A Novel MOGA-SVM Multinomial Classification for Organ Inflammation Detection
Abstract
Featured Application
Abstract
1. Introduction
2. Dataset and Overview of MOGA-SVM
2.1. Background of Organ Inflammation
2.1.1. Appendicitis
2.1.2. Acute Appendicitis
2.1.3. Duodenitis
2.1.4. Pancreatitis
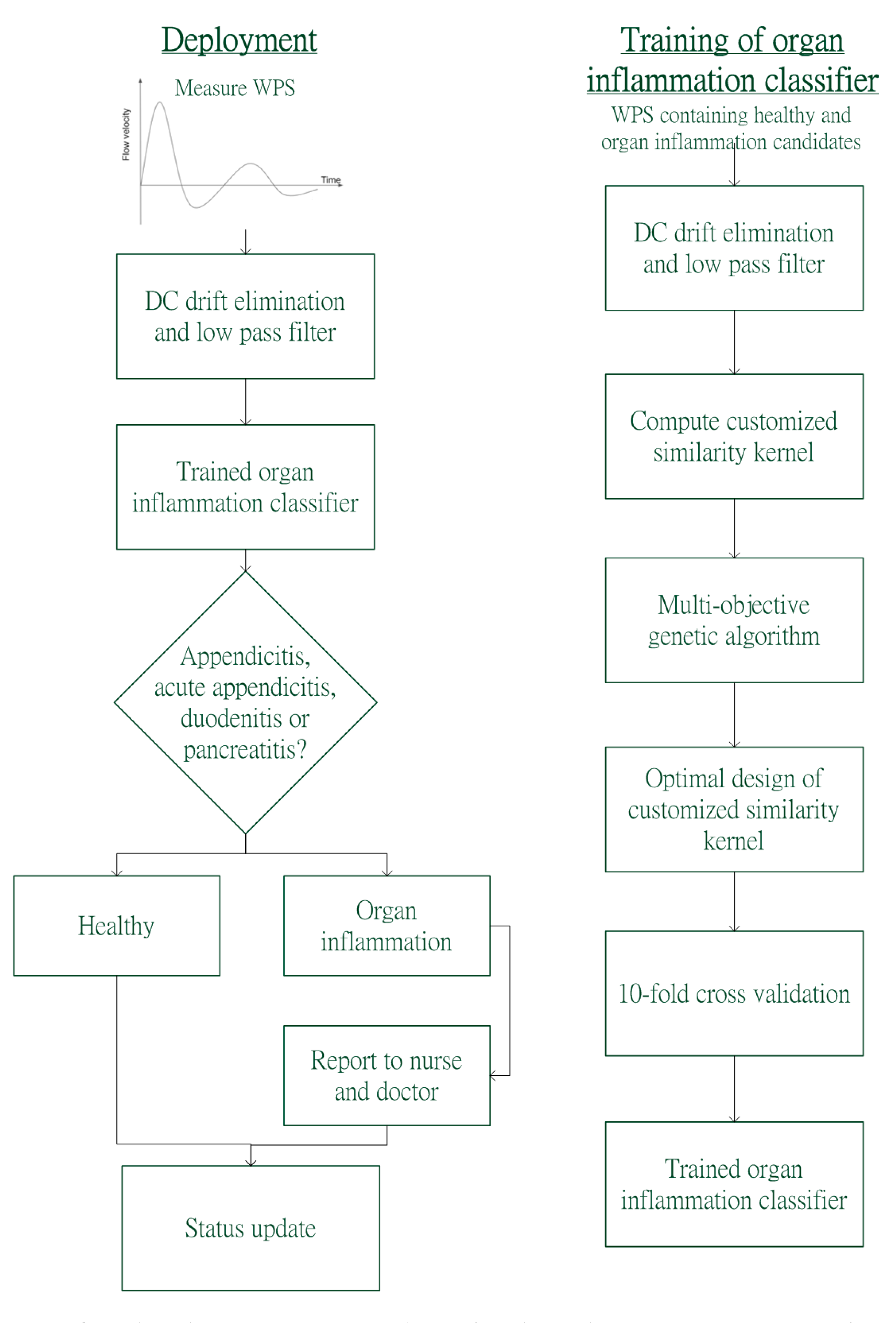
2.2. Overview of MOGA-SVM
3. Methodology
3.1. Datasets of Organ Inflammations Classifier
3.2. Data Preprocessing
3.3. Formulation of Optimal KCS and MOGA-SVM Classifier
| Algorithm 1 |
| Data: Organ inflammations of appendicitis, acute appendicitis, duodenitis and pancreatitis retrieved from 248 candidates [10], Xm Output: WPS samples Xi,j Step 1: dc drift elimination Step 2: Filter Xm using low pass filter Hlow Step 3: Locate local maxima and minima points of the Xm; Step 4: Locate two maxima points with interval of 120 sampling points; Xi,j (i = 1:4 = class label, j = length(Class))←Portion of signal between two maxima points with interval of 120 sampling points |
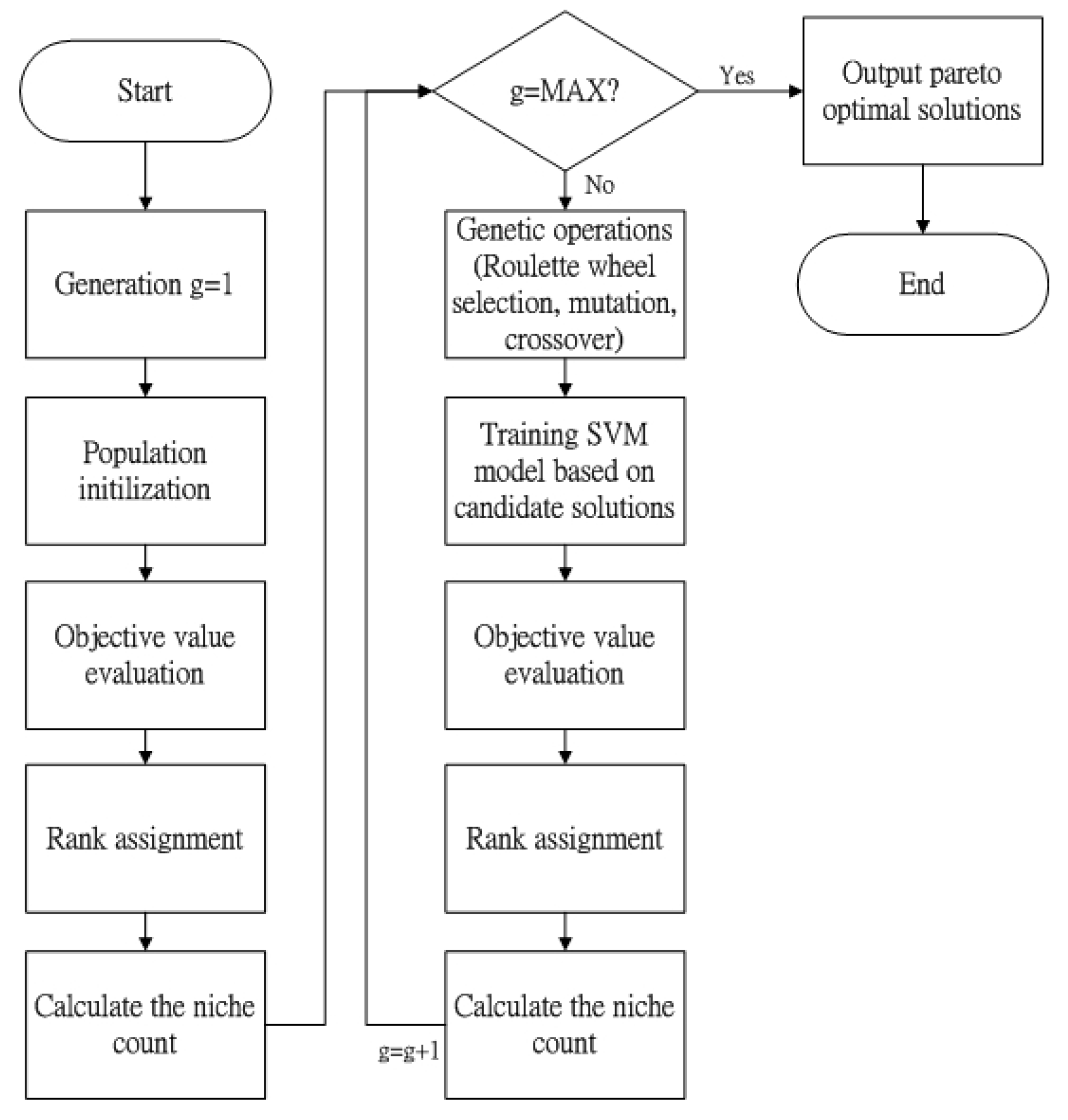
| Algorithm 2 |
| Data: Classlabel, Kc, Kcc Output: Model Step 1: generations = 1 Step 2: initialization (population) Step 3: Evaluate the individuals with the fitness function (F1 and F2) Step 4: rank the individuals by their fitness values by step 3 Step 5: do the Niche count calculation while generations <= max_generation do Step 6: Select two parents from the population Step 7: Create the offspring using Roulette wheel selection,crossover and mutation Step 8: Train SVM model for each individual Step 9: Evaluate the offspring with the fitness function (F1and F2) Step 10: rank the individuals by their fitness values by step 3 Step 11: do the Niche count calculation Step 12: Decide the new population based on the offspring Step 13: generations = generations + 1 End while Model←Pareto solutions |
4. Performance Evaluation and Comparison
4.1. Performance of Proposed MOGA-SVM Using KCS
4.2. Evaluation of Other Kernels Using Feature Extraction Approach
4.3. Comparison between Proposed and Related Work
