Chronic Disease Prediction Using Character-Recurrent Neural Network in The Presence of Missing Information
Abstract
1. Introduction
2. Related Work
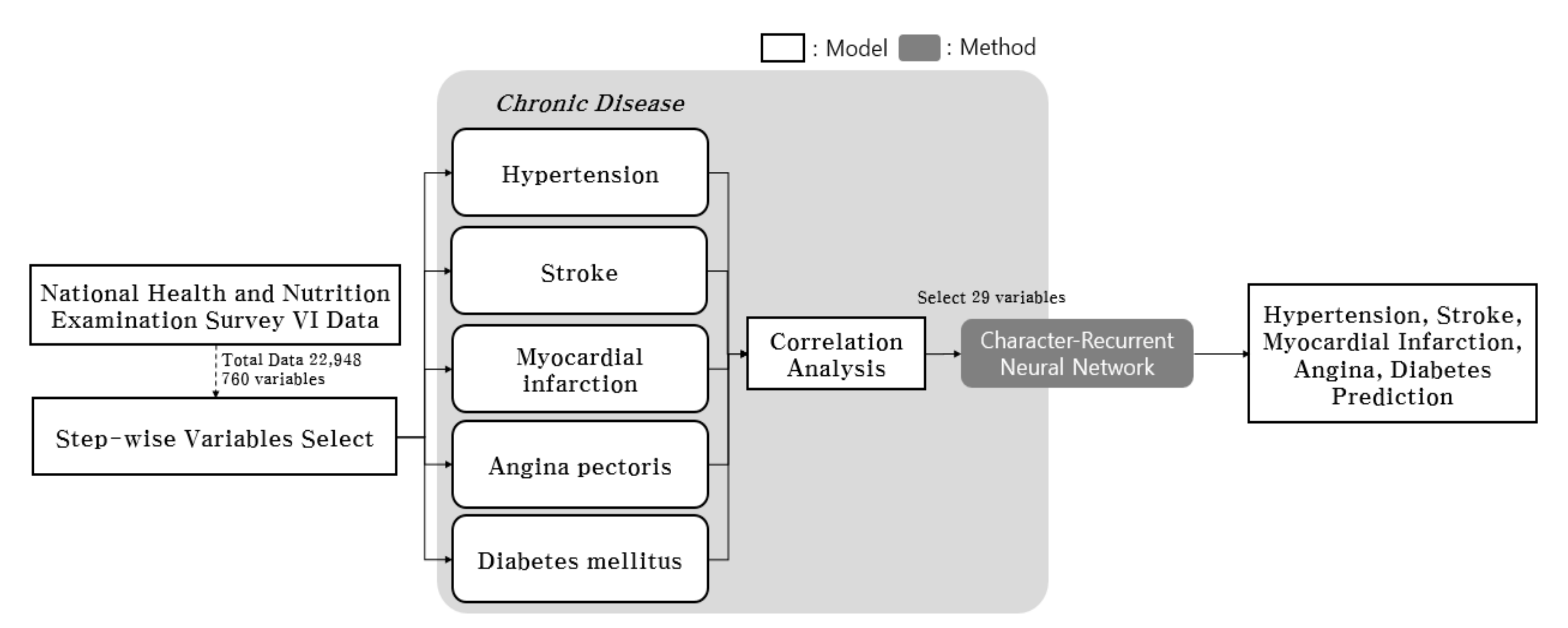
3. Materials and Methods
4. Data Description and Learning Procedure
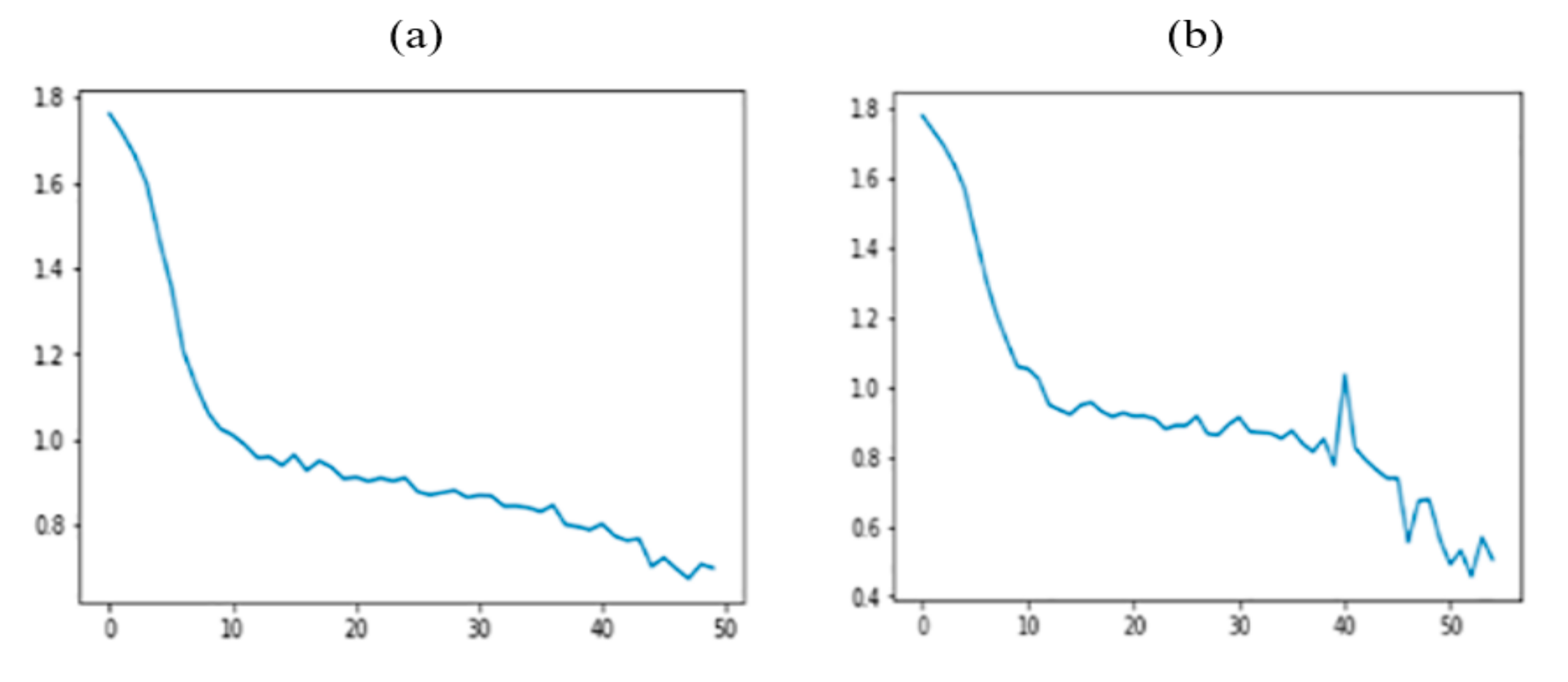
5. Experimental Results
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Chronic Disease | Variables | Pr(>|t|) |
|---|---|---|
| Hypertension | DI1_dg | <2e−16*** |
| DI1_pt | <2e−16*** | |
| DI1_2 | 0.003949** | |
| DE1_dg | 0.023553* | |
| DE1_pt | 0.024046* | |
| DE1_32 | 0.011176* | |
| DF2_ag | 0.024011* | |
| LQ4_06 | 0.003265** | |
| LQ4_07 | 0.048531* | |
| LQ4_08 | 0.002055** | |
| LQ4_14 | 6.13e−06*** | |
| EC1_2 | 0.022494* | |
| BS6_2_1 | 0.042053* | |
| BS6_2_2 | 0.035517* | |
| HE_HPdg | 0.000380*** | |
| HE_DMdr | 6.90e−05*** | |
| HE_STRfh1 | 0.000204*** | |
| Myocardial infarction | DI1_dg | 0.018140* |
| DI1_pt | 0.022980* | |
| DI1_2 | 0.048649* | |
| DI3_dg | 0.015375* | |
| DI3_ag | 0.003401** | |
| DI3_2 | 6.39e−07*** | |
| DI4_dg | <2e−16*** | |
| DI4_pr | <2e−16*** | |
| DI4_pt | 0.005583** | |
| DI5_dg | 9.58e−10*** | |
| DI5_ag | <2e−16*** | |
| DI5_pt | <2e−16*** | |
| DI6_dg | <2e−16*** | |
| DI6_ag | 6.88e−08*** | |
| DI6_pt | <2e−16*** | |
| DE1_ag | 0.036523* | |
| DE1_33 | 0.000187*** | |
| LQ4_04 | 0.000334*** | |
| LQ1_mn | 0.012614* | |
| educ | 0.000503*** | |
| BO3_07 | 0.002166** | |
| BP6_31 | 0.027196* | |
| HE_HPdg | 0.000907*** | |
| DE1_33 | 0.000187*** | |
| LQ4_04 | 0.000334*** | |
| LQ1_mn | 0.012614* | |
| educ | 0.000503*** | |
| BO3_07 | 0.002166** | |
| BP6_31 | 0.027196* | |
| HE_HPdg | 0.000907*** | |
| Diabetes mellitus | DI1_dg | <2e−16*** |
| DI1_pt | <2e−16*** | |
| DI1_2 | 1.54e−14*** | |
| DI5_dg | 0.008476** | |
| DI5_ag | 0.003172** | |
| DI6_dg | 0.018528* | |
| DI6_ag | 0.003206** | |
| DE1_dg | 0.000918*** | |
| DE1_pt | 0.002990** | |
| DE1_4 | 0.014432* | |
| DE2_dg | 0.030405* | |
| DF2_pr | 0.014291* | |
| DK4_pr | 0.010164* | |
| LQ4_15 | 0.001020** | |
| LQ4_22 | 0.000761*** | |
| EC_occp | 0.022475* | |
| EC_lgw_2 | 0.000365*** | |
| EC_lgw_4 | 0.000505*** | |
| EC_lgw_5 | 0.003918** | |
| BO3_04 | 0.005202** | |
| BD7_4 | 0.023864* | |
| BP5 | 0.045927* | |
| BS6_2_1 | 0.044478* | |
| BS6_3 | 0.007516** | |
| BS6_4 | 0.015076* | |
| HE_HPdg | 0.000262*** | |
| HE_DMdg | 9.46e−14*** | |
| HE_HLdg | 1.07e−05*** | |
| HE_fh | 0.000380 *** | |
| Stroke | DI3_dg | <2e−16*** |
| DI3_dg | <2e−16*** | |
| DI3_ag | 4.82e−15*** | |
| DI3_pt | <2e−16*** | |
| DI3_2 | 3.31e−09*** | |
| DI4_dg | 3.94e−07*** | |
| DI4_pt | 0.000148*** | |
| DI5_ag | 0.000691*** | |
| DI5_pt | 2.13e−09*** | |
| DI6_pt | 1.91e−05*** | |
| LQ4_04 | 0.001673** | |
| LQ4_06 | 7.47e−14*** | |
| BS3_1 | 0.031119* | |
| BS3_2 | 0.030441* | |
| BS3_3 | 0.028916* | |
| BS6_3 | 0.002509** | |
| HE_fst | 0.041026* | |
| HE_HPdg | 0.001564** | |
| HE_HLfh3 | 0.044872* | |
