Abstract
This study proposes HGBRCond, a machine learning model for conductivity prediction in controlled biodegradation processes. Eight regression algorithms were evaluated using experimental data (n = 424) from a micro-pilot treatment system. HGBRCond, based on Histogram-Gradient Boosting Regression (best performing ML model), achieved optimal performance (R2 = 0.877 ± 0.011, RMSE = 10.235 ± 0.54 µS/cm) through 10-fold cross-validation. Unlike standard HGBR and previous conductivity models that lack comprehensive validation frameworks, HGBRCond integrates rigorous statistical validation (cross-validation, sensitivity analysis, confidence intervals) with multi-level interpretability (Morris screening, SHAP analysis, feature importance), achieving a 6.8% performance improvement over standard gradient boosting approaches while addressing mechanistic interpretability gaps present in prior work. However, limitations constrain direct potential industrial applicability: limited dataset (n = 424), narrow conductivity range (285–360 µS/cm), strong dissolved oxygen dependence, sensitivity across two critical parameters, constant flowrate, and validation restricted to controlled conditions. These constraints require model recalibration for potential industrial application. Future work will focus on model validation across extended operational ranges using industrial samples and full-scale testing to establish applicability beyond controlled experimental settings.
Keywords:
algorithm; regression; validation; analysis; environment; conductivity; pollution; biodegradation 1. Introduction
In order to ensure compliance with regulatory standards, continuous monitoring of wastewater quality parameters is essential. In the wastewater treatment processes, especially in biological systems that use microbial suspensions for pollutant (phenols, hydrocarbons, sulphides, nitrites, phosphates, heavy metals) removal, real-time parameter monitoring is critical for process optimization. Conductivity is a key indicator in treatment processes because it reflects the concentration of dissolved ionic species, correlates with overall water quality, providing rapid feedback regarding treatment efficiency [1,2,3,4,5,6,7,8,9,10,11].
Machine Learning (ML)-based predictive models supply significant potential for improving monitoring capabilities and they ensure proactive process control in biological treatment systems.
Regarding key contributions of previous research, several ML approaches have been developed for conductivity monitoring and prediction, including LSTM networks for temporal dynamics in activated sludge process (R2 = 0.88) and Gaussian process regression (GPR) for membrane filtration processes with superior accuracy (MAE < 0.3 μS/cm) [12,13]. Also, ensemble methods (XGBoost, hybrid CNN-LSTM, MLP, KNN) have been developed for surface water parameters (R2 = 0.82; RMSE = 53.83 μS/cm), as has hybrid ANN-PCA combining neural networks with dimensionality reduction for industrial treatment plants (R2 = 0.88) [14,15].
Despite this progress, current ML approaches for conductivity prediction have certain gaps that motivate the present study, such as inadequate validation, as predictive accuracy metrics (R2 Score, RMSE, MAE, MAPE, MedAE) are prioritized while neglecting robust validation methods (cross-validation, sensitivity analysis, confidence intervals) or multi-level interpretability (Morris screening, SHAP analysis, feature importance). In addition, robustness evaluation is insufficient since sensitivity analysis is often neglected, therefore model stability across hyperparameter variations remains unassessed. In addition, the interpretability framework is limited, with narrow usage of feature importance, Morris screening, and SHAP analysis to identify critical process parameters. Finally, Histogram Gradient Boosting Regression (HGBR) is underutilized for water conductivity despite multiple advantages (fast training, native missing value handling), while related gradient boosting methods (XGBoost, GBR, LightGBM) are frequently applied [16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35].
The original contributions of the study address the mentioned gaps through an integrated framework:
- Implementation and customization of eight ML algorithms designed to address specific characteristics of water conductivity;
- Comparative analysis identifying the optimal approach using performance metrics;
- Development of HGBRCond model for water final conductivity (C2) prediction that combines gradient boosting speed and accuracy with hyperparameter optimization during biodegradation of synthetic wastewater;
- Rigorous statistical validation using 10-fold cross-validation;
- Sensitivity analysis and confidence intervals (CI) demonstrating model robustness and calibration;
- Multi-level interpretability using feature importance, Morris screening and SHAP analysis to quantify the contribution of water initial conductivity (C1), dissolved oxygen (O1) and flowrate (FR) features to final conductivity (C2) prediction.
This integrated approach links the gap between the modeling focused exclusively on accuracy and a more comprehensive evaluation that integrates performance, robustness and interpretability.
