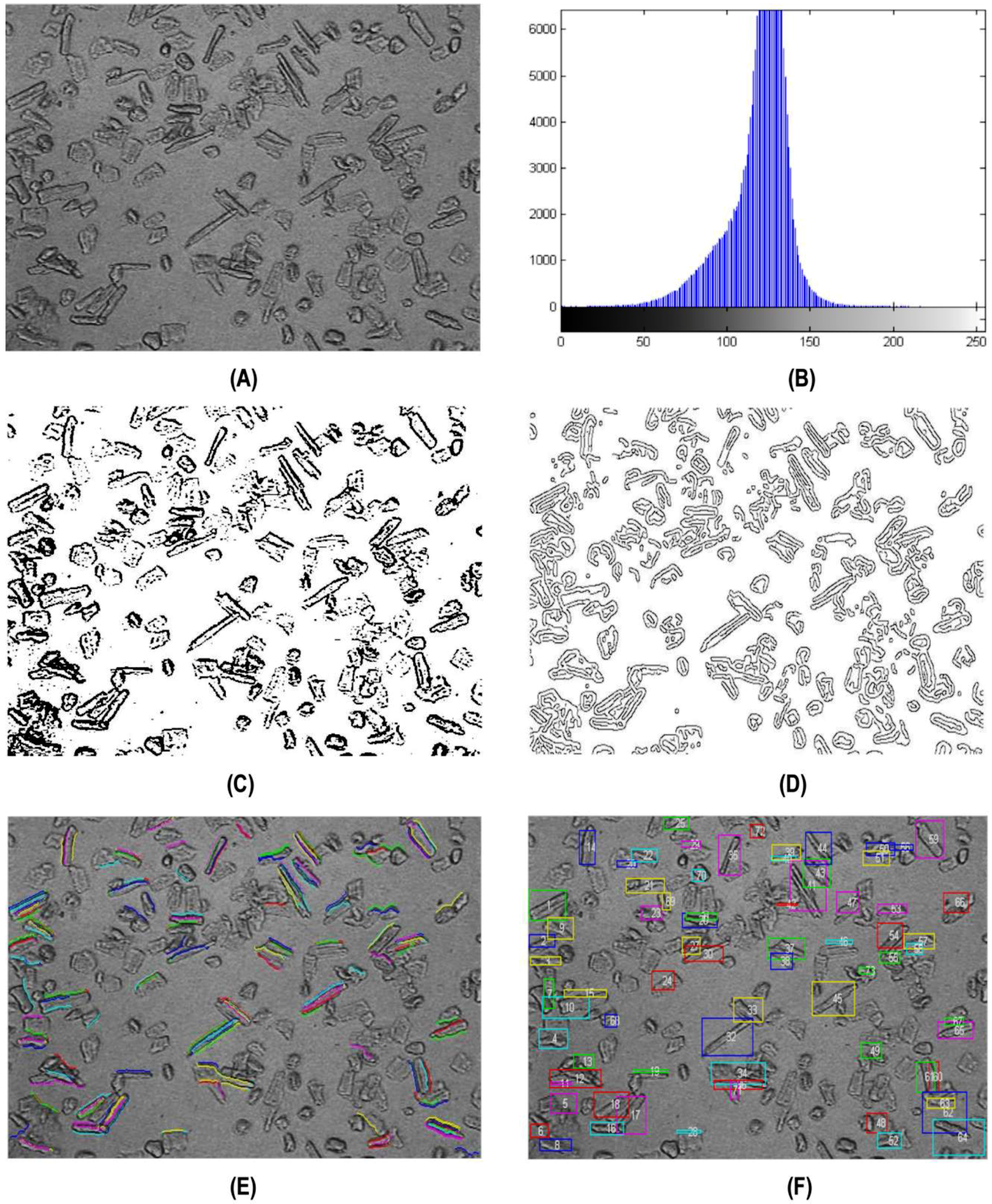
3.3. Automatic Counting for Cardiomyocytes
3.4. Hesitant Fuzzy Sets for Clustering
- 1.
- ;
- 2.
- ;
- 3.