| HE_IHDfh3 | 0.003670** | |
| HE_STRfh1 | 1.82e−05*** | |
| Angina pectoris | DI1_dg | 0.001704** |
| DI1_pt | 0.001441** | |
| DI1_2 | 0.000919*** | |
| DI3_dg | 0.002154** | |
| DI3_ag | 0.000437*** | |
| DI3_2 | 1.05e−08*** | |
| DI4_dg | <2e−16*** | |
| DI4_pr | <2e−16*** | |
| DI4_pt | <2e-16*** | |
| DI5_dg | 6.35e−11*** | |
| DI5_ag | 5.68e−11*** | |
| DI6_dg | <2e−16*** | |
| DI6_pt | <2e−16*** | |
| DE1_33 | 0.000111*** | |
| LQ4_04 | 0.003495** | |
| LQ4_06 | 0.008981** | |
| LQ1_mn | 0.038536* | |
| educ | 0.008716** | |
| BO3_07 | 0.047492* | |
| BD2_32 | 0.028725* | |
| BS6_3 | 0.049513* | |
| HE_STRfh1 | 0.036219* |
| Chronic Disease | Variables | Variable Description | |
|---|---|---|---|
| Hypertension | DI1_dg | Whether diagnosed with hypertension by a physician | |
| DI1_pt | Hypertension treatment | ||
| DI1_2 | Taking blood pressure regulator | ||
| DE1_dg | Whether diagnosed with diabetes mellitus by a physician | ||
| DE1_pt | Diabetes mellitus treatment | ||
| DE1_32 | Diabetes mellitus treatment_antidiabetics | ||
| DF2_ag | Time of depression diagnosis | ||
| LQ4_06 | (Adult) Reason for limited activity: stroke | ||
| LQ4_07 | (Adult) Reason for limited activity: diabetes mellitus | ||
| LQ4_08 | (Adult) Reason for limited activity: hypertension | ||
| LQ4_14 | (Adult) Reason for limited activity: dementia | ||
| EC1_2 | Reason for unemployment | ||
| BS6_2_1 | (Adult) Smoking duration of past smokers (years) | ||
| BS6_2_2 | (Adult) Smoking duration of past smokers (months) | ||
| Stroke | DI3_dg | Whether diagnosed with stroke by a physician | |
| DI3_ag | Time of stroke diagnosis | ||
| DI3_pt | Stroke treatment | ||
| DI3_2 | Sequelae of stroke | ||
| DI4_dg | Whether diagnosed with myocardial infarction, angina pectoris by a physician | ||
| DI4_pt | Myocardial infarction, angina pectoris treatment | ||
| DI5_ag | Time of myocardial infarction diagnosis | ||
| DI5_pt | Myocardial infarction treatment | ||
| DI6_pt | Angina pectoris treatment | ||
| LQ4_04 | Reason for limited activity: heart disease | ||
| LQ4_06 | (Adult) Reason for limited activity: stroke | ||
| BS3_1 | (Adult) Currently smoking or not | ||
| BS3_2 | (Adult) Average daily smoking amount of current smokers | ||
| BS3_3 | (Adult) Number of days smoking per month of occasional smokers | ||
| BS6_3 | (Adult) Average daily smoking amount of past smokers | ||
| HE_fst | Fasting duration | ||
| HE_HPdg | Whether diagnosed with hypertension by a physician | ||
| HE_HLfh3 | Whether diagnosed with hypercholesterolemia by a physician (siblings) | ||
| HE_IHDfh3 | Whether diagnosed with ischemic heart disease by a physician (siblings) | ||
| HE_STRfh1 | Whether diagnosed with stroke by a physician (father) | ||
| Myocardial infarction | DI1_dg | Whether diagnosed with hypertension by a physician | |
| DI1_pt | Hypertension treatment | ||
| DI1_2 | Taking blood pressure regulator | ||