The proposed HGBRCond model has several technical differences compared to XGBoost and traditional GBR models. Regarding the architecture, HGBRCond uses only two input features (O1 and C1, primary and secondary predictors) versus 4–6 parameters (DO, pH, BOD, COD, NH4) in XGBoost/GBR implementations [12,36]. Regarding the algorithm, it uses histogram-based splitting (63 s training time), while traditional GBR uses exact splitting (150–400 s). Concerning data handling, it integrates native missing data processing, while traditional GBR requires preprocessing. The model validation framework includes 10-fold cross-validation (SD = 0.011), sensitivity analysis, and confidence intervals, while the one for XGBoost and GBR models does not contain such elements [12,13]. Regarding interpretability, it explicitly integrates SHAP screening and Morris analysis for predictor contribution evaluation (O1 relative importance > 90%) that are missing from the analyzed XGBoost/GBR implementations. In terms of performance, HGBRCond achieves R 2 = 0.877 on pilot data versus R2 = 0.82 for XGBoost/GBR on industrial datasets [12,36].
The proposed HGBRCond model novelty consists of the integration of rigorous statistical validation (10-fold cross-validation), sensitivity analysis and confidence intervals (95% CI: [0.855, 0.899]), ensuring model robustness (SD = 0.011), an approach absent in previous studies [12,13,14,32]. In addition, a multi-level interpretability framework combining feature importance, Morris screening, and SHAP analysis was used to quantify predictor contributions. It is a computational efficiency model through histogram-based gradient boosting with systematic hyperparameter optimization, achieving competitive accuracy (R2 = 0.877) with faster training than traditional GBR and XGBoost models, while maintaining superior stability. Its operational simplicity requires only two measurable predictors (O1 and C1) compared with 4–6 parameters in existing models, reducing sensor infrastructure complexity by 67%. This integrated approach bridges the gap between accuracy-focused modeling and comprehensive evaluation, providing a validated, robust, and interpretable solution for pilot-scale water conductivity prediction.
2. Materials and Methods
2.1. Experimental Design and Wastewater Treatment System
2.1.1. Installation Description
The chemical–biological treatment tests were carried out in a glass installation (micro pilot) consisting of a continuously fed vessel throughout the experiment with wastewater prepared from mineral medium (MM). The operating principle of the micro-treatment station was based on the continuous circulation of the synthetic medium of polluted water at a continuous flowrate of 0.5 L/h for the feed and the same continuous flow rate of 0.5 L/h for the recirculation of the biological sludge from the decanter to the aerotank [37,38].
The monitoring was carried out using two identical multi-parameter devices, WTW Inolab MULTI 9630 IDS, with three galvanically isolated channels for conductivity, pH, and oxygen measurement, which were used for monitoring the continuous flow in the aerotank and decanter [39,40].
The components of the microstation were the following: the feed vessel with a capacity of 5 L mounted on a high support connected to a vessel called an aerotank (useful capacity 700 mL, dimensions diameter 7 cm, height 50 cm) (Figure 1). At the bottom, the aerotank was provided with a nozzle through which air was introduced into the system (via a compressor) to ensure the oxygen requirement for the microfauna in the biological sludge and to maintain the bacterial suspension in contact with the supplied water with impurities. At the top, the aerotank is provided with a nozzle for discharge into the decanter (which is a glass vessel identical to the aerotank in terms of construction). The decanter was kept at rest, without an air supply. The installation had a recirculation system, a recirculation pump, the biological sludge being collected through sedimentation in the decanter and returned to the circuit in the aerotank [41,42,43,44,45]. Throughout the experiment, conductivity, pH, and oxygen were continuously monitored with the WTW sensor for 20 days (2.4 days was the adaptation period and 17.6 days was the testing period). The tests were carried out in an aerotank and in a decanter. The purified water was captured in a collection vessel and was periodically analyzed. The connections between the vessels were made with silicone rubber tubing, and the throttling was performed with screw clamps. Both pilot installations were placed parallel on a metal support. During the experiment, microscopic visualizations were performed by optical microscopy and TEM microscopy [38,40].
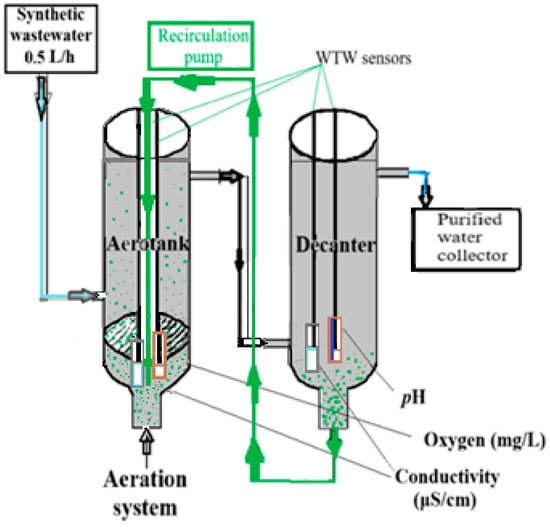
Figure 1.