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Health Workforce Requirements for Universal Health Coverage and the Sustainable Development Goals; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Chui, K.T.; Alhalabi, W.; Pang, S.S.H.; Pablos, P.O.D.; Liu, R.W.; Zhao, M. Disease Diagnosis in Smart Healthcare: Innovation, Technologies and Applications. Sustainability 2017, 9, 2309. [Google Scholar] [CrossRef]
- Spruit, M.; Lytras, M. Applied Data Science in Patient-centric Healthcare. Telemat. Inform. 2018, 35, 643–653. [Google Scholar] [CrossRef]
- Khaire, N.N.; Joshi, Y.V. Disgnosis of disease using wrist pulse signal for classification of pre-meal and post-meal samples. In Proceedings of the 2015 International Conference on Industrial Instrumentation and Control, Maharashtra, India, 28–30 May 2015; IEEE: Piscataway, NJ, USA. [Google Scholar]
- Reddy, R.K.; Pooni, R.; Zaharieva, D.P.; Senf, B.; El Youssef, J.; Dassau, E.; Castle, J.R. Accuracy of Wrist-Worn Activity Monitors during Common Daily Physical Activities and Types of Structured Exercise: Evaluation Study. JMIR mHealth uHealth 2018, 6, e10338. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, B.; Lu, G.; You, J.; Zhang, D. Body surface feature-based multi-modal Learning for Diabetes Mellitus detection. Inf. Sci. 2019, 472, 1–14. [Google Scholar] [CrossRef]
- He, D.; Wang, L.; Fan, X.; Yao, Y.; Geng, N.; Sun, Y.; Xu, L.; Qian, W. A new mathematical model of wrist pulse waveforms characterizes patients with cardiovascular disease—A pilot study. Med. Eng. Phys. 2017, 48, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.J.; Qi, Z.; Tu, L.P.; Zhang, Y.H.; Zhu, L.P.; Xu, J.T.; Zhang, Z.F. The Association of Radial Artery Pulse Wave Variables with the Pulse Wave Velocity and Echocardiographic Parameters in Hypertension. Evid. Based Complement. Altern. Med. 2018, 2018, 5291759. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Yao, L.; Song, H.; Kos, A. A sensor-based wrist pulse signal processing and lung cancer recognition. J. Biomed. Inform. 2018, 79, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, L.; Zhang, D.; Zhang, D. Computerized wrist pulse signal diagnosis using modified auto-regressive models. J. Med. Syst. 2011, 35, 321–328. [Google Scholar] [CrossRef]