| DI3_dg | Whether diagnosed with stroke by a physician | ||
| DI3_ag | Time of stroke diagnosis | ||
| DI3_2 | Sequelae of stroke | ||
| DI4_dg | Whether diagnosed with myocardial infarction, angina pectoris by a physician | ||
| DI4_pr | Current morbidity of myocardial infarction, angina pectoris | ||
| DI4_pt | Myocardial infarction, angina pectoris treatment | ||
| DI5_dg | Whether diagnosed with myocardial infarction by a physician | ||
| DI5_ag | Time of myocardial infarction diagnosis | ||
| DI5_pt | Myocardial infarction treatment | ||
| DI6_dg | Whether diagnosed with angina pectoris by a physician | ||
| DI6_ag | Time of angina pectoris diagnosis | ||
| DI6_pt | Angina pectoris treatment | ||
| DE1_ag | Time of diabetes mellitus diagnosis | ||
| DE1_33 | Diabetes mellitus treatment: non-pharmaceutical therapy | ||
| LQ4_04 | Reason for limited activity: heart disease | ||
| LQ1_mn | Number of days bedridden in the last month | ||
| educ | Education level | ||
| BO3_07 | Weight control method: health functional food | ||
| BP6_31 | Whether attempted suicide in the past year | ||
| HE_HPdg | Whether diagnosed with hypertension by a physician | ||
| Angina pectoris | DI1_dg | Whether diagnosed with hypertension by a physician | |
| DI1_pt | Hypertension treatment | ||
| DI1_2 | Taking blood pressure regulator | ||
| DI3_dg | Whether diagnosed with stroke by a physician | ||
| DI3_ag | Time of stroke diagnosis | ||
| DI3_2 | Sequelae of stroke | ||
| DI4_dg | Whether diagnosed with myocardial infarction, angina pectoris by a physician | ||
| DI4_pr | Current morbidity of myocardial infarction, angina pectoris | ||
| DI4_pt | Myocardial infarction, angina pectoris treatment | ||
| DI5_dg | Whether diagnosed with myocardial infarction by a physician | ||
| DI5_ag | Time of myocardial infarction diagnosis | ||
| DI6_dg | Whether diagnosed with angina pectoris by a physician | ||
| DI6_pt | Myocardial infarction treatment | ||
| DE1_33 | Diabetes mellitus treatment: non-pharmaceutical therapy | ||
| LQ4_04 | Reason for limited activity: heart disease | ||
| LQ4_06 | (Adult) Reason for limited activity: stroke | ||
| LQ1_mn | Number of days bedridden in the last month | ||
| educ | Education level | ||
| BO3_07 | Weight control method: health functional food | ||
| BD2_32 | (Adult) Frequency of heavy drinking | ||
| BS6_3 | (Adult) Average daily smoking amount of past smokers | ||
| HE_STRfh1 | Whether diagnosed with stroke by a physician (father) | ||
| Diabetes mellitus | DI1_dg | Whether diagnosed with hypertension by a physician | |
| DI1_pt | Hypertension treatment | ||
| DI1_2 | Taking blood pressure regulator | ||
| DI5_dg | Whether diagnosed with myocardial infarction by a physician | ||
| DI5_ag | Time of myocardial infarction diagnosis | ||
| DI6_dg | Whether diagnosed with angina pectoris by a physician | ||
| DI6_ag | Time of angina pectoris diagnosis | ||