- .
| Algorithm 1. Automatic classification algorithm of cardiomyocytes based on hesitating fuzzy set |
| 1. Cardiomyocytes to be evaluated were selected as the evaluation objects of the hesitating fuzzy set, and denoted by {}; |
| 2. According to the given attribute parameters, the correlation coefficient between them is calculated according to formula (3), and a matrix of correlation coefficients is constructed; |
| 3. If is an equivalent correlation matrix, then |
| Check whether it satisfies , where |
| Otherwise, construct the equivalent correlation matrix : |
| End if |
| 5. Using definition 5, construct a -cutting Matrix in order to classify the HFS’s s . |
| 6. If then |
| All the elements are in a class of their own |
| 7. Otherwise, classify these m cardiomyocytes according to |
| 8. End if |
4. Experimentation and Evaluation
4.1. Experimental Setup
4.2. Parameter Space
4.3. Clustering Cardiomyocyte Cells
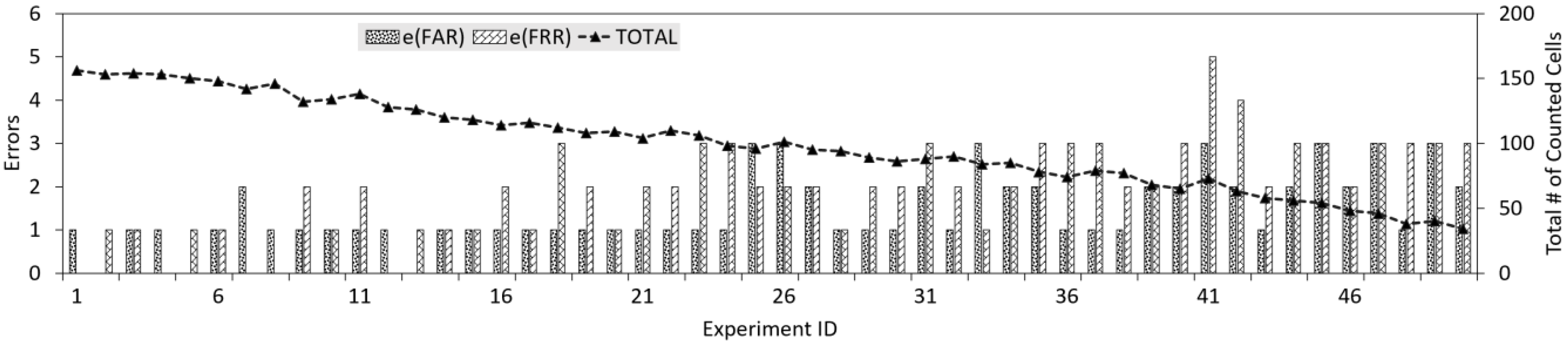
4.4. Evaluating Cell Counting
4.5. Comparison to the Conventional Approach
5. Conclusions and Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kasbe, T.; Pippal, R.S. Design of heart disease diagnosis system using fuzzy logic. In Proceedings of the 2017 International Conference on Energy, Communication, Data Analytics and Soft Computing (ICECDS), Chennai, India, 1–2 August 2017; pp. 3183–3187. [Google Scholar]
- Masaki, I.; Ji-Dong, F.; Paul, D.O.; Vasanth, V.; Yohei, H.; Bruneau, B.G.; Deepak, S. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Nihon Rinsho Jpn. J. Clin. Med. 2011, 142, 375–386. [Google Scholar] [CrossRef]
- Senyo, S.E.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Lei, C.; Mei, W.; Ting-Di, W.; Jean-Luc, G.K.; Lechene, C.P.; Lee, R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013, 493, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Ping, L.; Feng, L.; Lee, A.S.; Tingyu, G.; Veronica, S.F.; Yongming, W.; Sebastian, D.; Karim, S.; Knowles, J.W.; Wang, P.J. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation 2013, 127, 1677. [Google Scholar] [CrossRef]
- Li, Q.; Yu, H.; Ian, S.C.; Amy, F.; Vasanth, V.; Lei, L.; Conway, S.J.; Ji-Dong, F.; Deepak, S. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef]
- Benderskaya, E.N. Cluster analysis problems and bio-inspired clustering methods. In Proceedings of the 2017 IEEE International Conference on Soft Computing and Measurements, St. Petersburg, Russia, 24–26 May 2017. [Google Scholar] [CrossRef]
- Tas, M.; De Haan-Meulman, M.; Kabel, P.J.; Drexhage, H.A. Defects in monocyte polarization and dendritic cell clustering in patients with Graves’ disease. A putative role for a non-specific immunoregulatory factor related to retroviral p15E. Clin. Endocrinol. 2010, 34, 441–448. [Google Scholar] [CrossRef]
- Takaaki, M.; Siripong, T.; Tomoko, M.; Hisaya, K.; Takuji, N.; Yoshio, H.; Yoshikazu, H.; Hitoshi, O.; Yasumasa, B. Canopy1, a positive feedback regulator of FGF signaling, controls progenitor cell clustering during Kupffer’s vesicle organogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 9881–9886. [Google Scholar]
- Nosaka, R.; Fukui, K. HEp-2 cell classification using rotation invariant co-occurrence among local binary patterns. Pattern Recognit. 2014, 47, 2428–2436. [Google Scholar] [CrossRef]