- Chow, W.H.; Wu, C.K.; Tsang, K.F.; Li, B.Y.S.; Chui, K.T. Wrist pulse signal classification for inflammation of appendix, pancreas, and duodenum. In Proceedings of the 40th Annual Conference of the IEEE Industrial Electronics Society, Dallas, TX, USA, 30 October–1 November 2014; IEEE: Piscataway, NJ, USA. [Google Scholar]
- Garg, N.; Bisht, A.; Ryait, H.S.; Kumar, A. Identification of motion outliers in wrist pulse signal. Comput. Electr. Eng. 2018, 67, 776–790. [Google Scholar] [CrossRef]
- Liu, X.; Ji, Z.; Tang, Y. Recognition of pulse wave feature points and non-invasive blood pressure measurement. J. Signal Process. Syst. 2017, 87, 241–248. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, D.; Lu, G. Generalized Feature Extraction for Wrist Pulse Analysis: From 1-D Time Series to 2-D Matrix. IEEE J. Biomed. Health Inform. 2017, 21, 978–985. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Health Estimates 2015: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2015; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Chui, K.T.; Tsang, K.F.; Chi, H.R.; Ling, B.W.K.; Wu, C.K. An accurate ECG based transportation safety drowsiness detection scheme. IEEE Trans. Ind. Inform. 2016, 12, 1438–1452. [Google Scholar] [CrossRef]
- Montazeri, A.; West, C.; Monk, S.D.; Taylor, C.J. Dynamic modelling and parameter estimation of a hydraulic robot manipulator using a multi-objective genetic algorithm. Int. J. Control 2017, 90, 661–683. [Google Scholar] [CrossRef]
- Tseng, F.H.; Wang, X.; Chou, L.D.; Chao, H.C.; Leung, V.C. Dynamic Resource Prediction and Allocation for Cloud Data Center Using the Multiobjective Genetic Algorithm. IEEE Syst. J. 2018, 12, 1688–1699. [Google Scholar] [CrossRef]