Simplified diagram of the microtest biological treatment installation.
2.1.2. Materials, Reagents, and Chemicals
Synthetic wastewater (MM) with the following chemical composition, according to the protocol OECD-“Guideline for testing of chemicals, 302 B”: KH2PO4 (1 g/L), KNO3 (0.4 g), NH4Cl (5 g/L), FeSO4•7H2O (0.025 g), MgCl2•6H2O (0.5 g/L), MgSO4•7H2O (0.25 g/L), CaCl2•2H2O (0.25 g/L), NaH2PO4•2H2O (0.3 g/L), NaHCO3 (0.3 g/L), FeCl3• 6H2O (0.025 g/L). The pH was adjusted to 8.5.
The COD analysis was interpreted based on a calibration curve previously constructed by analyzing a standard solution according to OECD regulations (Annex V C.9. “Chemical oxygen demand degradation”, Directive 84/449/EEC). The standard solution was prepared from the following mineral components: KH2PO4 8.50 g/L, K2HPO4 21.75 g/L, Na2HPO4•2H2O 33.40 g/L, NH4Cl 0.50 g/L, CaCl2 27.50 g/L, MgSO4•7H2O 22.50 g/L, FeCl3•6H2O 0.25 g/L.
The viability of the microorganisms was monitored throughout the experiments by microscopic visualization using a Celestron Microscope, model 4434.
2.1.3. Operating Conditions and Experimental Protocol
The tests carried out aimed to monitor chemical parameters correlated with the ability of microorganisms to biodegrade chemical pollutants. The identification of strains specific to the degradation of each pollutant helps to restore the biocenosis in a treatment plant that could be affected by accidental pollution and can intervene in the efficient biodegradation of wastewater [39,40,41,42]. The conductivity (or electrical conductivity) of water is an important indicator used to evaluate the level of water pollution and is a parameter used to control wastewater treatment processes that describes the degree of chemical load of a water. According to the United States Environmental Protection Agency, conductivity is quantified as the concentration of dissolved ions (representing the totality of cations and anions present in water: Ba2+, Ca2+, Cu2+, Fe2+, HCO−, K+, Li+, Mg2+, Mn2+, Na+, Ni2+, SO42−, Zn2+), originating from industrial pollution. Water conductivity is an important indicator used to assess the level of water pollution and is a parameter used to control wastewater treatment processes that describes the degree of chemical loading of water, which comes from industrial pollution, agriculture, or domestic discharges (chemical fertilizers-nitrates, phosphates, domestic water—detergents, salts, industrial discharges—heavy metals, acids. Conductivity depends on the concentration in ions, the nature of the ions, temperature, the concentration of dissolved oxygen (the weight of oxygen consumed by microorganisms per unit of time), and the viscosity of the solution, which is why it is a parameter for controlling the degree of mineralization of water [45,46].
In the present study, the evolution of chemical parameters in pollution was monitored with chemical substances obtained from a synthetic mineral medium. The present study is carried out on a continuous feed process with mineral medium while the microorganisms in the biological sludge are aerobic, maintained in suspension by continuous agitation provided by an air compressor. Dissolved oxygen is an important water quality parameter because it influences the existence of most microorganisms in the biological sludge.
In the study carried out, the evolution of the control parameters-conductivity (μS/cm) and dissolved oxygen (mg/L)—was correlated with the variation in pH. The biopurification process was ensured by a population of protozoan and metazoan microorganisms adapted on a zoogleal mass forming a biological sludge that was kept in suspension by continuous agitation with a compressor. The purification system was continuously fed with a synthetic mineral medium, artificially prepared, using chemical substances that can be found in practice in a purification plant originating from wastewater from the chemical industry [41,42,43,44,45]. Throughout the experiment, conductivity, pH, and oxygen was continuously monitored with the WTW sensor for 20 days (2.4 days was the adaptation period and 17.6 days was the testing period). The tests were carried out in an aerotank and in a decanter.
2.2. Machine Learning Framework
Eight regression algorithms were evaluated to identify the optimal approach for conductivity prediction: Histogram-based Gradient Boosting Regression (HGBR), Gradient Boosting Regression (GBR), Random Forest Regression (RFR), Support Vector Regression (SVR), K-Neighbors Regression (KNR), Decision Tree Regression (DTR), Ridge Regression (RR), and Linear Regression (LR). All algorithms were customized and implemented in Python 3.9 (Python Software Foundation, Wilmington, DE, USA).