- Chen, C.L.; Mahjoubfar, A.; Tai, L.C.; Blaby, I.K.; Huang, A.; Niazi, K.R.; Jalali, B. Deep Learning in Label-free Cell Classification. Sci. Rep. 2016, 6, 21471. [Google Scholar] [CrossRef]
- Frankel, D.S.; Frankel, S.L.; Binder, B.J.; Vogt, R.F. Application of neural networks to flow cytometry data analysis and real-time cell classification. Cytometry 2015, 23, 290–302. [Google Scholar] [CrossRef]
- Wu, S. Survey on K-means Algorithm. New Technol. Libr. Inf. Serv. 2011, 29, 433–439. [Google Scholar]
- Pérez-Suárez, A.; Martńez-Trinidad, J.F.; Carrasco-Ochoa, J.A.; Medina-Pagola, J.E. A New Overlapping Clustering Algorithm Based on Graph Theory. In Proceedings of the Mexican International Conference on Artificial Intelligence, San Luis Potosí, Mexico, 27 October–4 November 2012. [Google Scholar]
- He, Y.; Tan, H.; Luo, W.; Mao, H.; Ma, D.; Feng, S.; Fan, J. MR-DBSCAN: An Efficient Parallel Density-Based Clustering Algorithm Using MapReduce. In Proceedings of the IEEE International Conference on Parallel & Distributed Systems, Tainan, Taiwan, 7–9 December 2011. [Google Scholar]
- Tsai, W.K.; Sheu, M.H. An Efficient Foreground Object Detection Method Using a Color Cluster-Based Background Modeling Algorithm. In Proceedings of the International Symposium on Computer, Xi’an, China, 4–6 July 2016. [Google Scholar]
- Buck, R. Cluster-Based Salient Object Detection Using K-Means Merging and Keypoint Separation with Rectangular Centers. Master’s Thesis, Utah State University, Logan, UT, USA, 2016. [Google Scholar]
- Chen, T.; Lu, S.; Fan, J. S-CNN: Subcategory-aware convolutional networks for object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 2522–2528. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Huang, Z.; Wang, S.; Ji, X. A Scalable Object Detection Framework Based on Embedded Manycore Cluster. In Proceedings of the International Conference on Cyber-enabled Distributed Computing & Knowledge Discovery, Chengdu, China, 13–15 October 2016. [Google Scholar]
- Torra, V.; Narukawa, Y. On Hesitant Fuzzy Sets and Decision. In Proceedings of the IEEE International Conference on Fuzzy Systems, Jeju Island, South Korea, 20–24 August 2009. [Google Scholar]
- Du, X. Correlation coefficient of hesitant fuzzy sets. In Proceedings of the 2017 32nd Youth Academic Annual Conference of Chinese Association of Automation (YAC), Hefei, China, 19–21 May 2017. [Google Scholar]
- Torra, V. Hesitant fuzzy sets. Int. J. Intell. Syst. 2010, 25, 529–539. [Google Scholar] [CrossRef]
- Atanassov, K.T. Intuitionistic fuzzy sets. Fuzzy Sets Syst. 1986, 20, 87–96. [Google Scholar] [CrossRef]
- Dubois, D. Fuzzy Sets and Systems: Theory and Applications; Academic Press, Inc.: Orlando, FL, USA, 1980. [Google Scholar]
- Yager, R.R. On the theory of bags. Int. J. Gen. Syst. 1986, 13, 23–37. [Google Scholar] [CrossRef]
- Ye, Y.; Zhang, Z.; Yang, X.; Yeo, J.H.; Jiang, L.J.; Jiang, D. Blood cell counting and classification by nonflowing laser light scattering method. J. Biomed. Opt. 2004, 3897, 995–1001. [Google Scholar]
- Serafini, R.; Longobardi, V.; Spadetta, M.; Neri, D.; Ariota, B.; Gasparrini, B.; Di, P.R. Trypan blue/giemsa staining to assess sperm membrane integrity in salernitano stallions and its relationship to pregnancy rates. Reprod. Domest. Anim. 2014, 49, 41–47. [Google Scholar] [CrossRef]