- Chui, K.T.; Tsang, K.F.; Wu, C.K.; Hung, F.H.; Chi, H.R.; Chung, H.S.H.; Man, K.F.; Ko, K.T. Cardiovascular diseases identification using electrocardiogram health identifier based on multiple criteria decision making. Expert Syst. Appl. 2015, 42, 5684–5695. [Google Scholar] [CrossRef]
- Fuchida, M.; Pathmakumar, T.; Mohan, R.E.; Tan, N.; Nakamura, A. Vision-based perception and classification of mosquitoes using support vector machine. Appl. Sci. 2017, 7, 51. [Google Scholar] [CrossRef]
- Wu, J.L.; Chang, P.C.; Tsao, C.C.; Fan, C.Y. A patent quality analysis and classification system using self-organizing maps with support vector machine. Appl. Soft Comput. 2016, 41, 305–316. [Google Scholar] [CrossRef]
- Ryan, W.L. Digestive Diseases—Research and Clinical Developments: Appendicitis: Symptoms, Diagnosis, and Treatments; Nova Science: New York, NY, USA, 2011. [Google Scholar]
- Keyzer, C.; Gevenois, P.A. Imaging of Acute Appendicitis in Adults and Children; Springer: Berlin, Germany, 2011. [Google Scholar]
- Serra, S.; Jani, P.A. An approach to duodenal biopsies. J. Clin. Pathol. 2006, 59, 1133–1150. [Google Scholar] [CrossRef]
- Adams, D.B.; Cotton, P.B.; Zyromski, N.J.; Windsor, J. Pancreatitis: Medical and Surgical Management; Wiley Blackwell: Chichester, UK, 2017. [Google Scholar]
- Yadav, D.; Timmons, L.; Benson, J.T.; Dierkhising, R.A.; Chari, S.T. Incidence, prevalence, and survival of chronic pancreatitis: A population-based study. Am. J. Gastroenterol. 2011, 106, 2192. [Google Scholar] [CrossRef]
- De Haan, R.R.; Visser, J.B.; Pons, E.; Feelders, R.A.; Kaymak, U.; Hunink, M.M.; Visser, J.J. Patient-specific workup of adrenal incidentalomas. Eur. J. Radiol. Open 2017, 4, 108–114. [Google Scholar] [CrossRef]
- Roberts, D.R.; Bahn, V.; Ciuti, S.; Boyce, M.S.; Elith, J.; Guillera-Arroita, G.; Hauenstein, S.; Lahoz-Monfort, J.J.; Schroder, B.S.; Thuiller, W.; et al. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]
- Deb, K. Multi-Objective Optimization Using Evolutionary Algorithms; John Wiley & Sons, Inc.: New York, NY, USA, 2001. [Google Scholar]
- Herbrich, R. Learning Kernel Classifiers Theory and Algorithms; The MIT Press: London, UK, 2002. [Google Scholar]