This framework addresses three modeling requirements [21,22,47,48,49,50,51,52,53]: (a) non-linear relationship capture through ensemble (HGBR, GBR, RFR) and kernel-based methods (SVR); (b) process interpretability through tree-based feature importance rankings (HGBR, RFR, DTR) and linear baselines (LR, RR); and (c) computational efficiency through HGBR’s histogram-based optimization and KNR’s local pattern modeling.
This comparative evaluation determines whether conductivity prediction requires non-linear modeling, critical aspect regarding control strategy selection.
The best-performing ML algorithm was HGBR used to develop HGBRCond, a hybrid model. The model was validated through 10-fold cross-validation, sensitivity analysis and CI and a multi-level interpretability (feature importance, Morris screening, SHAP analysis). The model implementation used optimized HGBR hyperparameters (max_bins = 128, min_samples_leaf = 20). A selection of the Python code used for HGBRCond model implementation, cross-validation, sensitivity analysis, feature importance, Morris screening, SHAP analysis and confidence intervals is provided in the Supplementary Materials file.
The complete workflow, from data collection to model training, selection and validation, is presented in Figure 2.
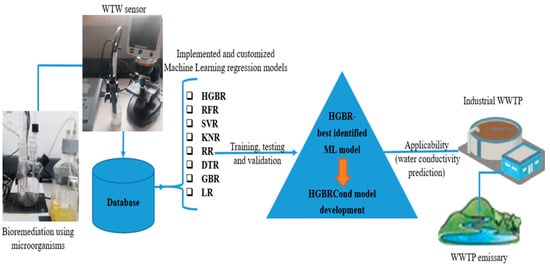
Figure 2.
The materials and methods used.
3. Results
3.1. Biodegradability Evolution
3.1.1. Biocenosis Evolution Analysis
The initial volume of sludge introduced into the system was 150 mL/L, measured in the Imhoff cone after 30 min of rest. At the end of the 20 days, the volume of sludge collected from the system was 160 mL/L. We considered the fact that MM did not have an inhibition on the biological mass, but on the contrary, the volume of sludge increased by 6.6%. The tests were based on the activity of microorganisms in water with MM in the presence of oxygen. The biological sludge was used in the aerotank, consisting of a mixed population of microorganisms, composed of protozoa represented by Paramecium caudatum, Aspidisca polistila, Colpidium colpoda, Stentor, Vorticella microstoma, Litonotus setigerum (Table 1) [37,38,39,40]. Towards the end of the experiment, metazoans such as rotifers sp. were observed, which indicates that he biological sludge is in stable and optimal viability conditions, the synthetic environment being non-toxic for them. The microscopic examination was performed using a 40 × 12.5 objective lens and was accomplish periodically [54,55,56,57].

Table 1.
Cellular abundance in the biological sludge.
The microbiological analyzes regarding the number of bacterial cells were quantified from a diluted aliquot sample that was pipetted onto the Petri plate using agar medium, subsequently incubated for 48 h at 37 °C in a thermostat. Total aerobic bacteria were quantified as “Total bacterial colony count”, CFU/mL [54,55,56]. Abundance was examined by counting live ciliated cells using a Thoma counting chamber by examining samples from an aliquot volume of biological sludge collected from the aeration tank [38,40].
As a result of the tests carried out, it was observed that the number of bacterial cells in the tested samples had values ranging between 2 × 105 and 2 × 106 CFU/mL in the stabilization stage, and between 5 × 107 and 2 × 105 and 2 × 106 CFU/mL in the stabilization stage, and between 5 × 107 and 2 × 108 in the testing stage. The abundance of Paramecium caudatum cells was between 4 and 15% during the biological sludge adaptation period and between 10 and 18% during the testing period. The biological mass of Zooglee ramigera was in abundance between 5 and 25% during the adaptation period, respectively, 10–50% during the testing period.
The biological treatment of biodegradation of chemical pollutants used the metabolic activity of groups of microorganisms that developed in the bacterial mass Zooglee ramigera.
In the microscopically examined samples, the ciliates Paramecium caudatum were observed throughout the experiment. Aspidisca polystyle had a low abundance, being present only in the second part of the adaptation phase with 2% and between 4 and 7% in the second part of the test experiment.