- Liu, T.X.; Chu, C.C. Comparison of absolute estimates of Thrips tabaci (Thysanoptera: Thripidae) with field visual counting and sticky traps in onion field in south Texas. Southwest. Entomol. 2004, 29, 83–89. [Google Scholar]
- Liu, X.; Iyengar, S.G.; Rittscher, J. Monitoring cardiomyocyte motionin real time through image registration and time series analysis. In Proceedings of the IEEE International Symposium on Biomedical Imaging, Barcelona, Spain, 2–5 May 2012. [Google Scholar]
- Hu, N.; Fang, J.; Li, H.; Su, K.; Wang, P. Dual-function microelectrode array system for simultaneously monitoring electromechanical integration status of cardiomyocytes. In Proceedings of the Transducers-International Conference on Solid-state Sensors, Anchorage, AK, USA, 21–25 June 2015. [Google Scholar]
- Piccinini, F.; Tesei, A.; Paganelli, G.; Zoli, W.; Bevilacqua, A. Improving reliability of live/dead cell counting through automated image mosaicing. Comput. Methods Progr. Biomed. 2014, 117, 448–463. [Google Scholar] [CrossRef]
- Cibej, U.; Lojk, J.; Pavlin, M.; Sajn, L. Automatic adaptation of filter sequences for cell counting. In Proceedings of the International Convention on Information & Communication Technology, Electronics & Microelectronics, Opatija, Croatia, 25–29 May 2015. [Google Scholar]
- Maji, P.; Mandal, A.; Ganguly, M.; Saha, S. An automated method for counting and characterizing red blood cells using mathematical morphology. In Proceedings of the 2015 Eighth International Conference on Advances in Pattern Recognition (ICAPR), Kolkata, India, 4–7 January 2015; pp. 1–6. [Google Scholar]
- Sarrafzadeh, O.; Dehnavi, A.M.; Rabbani, H.; Ghane, N.; Talebi, A. Circlet based framework for red blood cells segmentation and counting. In Proceedings of the 2015 IEEE Workshop on Signal Processing Systems (SiPS), Hangzhou, China, 14–16 October 2015. [Google Scholar]
- Loddo, A.; Putzu, L.; Ruberto, C.D.; Fenu, G. A Computer-Aided System for Differential Count from Peripheral Blood Cell Images. In Proceedings of the International Conference on Signal-image Technology & Internet-based Systems, Naples, Italy, 28 November–1 December 2016. [Google Scholar]
- Ji, G.; Zheng, G.; Jun, C.; Jun, L.; John, N.; Zongyi, Y.; Chen, W.; Yu, S. A system for counting fetal and maternal red blood cells. IEEE Trans. Biomed. Eng. 2014, 61, 2823–2829. [Google Scholar] [CrossRef]
- Lou, J.; Mei, Z.; Li, Q.; Chen, Y.; Liu, H.; Lou, J.; Mei, Z.; Li, Q.; Chen, Y.; Liu, H. An automatic red blood cell counting method based on spectral images. In Proceedings of the International Congress on Image & Signal Processing, Datong, China, 15–17 October 2016. [Google Scholar]
- Yellin, F.; Haeffele, B.D.; Vidal, R. Blood cell detection and counting in holographic lens-free imaging by convolutional sparse dictionary learning and coding. In Proceedings of the IEEE International Symposium on Biomedical Imaging, Melbourne, Australia, 18–21 April 2017. [Google Scholar]
- Huang, X.; Jiang, Y.; Liu, X.; Xu, H.; Han, Z.; Rong, H.; Yang, H.; Yan, M.; Yu, H. Machine Learning Based Single-Frame Super-Resolution Processing for Lensless Blood Cell Counting. Sensors 2016, 16, 1836. [Google Scholar] [CrossRef]
- Ghosh, P.; Bhattacharjee, D.; Nasipuri, M. Blood smear analyzer for white blood cell counting: A hybrid microscopic image analyzing technique. Appl. Soft Comput. 2016, 46, 629–638. [Google Scholar] [CrossRef]