- Smits, G.F.; Jordan, E.M. Improved SVM regression using mixtures of kernels. In Proceedings of the 2002 International Joint Conference on Neural Networks, Honolulu, HI, USA, 12–17 May 2002; IEEE: Piscataway, NJ, USA. [Google Scholar]
- Rangaprakash, D.; Dutt, D.N. Study of wrist pulse signals using time domain spatial features. Comput. Electr. Eng. 2015, 45, 100–107. [Google Scholar] [CrossRef]
| GHE Code | GHE Cause | Number of Deaths (Annual) | |||
|---|---|---|---|---|---|
| 2000 | 2005 | 2010 | 2015 | ||
| 1240 | Appendicitis | 34,800 | 39,400 | 43,300 | 45,000 |
| 1241 | Duodenitis | 37,900 | 40,400 | 43,800 | 47,000 |
| 1248 | Pancreatitis | 64,400 | 77,800 | 93,900 | 103,500 |
| Class | Name | Age | Total | |||
|---|---|---|---|---|---|---|
| [0,20) | [20,40) | [40,60) | [60,100) | |||
| 0 | Healthy | 8 | 26 | 30 | 16 | 100 |
| 1 | Appendicitis | 0 | 22 | 0 | 0 | 22 |
| 2 | Acute Appendicitis | 20 | 8 | 10 | 0 | 38 |
| 3 | Duodenitis | 4 | 26 | 6 | 6 | 42 |
| 4 | Pancreatitis | 16 | 26 | 4 | 0 | 46 |
| Kernel | Performance | ||
|---|---|---|---|
| Scenario (i) (Se,Sp,Acc)% | Scenario (ii) (Se,Sp,Acc)% | Scenario (iii) (Se,Sp,Acc)% | |
| k1(xi,xj) | (57.6, 58.2, 57.9) | (57.7, 57.1, 57.4) | (58.8, 60.4, 59.6) |
| k2(xi,xj) | (76.7, 77.5, 77.1) | (76.8, 76.6, 76.7) | (77.3, 78.3, 77.8) |
| k3(xi,xj) | (77.6, 78.2, 77.9) | (78.3,78.9, 78.6) | (78.7, 80.1, 79.4) |
| k4(xi,xj) | (73.8, 74.6, 74.2) | (73.2, 73.0, 73.1) | (74.8, 75.8, 75.3) |
| k5(xi,xj) | (79.9, 80.3, 80.1) | (82.0, 81.0, 81.5) | (83.8, 84.4, 84.1) |
| Work | Method | Feature Extraction | Dataset (Samples) | Cross Validation | Class Labels | Se (%) | Sp (%) | Acc (%) |
|---|---|---|---|---|---|---|---|---|
| [10] | Binary Classification using modified auto-regressive model and linear kernel SVM | Mean and standard deviation of prediction error | Healthy (100), appendicitis (22), acute appendicitis (38), duodenitis (42) and pancreatitis (46) | No | Class 0: healthy; Class 1: appendicitis | 81.8 | 93.3 | 91.2 |