| DE1_dg | Whether diagnosed with diabetes mellitus by a physician | ||
| DE1_pt | Diabetes mellitus treatment | ||
| DE1_4 | Ophthalmoscopy | ||
| DE2_dg | Whether diagnosed with thyroid disease by a physician | ||
| DF2_pr | Current morbidity of depression | ||
| DK4_pr | Current morbidity of cirrhosis | ||
| LQ4_15 | Reason for limited activity: depression/anxiety/emotional problem | ||
| LQ4_22 | (Adult) Reason for limited activity: old age | ||
| EC_occp | (If employed) Occupation type | ||
| EC_lgw_2 | (Adult) Longest occupation: occupational code + unemployment/non-economic activity status | ||
| EC_lgw_4 | (Adult) Longest occupation: worker title | ||
| EC_lgw_5 | (Adult) Longest occupation: worker title wage workers in detail | ||
| BO3_04 | (Adult) Weight control method: prescription weight loss pills | ||
| BD7_4 | (Adult) Whether family/physician recommended to quit drinking | ||
| BP5 | Whether feeling depressed for two 2 weeks or more at a time | ||
| BS6_2_1 | (Adult) Smoking duration of past smokers (years) | ||
| BS6_3 | (Adult) Average daily smoking amount of past smokers | ||
| BS6_4 | (Adult) Smoking cessation period of past smokers converted to months | ||
| HE_HPdg | Whether diagnosed with hypertension by a physician | ||
| HE_DMdg | Whether diagnosed with diabetes mellitus by a physician | ||
| HE_HLdg | Whether diagnosed with hypercholesterolemia by a physician | ||
| HE_fh | Family history of diagnosis of chronic disease by a physician | ||
References
- Beratarrechea, A.; Lee, A.G.; Willner, J.M.; Jahangir, E.; Ciapponi, A.; Rubinstein, A. The impact of mobile health interventions on chronic disease outcomes in developing countries: A systematic review. Telemed. J. E Health 2014, 20, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Sumner, M.D.; Elliott-Eller, M.; Weidner, G.; Daubenmier, J.J.; Chew, M.H.; Marlin, R.; Raisin, C.J.; Ornish, D. Effects of pomegranate juice consumption on myocardial perfusion in patients with coronary heart disease. Am. J. Cardiol. 2005, 96, 810–814. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Takehara, I.; Masuzawa, T.; Saito, T.; Naoki, Y. Nutrigenomic studies of effects of Chlorella on subjects with high-risk factors for lifestyle-related disease. J. Med. Food 2008, 11, 395–404. [Google Scholar] [CrossRef]
- Liu, S.H.; Erion, G.; Novitsky, V.; De Gruttola, V. Viral genetic linkage analysis in the presence of missing data. PLoS ONE 2015, 10, e0135469. [Google Scholar] [CrossRef] [PubMed]
- García-Laencina, P.J.; Sancho-Gómez, J.L.; Figueiras-Vidal, A.R. Pattern classification with missing data: A review. Neural Comput. Appl. 2010, 19, 263–282. [Google Scholar] [CrossRef]
- Jerez, J.M.; Molina, I.; García-Laencina, P.J.; Alba, E.; Ribelles, N.; Martín, M.; Franco, L. Missing data imputation using statistical and machine learning methods in a real breast cancer problem. Artif. Intell. Med. 2010, 50, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Polom, K.; Adamczyk, B.; Afshar, M.; D’Ignazio, A. Machine learning methodology applied to characterize subgroups of gastric cancer patients using an integrated large biomarker dataset. Eur. J. Surg. Oncol. 2019, 45, e79. [Google Scholar] [CrossRef]