Microscopic observations on the activity of the biological sludge highlighted the presence of microorganisms formed by mixed populations of bacteria that formed the zoogleal mass in which protozoan microorganisms (Paramecium caudatum, Colpidium colpoda, Stentor, Aspidisca polystila) were active. Ciliates cells are microbiological indicators for a stable biocenosis, with adequate nutritional values, without toxic contaminants and with optimal oxygenation. The stability of the biocenosis after the 12-day period was observed by the appearance of cells of Litonotus setigerum and after 15 days of the test period. The ciliates Stentor and Vorticella microstoma appeared, both ciliates being indicators for good biodegradation and stability of the biocenosis. Rotifers appeared in the biocenosis in the last part of the test, with an abundance of maximum 10% at the end of the experiment.
The biological sludge produced in the micropilot station has a high degree of stability due to the continuous feeding, with a constant flow rate, with a balanced nutrient medium with nitrogen, potassium, and phosphorus. These elements provide stability to the biocenosis and support the development of the microbial community responsible for the biodegradation of mineral substances introduced into the microstation feed container. The slight increase in sludge volume from 150 mL/L to 160 mL/L over a period of 20 days is due to the formation of flocs with a stable settling behavior. Overall, these observations indicate an efficient performance of the biodegradation process correlated with an adequate operational regime.
3.1.2. Analysis of pH, BOD and COD Parameters Evolution
The pH parameter was monitored in the supernatant from the decanter in order to monitor the viability of microorganisms in the biocenosis. The acclimation period (2.4 days) to laboratory conditions in which a mineral medium feed was carried out was the time required to reach a reasonable stability of the system, reflected by the pH. The pH value (Figure 3) was located during the acclimation period between 4.2 and 5.5 after which it reached almost 6, continuing throughout the experiment at a stable pH range, in the neutral range of 6.2–7.
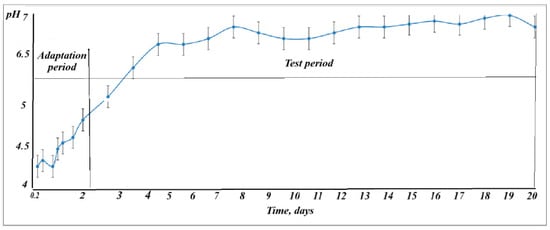
Figure 3.
The pH variation during the experiment (acclimatization and testing phase). Error bars represent standard deviation (n = 5).
The graphic representation indicates that during the adaptation period of the biological sludge it had a minimum between 4.2 and a maximum of 4.6 after which the pH value was increasing until day 5 when it reached the minimum value of the test period (6.5). The maximum of the test period had an ascending peak on day 8 and day 18 reaching a value of approximately 7.
The purified water was captured in a collection vessel and was periodically analyzed. Because the study conducted followed the evolution of microorganisms in the biological sludge in the installation with constant flow and continuous dosing of mineral medium, was analyzed every 5 days the BOD/COD rate in the purified water collector vessel. The biological oxygen demand (BOD, mg/L) from the analyzed samples was quantified by measuring with the WTW sensor. The BOD was expressed by calculating the difference between the amount of oxygen (mg/L) present in the sample at the initial time of collection and the amount of oxygen (mg/L) present in the sample after 5 days. Oxygen monitoring is necessary because it intervenes in metabolic processes in biological environments.
COD analysis was performed in the laboratory by determining the oxygen resulting from the oxidation of organic substances in water in an oxidizing medium (K2Cr2O7 0.25 N, H2SO4 98%, molar ratio (1:3). The reaction was catalyzed by (HgSO4) and (Ag2SO4) (1:1, 0.5 g/L) to eliminate chloride interference. Glass test tubes with hermetically sealed screw caps were used as follows: 1 mL of sample collected from the collection vessel was treated with 1 mL of oxidizing mixture and 0.5 g of catalyst (0.3g) and 8 mL of distilled water. The samples were kept in the oven at 150 °C for 2 h and then 30 min maintained at room temperature. The samples were photometered using a spectrophotometer by measuring the absorbance at a wavelength of 600 nm. by comparison with the control sample (with distilled water). The COD expression (mg O2/L) was made relative to the calibration curve.
Microorganisms, in an aerobic environment, with continuous aeration, through biochemical reactions, with the help of cellular enzymes catalyze chemical reactions with the transformation of the chemical substrate into metabolites that can go up to CO2 and H2O. The reactions in which microorganisms are involved are hydrolysis, nitrification, denitrification, oxidation, phosphorylation reactions, which lead to transformations of the chemi cal composition of the purified water [3,5,11].