| Class 0: healthy; Class 1: acute appendicitis | 76.5 | 82.4 | 80.8 | |||||
| Class 0: healthy; Class 1: duodenitis | 80.0 | 91.4 | 88.0 | |||||
| Class 0: healthy; Class 1: pancreatitics | 83.3 | 94.4 | 90.9 | |||||
| Class 0: healthy; Class 1: All inflammations | 80.4 | 89.7 | 87.3 | |||||
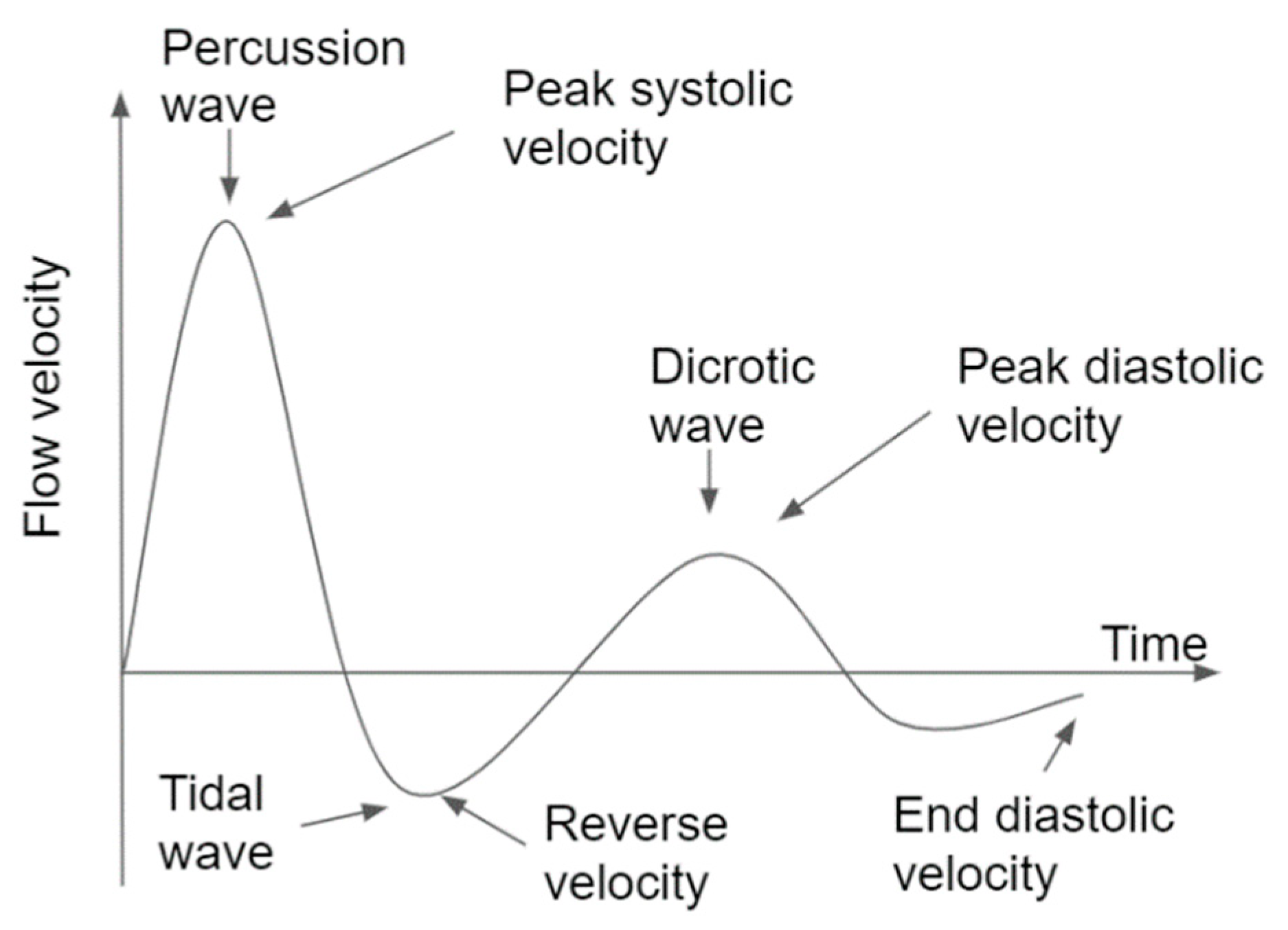
| [11] | Binary Classification using RBF SVM | peak systolic velocity; reverse velocity; peak diastolic velocity; end diastolic velocity; duration of systole; and duration of diastole | Healthy (100), appendicitis (100), acute appendicitis (100), duodenitis (100) and pancreatitis (100) | 10-fold | Class 0: healthy; Class 1: appendicitis | N/A | N/A | 92.8 |
| Class 0: healthy; Class 1: acute appendicitis | N/A | N/A | 88.1 | |||||
| Class 0: healthy; Class 1: duodenitis | N/A | N/A | 88.6 | |||||
| Class 0: healthy; Class 1: pancreatitics | N/A | N/A | 98.4 | |||||
| Our work | Multinomial Classification using customized kernel | Cross-correlation and convolution coefficients | Healthy (1800), Appendicitis (630), Acute Appendicitis (970), Duodenitis (1380) and Pancreatitis (820) | 10-fold | Class 0: health; Class 1: appendicitis; Class 2: acute appendicitis; Class 3: duodenitis; Class 4: pancreatitis | 92.0 | 91.2 | 91.6 |
| Work | Method | Feature Extraction | Dataset (Samples) | Cross Validation | Class Labels | Se (%) | Sp(%) | Acc (%) |
|---|---|---|---|---|---|---|---|---|
| [10] | Binary Classification using modified auto-regressive model and linear kernel SVM | Mean and standard deviation of prediction error | Healthy (1800), appendicitis (630), acute appendicitis (970), duodenitis (1380) and pancreatitis (820) | 10-fold | Class 0: healthy; Class 1: appendicitis; Class 2: acute appendicitis; Class 3: duodenitis; Class 4: pancreatitis | 81.3 | 80.3 | 80.8 |
| [11] | Binary Classification using RBF SVM | peak systolic velocity; reverse velocity; peak diastolic velocity; end diastolic velocity; duration of systole; and duration of diastole | Healthy (1800), appendicitis (630), acute appendicitis (970), duodenitis (1380) and pancreatitis (820) | 10-fold | Class 0: healthy; Class 1: appendicitis; Class 2: acute appendicitis; Class 3: duodenitis; Class 4: pancreatitis | 81.7 | 82.9 | 82.3 |