- Schuster, T.L.; Dobson, M.; Jauregui, M.; Blanks, R.H. Wellness lifestyles I: A theoretical framework linking wellness, health lifestyles, and complementary and alternative medicine. J. Altern. Complement. Med. 2004, 10, 349–356. [Google Scholar] [CrossRef]
- Razzaghi, T.; Roderick, O.; Safro, I.; Marko, N. Multilevel weighted support vector machine for classification on healthcare data with missing values. PLoS ONE 2016, 11, e0155119. [Google Scholar] [CrossRef]
- Tian, T.; McLachlan, G.J.; Dieters, M.J.; Basford, K.E. Application of multiple imputation for missing values in three-way three-mode multi-environment trial data. PLoS ONE 2015, 10, e0144370. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Xu, Q.; Wu, C.; Gao, Y.; Hua, T.; Xu, C. Performance Evaluation of missing-value imputation clustering based on a multivariate Gaussian mixture model. PLoS ONE 2016, 11, e0161112. [Google Scholar] [CrossRef]
- Fine, L.J.; Philogene, G.S.; Gramling, R.; Coups, E.J.; Sinha, S. Prevalence of multiple chronic disease risk factors: 2001 National Health Interview Survey. Am. J. Prev. Med. 2004, 27, 18–24. [Google Scholar] [CrossRef]
- Gupta, S.; Kumar, D.; Sharma, A. Performance analysis of various data mining classification techniques on healthcare data. Perform. J. Comput. Sci. Inf. Technol. 2011, 3, 155–169. [Google Scholar]
- Casaburi, R.; Mahler, D.A.; Jones, P.W.; Wanner, A.; San Pedro, G.; ZuWallack, R.L.; Menjoge, S.S.; Serby, C.W.; Witek, T. A long-term evaluation of once-daily inhaled tiotropium in chronic obstructive pulmonary disease. Eur. Respir. J. 2002, 19, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yu, M.; Graubard, B.I.; Troiano, R.P.; Schenker, N. Multiple imputation of completely missing repeated measures data within person from a complex sample: Application to accelerometer data in the National Health and Nutrition Examination Survey. Stat. Med. 2016, 35, 5170–5188. [Google Scholar] [CrossRef]
- Beaulieu-Jones, B.K.; Moore, J.H. Missing data imputation in the electronic health record using deeply learned autoencoders. In Proceedings of the Pacific Symposium Pacific Symposium on Biocomputing 2017, Kohala Coast, HI, USA, 4–8 January 2017; pp. 207–218. [Google Scholar]
- Youm, S.; Park, S. How the awareness of u-Healthcare service and health conditions affect healthy lifestyle: An empirical analysis based on a u-Healthcare service experience. Telemed. J. e-Health 2015, 21, 286–295. [Google Scholar] [CrossRef]
- Azimi, I.; Pahikkala, T.; Rahmani, A.M.; Niela-Vilén, H.; Axelin, A.; Liljeberg, P. Missing data resilient decision-making for healthcare IoT through personalization: A case study on maternal health. Future Gener. Comput. Syst. 2019, 96, 297–308. [Google Scholar] [CrossRef]