In order to observe the degree of purification of the microbiologically treated water, we followed the value of the BOD/COD ratio, which to be considered purified water must have a value greater than 0.3. Theoretically, a BOD/COD ratio in the range of 0.0–0.3 would indicate that the mineral medium introduced into the installation is toxic to the biological species existing in the biological sludge or indicates that the biocenosis is not sufficiently adapted to the concentrations of chemical species in the mineral medium.
The analysis of experimental values obtained from measuring BOD/COD ratio in the analyzed samples is presented in Figure 4 that shows that in the first part of the test, during the adaptation period of the biological sludge, the ratio was slightly increased in the range of 0.69–0.65. During the purification process of the chemical medium introduced into the system, the BOD/COD ratio was decreasing, during the period 3–9 days it had values between 0.65 and 0.55. During the period of 9–12 days, the ratio was maintained with a trend of stability at a value close to 0.55 with a slight increase to 0.57 on day 12, and in the following period it was to decrease slightly, reaching the value of 0.47 at the end of the experiment [39]. The average of the values obtained is 0.593 and the standard deviation is 0.073. The minimum value is 0.480 and the maximum value is 0.68. The coefficient of variation (RDS) is 12.24%
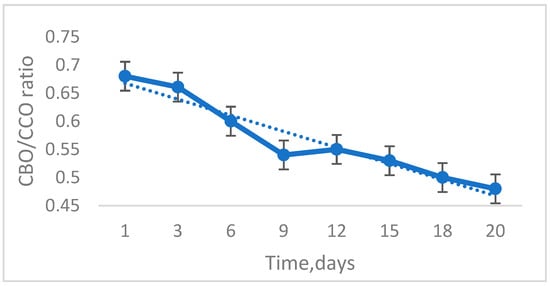
Figure 4.
BOD/COD ratio analysis conducted throughout the experiment. Error bars represent standard deviation (n = 4) and trendlines.
The BOD/COD ratio monitoring process was to observe the biodegradability of the chemical medium introduced into the system by the microbial population in the biological sludge.
In Figure 5 are presented the microscopic observations obtained during the experiment through optical microscopy and TEM microscopy.
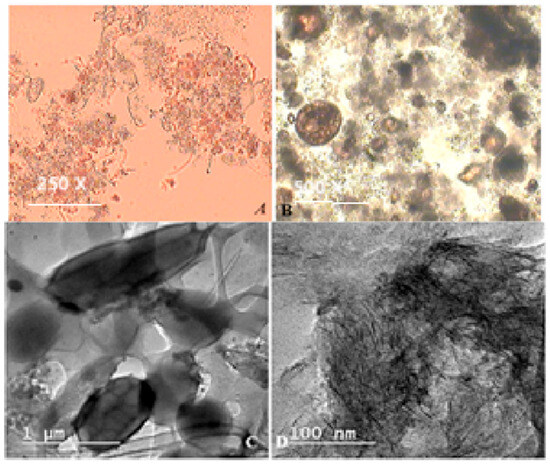
Figure 5.
Optical light microscope imagery of Zooglee ramigera, after 10 days of experiment magnifications of 250× (A) Paramecium caudatum imagery after 20 days of experiment, magnifications of 500× (B), respectively, TEM imagery after 20 days of experiment (C,D).
3.2. HGBRCond Mathematical Model Development
For testing the custom Python ML-mentioned regression algorithms, a number of 424 distinct scenarios (during the 17.6-day test period, 424 distinct scenarios were obtained at a rate of one recording per hour through continuous monitoring) assigned to the analyzed process were used. The final goal was the identification of an ML method that is more suitable for water conductivity prediction (C2-output [μS/cm]), when the inputs FR (water flowrate-[L/h]), C1 (initial conductivity-[μS/cm]) and O1 (oxygen-[mg/L]) are known. In this sense, a much-needed primary data processing (missing values removal, normalization, data splitting into training, evaluation, and validation sets, etc.) was achieved, followed by the configuration of each algorithm’s parameters. In Table 2 is a selection of the data that compose the .csv type file necessary for the analysis of the customized ML methods, using Python 3.9 software, while Table 3 presents a brief descriptive statistics (range, mean, SD and mean ± SD) for the used dataset.

Table 2.
The structure of .csv developed file.

Table 3.
Dataset descriptive statistics.
To compare the analyzed custom Python ML algorithms, a set of evaluation metrics, such as the explained variance score (EVS), the coefficient of determination (R2 Score), mean absolute error (MAE), mean absolute percentage error (MAPE), and median absolute error (MedAE) was used, and the metrics are defined in the paper [53].