| [32] | A recursive cluster elimination based SVM | spatial features obtained from a bi-modal Gaussian model | Healthy (1800), appendicitis (630), acute appendicitis (970), duodenitis (1380) and pancreatitis (820) | 10-fold | Class 0: healthy; Class 1: appendicitis; Class 2: acute appendicitis; Class 3: duodenitis; Class 4: pancreatitis | 84.7 | 84.1 | 84.4 |
| [14] | RBF SVM | Periodic and non-periodic feature extension | Healthy (1800), appendicitis (630), acute appendicitis (970), duodenitis (1380) and pancreatitis (820) | 10-fold | Class 0: healthy; Class 1: appendicitis; Class 2: acute appendicitis; Class 3: duodenitis; Class 4: pancreatitis | 85.3 | 86.1 | 85.7 |
| Our work | Multinomial Classification using customized kernel | Cross-correlation and convolution coefficients | Healthy (1800), appendicitis (630), acute appendicitis (970), duodenitis (1380) and pancreatitis (820) | 10-fold | Class 0: healthy; Class 1: appendicitis; Class 2: acute appendicitis; Class 3: duodenitis; Class 4: pancreatitis | 92.0 | 91.2 | 91.6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chui, K.T.; Lytras, M.D. A Novel MOGA-SVM Multinomial Classification for Organ Inflammation Detection. Appl. Sci. 2019, 9, 2284. https://doi.org/10.3390/app9112284
Chui KT, Lytras MD. A Novel MOGA-SVM Multinomial Classification for Organ Inflammation Detection. Applied Sciences. 2019; 9(11):2284. https://doi.org/10.3390/app9112284
Chicago/Turabian StyleChui, Kwok Tai, and Miltiadis D. Lytras. 2019. "A Novel MOGA-SVM Multinomial Classification for Organ Inflammation Detection" Applied Sciences 9, no. 11: 2284. https://doi.org/10.3390/app9112284
APA StyleChui, K. T., & Lytras, M. D. (2019). A Novel MOGA-SVM Multinomial Classification for Organ Inflammation Detection. Applied Sciences, 9(11), 2284. https://doi.org/10.3390/app9112284