- Relli, C.D.S.; Facon, J.; Ayala, H.L.; Britto, A.D.S. Automatic counting of trypanosomatid amastigotes in infected human cells. Comput. Biol. Med. 2017, 89, 222–235. [Google Scholar] [CrossRef]
- Melo, G.J.A.D.; Gomes, V.; Baccili, C.C.; Almeida, L.A.L.D.; Lima, A.C.D.C. A robust segmentation method for counting bovine milk somatic cells in microscope slide images. Comput. Electron. Agric. 2015, 115, 142–149. [Google Scholar] [CrossRef]
- Fuyong, X.; Hai, S.; Janna, N.; Lin, Y. Automatic Ki-67 counting using robust cell detection and online dictionary learning. IEEE Trans. Biomed. Eng. 2014, 61, 859. [Google Scholar] [CrossRef]
- Ding, L.; Goshtasby, A. On the Canny edge detector. Pattern Recognit. 2001, 34, 721–725. [Google Scholar] [CrossRef]
- Na, C.; Xu, Z.; Xia, M. Correlation coefficients of hesitant fuzzy sets and their applications to clustering analysis. Appl. Math. Model. 2013, 37, 2197–2211. [Google Scholar]
- Xu, Z.; Jian, C.; Wu, J. Clustering algorithm for intuitionistic fuzzy sets. Inf. Sci. 2008, 178, 3775–3790. [Google Scholar] [CrossRef]
- Hassen, R.; Zhou, W.; Salama, M. No-reference image sharpness assessment based on local phase coherence measurement. In Proceedings of the IEEE International Conference on Acoustics Speech & Signal Processing, Dallas, TX, USA, 14–19 March 2010. [Google Scholar]
- Guo, J.; Hong, H.; Wang, L.; Peng, Q.; Li, X. Design of Focusing Window Based on Energy Function of Gradient. Infrared Technol. 2016, 38, 197–202. [Google Scholar]

| Type | Parameters | Scores |
|---|---|---|
| l:d | (0,1]:1 | 0 |
| l:d | (1,2]:1 | (0.3,0.6] |
| l:d | (2,4]:1 | (0.6,0.8] |
| l:d | (4,6]:1 | (0.8,1] |
| [0,10] | [0,0.2] | |
| (10,20] | (0.2,0.5] | |
| (20,30] | (0.5,0.6] | |
| (30,40] | (0.6,0.7] | |
| (40,50] | (0.7,0.8] | |
| (50,60] | (0.8,0.9] | |
| (60,70] | (0.9,1] | |
| [1,1.25]:1 | [0,0.2] | |
| (1.25,1.8]:1 | (0.2,0.4] | |
| (1.8,2]:1 | (0.4,0.6] | |
| (2,3]:1 | (0.6,0.9] | |
| (3,5]:1 | (0.9,1] |
| Parameter Types | X1 | X2 | X3 |
|---|---|---|---|
| A1 | {0.7,0.8,0.9} | {0.7,0.8} | {0.7,0.8,0.9} |
| A2 | {0.6,0.7} | {0.6,0.7} | {0.5,0.6} |
| A3 | {0.2,0.3} | {0.2,0.3} | {0.2,0.4} |
| A4 | {0.3,0.4} | {0.2,0.3} | {0.2,0.5} |
| A5 | {0.2,0.3} | {0.3,0.4} | {0.2} |
| A6 | {0.5} | {0.4,0.6} | {0.4} |
| A7 | {0.6,0.8} | {0.7,0.8} | {0.7,0.8,0.9} |
| A8 | {0.6,0.7} | {0.4,0.5} | {0.6} |
| A9 | {0.5,0.7} | {0.5,0.6} | {0.4,0.5} |
| A10 | {0.7,0.8} | {0.6,0.7} | {0.5,0.6} |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Tawose, O.T.; Jiang, L.; Zhao, D. Toward Automatic Cardiomyocyte Clustering and Counting through Hesitant Fuzzy Sets. Appl. Sci. 2019, 9, 2875. https://doi.org/10.3390/app9142875
Wang J, Tawose OT, Jiang L, Zhao D. Toward Automatic Cardiomyocyte Clustering and Counting through Hesitant Fuzzy Sets. Applied Sciences. 2019; 9(14):2875. https://doi.org/10.3390/app9142875
Chicago/Turabian StyleWang, Jiayao, Olamide Timothy Tawose, Linhua Jiang, and Dongfang Zhao. 2019. "Toward Automatic Cardiomyocyte Clustering and Counting through Hesitant Fuzzy Sets" Applied Sciences 9, no. 14: 2875. https://doi.org/10.3390/app9142875
APA StyleWang, J., Tawose, O. T., Jiang, L., & Zhao, D. (2019). Toward Automatic Cardiomyocyte Clustering and Counting through Hesitant Fuzzy Sets. Applied Sciences, 9(14), 2875. https://doi.org/10.3390/app9142875