The dataset presented in Table 3 shows the experimental measurements obtained under controlled operating conditions. The input variables are water flowrate (FR [L/h]), water initial conductivity (C1 [μS/cm]) and dissolved oxygen (O1 [mg/L], with the ranges, mean values, standard deviations, and mean ± SD intervals presented in Table 3. The output variable is water final conductivity (C2 [μS/cm]) with the associated descriptive statistics presented also in the same table. The selection of these variables was based on their relevance on the modeled process and on their observed variability across experiments.
The flowrate was maintained constant at 0.5 L/h (Table 2) throughout the experiment to ensure controlled baseline conditions and isolate biological process variables for model development; therefore, it does not contribute information to the model. The chosen dataset structure ensures the fact that the model is trained using only variables with meaningful variability; therefore, it is improved the model predictive interpretability and robustness. The limited conductivity (C2) (Table 3) range of 285–360 (μS/cm) reflects the conditions observed during the experimental setup and the limitations of the current laboratory configuration. In order to solve these limitations, future work should include validation across multiple flowrates, expansion of the conductivity range, testing with real industrial wastewater, and validation in operational industrial plants.
Table 4 shows the identification of the analyzed Machine Learning models’ parameters, manually tuned through an intensive trial-and-error testing process (five tuning stages were performed for each analyzed ML model).

Table 4.
Hyperparameters tuning for ML Models.
In Table 5, the analyzed ML models’ performance metrics (EVS, R2 Score, MAE, MAPE, and MedAE) are presented.

Table 5.
Performance results of water conductivity (C2) prediction algorithms on training and evaluation sets.
As observed in Table 5, from the implemented and customized ML algorithms for water conductivity (C2) prediction, HGBR method proves to be the most suitable for this purpose. Its precision (for training and validation data obtained in test no. 3 from five achieved tests, Table 6), highlighted in Figure 6, highlights an EVS and R2 Score of 0.9468, indicating that it explains approximatively 95% of the overall variance.

Table 6.
Optimized hyperparameters (manually tuned) for the HGBR model for water conductivity prediction.
Figure 6.
HGBR precision for training and validation data (Test no. 3).
In addition, has the lowest values for MAE (3.6718 μS/cm) and MAPE (1.19%), a fact that suggests minimal error and highly accurate predictions.
In contrast, algorithms such as LR and RR are inadequate for the analyzed problem (conductivity C2 prediction), as EVS and R2 Score have values of 0.76 and high values for MAE, MAPE, and MedAE (as it can be observed in Table 5).
The results presented in Table 6 suggests the importance of parameters tuning in order to increase the HGBR model performance.
Therefore, learning_rate indicates how much a new model contributes to the final prediction, while max_iter controls the total number of iterations (how many decision trees were trained and sequential added to the final model).
In addition, max_depth indicates the maximum depth of each decision tree, min_samples_leaf controls how much data can be partitioned in the tree, max_bins represents the intervals in which the algorithm splits each features value and, L2_regularization reduces the amplitude of the leaf values.
In Figure 7, the metrics performance for each tested ML model highlights the HGBR model’s superiority.
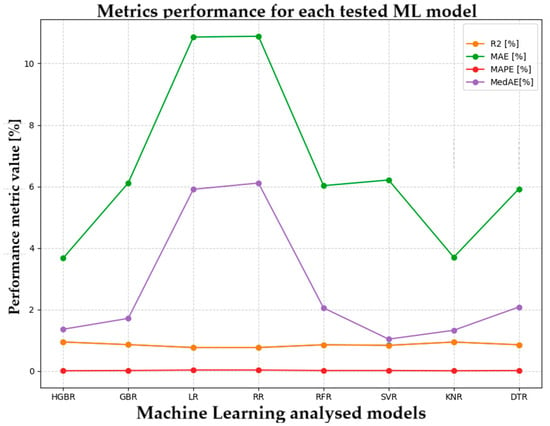
Figure 7.
Metrics performance for each tested ML model.
Next, a mathematical model (referred to as HGBRCond) is proposed, based on HGBR, a method that has been identified as the best one for the discussed problem.