- Kapourani, C.A.; Sanguinetti, G. Melissa: Bayesian clustering and imputation of single-cell methylomes. Genome Biol. 2019, 20, 61. [Google Scholar] [CrossRef]
- Delaporte, G.; Cladière, M.; Camel, V. Missing value imputation and data cleaning in untargeted food chemical safety assessment by LC-HRMS. Chemom. Intell. Lab. Syst. 2019, 188, 54–62. [Google Scholar] [CrossRef]
- Lin, Y.; Lv, F.; Zhu, S.; Yang, M.; Cour, T.; Yu, K.; Cao, L.; Huang, T. Large-scale image classification: Fast feature extraction and SVM training. In Proceedings of the 2011 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Colorado Springs, CO, USA, 20–25 June 2011. [Google Scholar]
- Lin, X.; Yang, J.; Zhao, J. The text analysis and processing of Thai language text to speech conversion system. In Proceedings of the 2014 9th International Symposium on Chinese Spoken Language Processing (ISCSLP), Singapore, 12–14 September 2014. [Google Scholar]
- Molina, C.; Yoma, N.B.; Huenupan, F.; Garretón, C.; Wuth, J. Maximum entropy-based reinforcement learning using a confidence measure in speech recognition for telephone speech. IEEE Trans. Audio Speech Lang. Process. 2010, 18, 1041–1052. [Google Scholar] [CrossRef]
- Kwak Müller, K.; Lee, S. A convolutional neural network for steady state visual evoked potential classification under ambulatory environment. PLoS ONE 2017, 12, e0172578. [Google Scholar]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Brief. Bioinform. 2017, 19, 1236–1246. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115. [Google Scholar] [CrossRef]
- Ravì, D.; Wong, C.; Deligianni, F.; Berthelot, M.; Andreu-Perez, J.; Lo, B.; Yang, G.Z. Deep learning for health informatics. J. Biomed. Health Inform. 2017, 21, 4–21. [Google Scholar] [CrossRef]
- Banaee, H.; Ahmed, M.; Loutfi, A. Data mining for wearable sensors in health monitoring systems: A review of recent trends and challenges. Sensors 2013, 13, 17472–17500. [Google Scholar] [CrossRef]
- Kaur, G.; Chhabra, A. Improved J48 classification algorithm for the prediction of diabetes. Int. J. Comput. Appl. 2014, 98, 13–17. [Google Scholar] [CrossRef]
- Mir, A.; Dhage, S.N. Diabetes Disease Prediction Using Machine Learning on Big Data of Healthcare. In Proceedings of the 2018 Fourth International Conference on Computing Communication Control and Automation (ICCUBEA), Pune, India, 16–18 August 2018; pp. 1–6. [Google Scholar]
- Chen, W.; Yan, X.; Zhao, Z.; Hong, H.; Bui, D.T.; Pradhan, B. Spatial prediction of landslide susceptibility using data mining-based kernel logistic regression, naive Bayes and RBFNetwork models for the Long County area (China). Bull. Eng. Geol. Environ. 2019, 78, 247–266. [Google Scholar] [CrossRef]
- Michielli, N.; Acharya, U.R.; Molinari, F. Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput. Biol. Med. 2019, 106, 71–81. [Google Scholar] [CrossRef]
- Yuan, W.; Jiang, D.; Nambiar, D.K.; Liew, L.P.; Hay, M.P.; Bloomstein, J.; Lu, P.; Turner, B.; Le, Q.T.; Tibshirani, R.; et al. Chemical space mimicry for drug discovery. J. Chem. Inf. Model. 2017, 57, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Bojanowski, P.; Joulin, A.; Mikolov, T. Alternative structures for character-level RNNs. arXiv 2015, arXiv:1511.06303. [Google Scholar]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder–decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Korea Centers for Disease Control & Prevention. Available online: https://knhanes.cdc.go.kr/knhanes/eng/index.do (accessed on 6 November 2018).
- Zhang, S. Nearest neighbor selection for iteratively kNN imputation. J. Syst. Softw. 2012, 85, 2541–2552. [Google Scholar] [CrossRef]
- Royston, P. Multiple imputation of missing values: Update of ice. Stata J. 2005, 5, 527–536. [Google Scholar] [CrossRef]

| Age | 1–9: 2489 (10.8%) 10–18: 2425 (10.6%) 19–29: 2250 (9.8%) 30–39: 2946 (12.8%) 40–49: 3283 (14.3%) 50–59: 3499 (15.2%) 60–69: 3014 (13.1%) 70 and older: 3042 (13.3%) |
|---|---|
| Gender variable | Male: 10,411 (45.4%) Female: 12,537 (54.6%) |
| Type of residential area | Neighborhood: 18,551 (80.8%) Town/township: 4397 (19.2%) |
| Marital status (for those aged 30 or older) | Not married: 904 (5.7%) Married (with a spouse): 12,298 (78.1%) Married (widowed, divorced, or separated): 2549 (16.2%) |
| Education level (for those aged 30 or older) | Graduated from elementary school or less: 3670 (27.2%) Graduated from middle school: 1609 (11.9%) Graduated from high school: 4135 (30.6%) Graduated from college or higher: 4084 (30.3%) |
| DF2_ag | DI4_pr | DI6_ag |
|---|---|---|
| 888 | 8 | 888 |
| 888 | 8 | 888 |
| 888 | 8 | 888 |
| 888 | 1 | 888 |
| 888 | 8 | 888 |
| 888 | 8 | 888 |
| 888 | 8 | 888 |
| 888 | 8 | 888 |
| 888 | 8 | 888 |
| MLP | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of Variables | Accuracy (100) | Accuracy (200) | Accuracy (300) | ||||||
| KNN | Mode | MI | KNN | Mode | MI | KNN | Mode | MI | |
| 10 | 58% | 54% | 51% | 57.3% | 56.1% | 53.1% | 60.9% | 58.2% | 64.4% |
| 20 | 67% | 65% | 62% | 65.5% | 68.6% | 64.5% | 68.8% | 64% | 62.5% |
| 30 | 84% | 75% | 87% | 79.2% | 71.7% | 73.7% | 72.5% | 77.8% | 78.5% |
| Naïve Bayes | |||||||||
| Number of Variables | Accuracy (100) | Accuracy (200) | Accuracy (300) | ||||||
| KNN | Mode | MI | KNN | Mode | MI | KNN | Mode | MI | |
| 10 | 42% | 45% | 48% | 48.2% | 51.8% | 42.2% | 68.1% | 62.5% | 64.2% |
| 20 | 62% | 69% | 63% | 65.2% | 62.4% | 64.6% | 69.3% | 65.6% | 66.7% |
| 30 | 76% | 74% | 77% | 79.5% | 71.7% | 72.6% | 74.8% | 71.8% | 76.8% |
| SVM | |||||||||
| Number of Variables | Accuracy (100) | Accuracy (200) | Accuracy (300) | ||||||
| KNN | Mode | MI | KNN | Mode | MI | KNN | Mode | MI | |
| 10 | 55% | 57% | 51% | 52.7% | 57.4% | 55.5% | 62.3% | 64.2% | 62.6% |
| 20 | 65% | 67% | 59% | 65.4% | 68.2% | 62.1% | 64.2% | 68.1% | 67.6% |
| 30 | 77% | 81% | 82% | 84.1% | 82.7% | 76.4% | 72.5% | 79.5% | 72.8% |
| LSTM | |||||||||
| Number of Variables | Accuracy (100) | Accuracy (200) | Accuracy (300) | ||||||
| KNN | Mode | MI | KNN | Mode | MI | KNN | Mode | MI | |
| 10 | 60% | 62% | 64% | 60.5% | 58.2% | 57.4% | 56.8% | 54.5% | 65.8% |
| 20 | 67% | 68% | 74% | 72.8% | 71.5% | 78.5% | 73.5% | 75.4% | 77.1% |
| 30 | 82% | 84% | 86% | 80.9% | 82.8% | 87.8% | 90.9% | 91.4% | 85.5% |
| Char-RNN | |||||||||
| Number of Variables | Accuracy (100) | Accuracy (200) | Accuracy (300) | ||||||
| 10 | 64% | 66.3% | 62.5% | ||||||
| 20 | 81% | 84.2% | 79.4% | ||||||
| 30 | 92% | 91.5% | 91.7% | ||||||
| MLP | Naïve Bayes | |||||
|---|---|---|---|---|---|---|
| Variables | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| Normal | 74.4% | 77.9% | 50.7% | 62.2% | 75.5% | 42.5% |
| Hypertension | 46.2% | 48.9% | 45.7% | 53.2% | 82.2% | 47.4% |
| Stroke | 35.7% | 46.2% | 51.4% | 48.7% | 84.6% | 56.4% |
| Myocardial infarction | 80.3% | 86.2% | 49.7% | 71.4% | 73% | 64.4% |
| Angina pectoris | 92.1% | 97.9% | 50.3% | 68.4% | 86.4% | 61.2% |
| Diabetes mellitus | 93.6% | 94.9% | 50.1% | 74.4% | 76.2% | 66.6% |
| SVM | LSTM | |||||
| Variables | Accuracy | Precision | Recall | Accuracy | Precision | Recall |
| Normal | 72.3% | 82.8% | 62.5% | 79.5% | 82.4% | 51.7% |
| Hypertension | 74.2% | 86.4% | 45.2% | 75.2% | 84.1% | 46.2% |
| Stroke | 81.4% | 96.5% | 47.6% | 80.1% | 97.1% | 48.5% |
| Myocardial infarction | 75.8% | 89.1% | 65.4% | 79.8% | 92.4% | 47.2% |
| Angina pectoris | 74.3% | 84.2% | 67.5% | 84.5% | 94.2% | 56.4% |
| Diabetes mellitus | 81.1% | 78.5% | 57.2% | 89.7% | 96.6% | 65.6% |
| Char-RNN | ||||||
| Variables | Accuracy | Precision | Recall | |||
| Normal | 77.6% | 78.4% | 49.8% | |||
| Hypertension | 82.6% | 86.0% | 50.7% | |||
| Stroke | 80.6% | 88.6% | 48.4% | |||
| Myocardial infarction | 82.5% | 94.4% | 48.6% | |||
| Angina pectoris | 96.5% | 98.0% | 49.7% | |||
| Diabetes mellitus | 95.2% | 96.9% | 47.5% | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, C.; Son, Y.; Youm, S. Chronic Disease Prediction Using Character-Recurrent Neural Network in The Presence of Missing Information. Appl. Sci. 2019, 9, 2170. https://doi.org/10.3390/app9102170
Kim C, Son Y, Youm S. Chronic Disease Prediction Using Character-Recurrent Neural Network in The Presence of Missing Information. Applied Sciences. 2019; 9(10):2170. https://doi.org/10.3390/app9102170
Chicago/Turabian StyleKim, Changgyun, Youngdoo Son, and Sekyoung Youm. 2019. "Chronic Disease Prediction Using Character-Recurrent Neural Network in The Presence of Missing Information" Applied Sciences 9, no. 10: 2170. https://doi.org/10.3390/app9102170
APA StyleKim, C., Son, Y., & Youm, S. (2019). Chronic Disease Prediction Using Character-Recurrent Neural Network in The Presence of Missing Information. Applied Sciences, 9(10), 2170. https://doi.org/10.3390/app9102170