Using the parameters supplied by HGBRCond model (Narb, LearnR, Predi), the model iteratively builds an ensemble in which each new estimator is trained on the negative gradients (residuals) of the loss function supplied from previous predictions, rather than directly predicting the target variable (it does not imply direct summation of predictions). The proposed model referred to as HGBRCond is given by Formula (1):
where —conductivity C2 final prediction for z observation;
- —the initial prediction obtained through loss function minimization over the training data set (in this case, 323.7071, representing the mean of C2 training values),
- —the learning rate (in this case, 0.01), which controls each estimator contribution and, it prevents overfitting;
- —the number of boosting iterations (sequential estimators), in this case, set to 5000 estimators;
- —histogram-based estimators trained on pseudo-residuals (negative gradients). It represents the prediction of the j-th histogram-based decision tree fitted on the negative gradient of the loss function (residuals) at iteration j-1. Therefore, it is not the direct prediction of the target value (conductivity) but the prediction of the histogram-based tree j trained on the negative gradient (residuals) of the loss function from the previous iteration;
- —observation .
Equation (1) was developed starting from the standard form of Gradient Boosting [57], implemented with histogram-based decision tree according LightGBN approach [58].
Regarding the residual fitting and loss minimization process, at each j iteration, the estimator approximates the negative gradient (given by Formula (2), which explicitly demonstrates the residual fitting process) of the loss function, rather than directly predicting the target conductivity C2 value (it does not imply predictions direct summation). Equation (2) shows that each estimator is trained to approximate the negative gradient of the loss function evaluated at the previous cumulative prediction (not the target value C2 directly).
where LF is the loss function, in this case the Mean Squared Error (MSE) for regression, given by Formula (3), where the minimization of the loss function is explicit and where the factor is a Gradient Boosting standard convention that simplifies the gradient calculation;
- represents the cumulative prediction after j-1 iterations;
This approach ensures that each new estimator (decision tree) corrects the ensemble residual errors, iteratively minimizing the overall loss. The learning rate (0.01) prevents overfitting by limiting each decision tree contribution, acting as a regularization parameter. The cumulative effect of 5000 sequential estimators acts as small corrections that finally leads to an optimal prediction (iteratively minimizing the overall loss).
Equation (1) aggregates gradient-based corrections (negative gradients scaled by ), not raw predictions—the core principle of Histogram Gradient Boosting [57,58,59,60,61] where models learn from predecessor errors.
The optimal hyperparameters (n_estimators = 5000, learning_rate = 0.01), configuration follows established gradient boosting principles where lower learning rates combined with higher numbers of estimators enhance generalization by enabling more gradual learning [57,58,61]. The computational efficiency of Histogram-based Gradient Boosting, which uses histogram-based splitting and reduces training complexity, makes this configuration practically feasible for real-time applications [58].
In Figure 8, the HGBRCond model performance is assessed by comparing the test data with the predicted values, which suggests that overall the proposed model performs well for the majority of the samples, presenting some deviations for other samples.
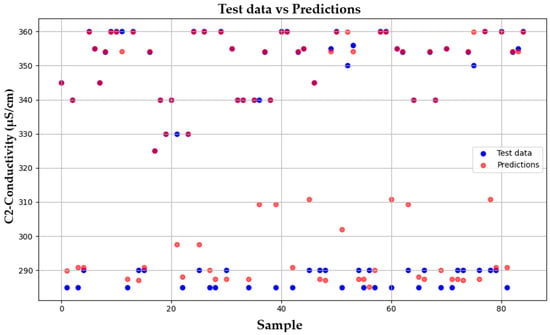
Figure 8.
Test data vs. Predictions-HGBRCond model.
The samples in Figure 8 that deviate the most from the predicted trend are statistical outliers reflecting the inherent variability of experimental micro-pilot biodegradation systems. These deviations can be generated by the dynamic behavior of microbial communities, WTW sensor instrumental measurement uncertainty (potential sensor fault, probe calibration errors) and minor fluctuations in operating conditions. Such variability is common in laboratory and pilot-scale experiments, especially when dealing with biological processes with non-linear behavior.
Rather than being limitations, these deviations provide essential information about the natural complexity of biological systems and real wastewater treatment processes variability. The information provided by these deviations may be applied to assess the model robustness under suboptimal experimental conditions, to provide early warnings regarding potential operational issues, as guidance for process monitoring improvements, and for control strategy tuning. The model’s ability to maintain a good predictive performance despite these outliers demonstrates its potential practical applicability to real industrial scale systems under non-ideal experimental conditions.
Therefore, the deviation from a linear correlation does not indicate the model low performance capacity, rather it reflects the chemical pollutants biodegradation process’s non-linear behavior, complexity, and may result from to the inherent variability of the chemical data obtained through laboratory instrumental measurements.
In Table 7 are presented the evaluation metrics obtained for HGBRCond (implemented in Python 3.9) proposed model, such as R2 Score, adjusted R2, MAE and Root Mean Square Error (RMSE).

Table 7.
Performance results for HGBRCond proposed model.
The evaluation metrics values for HGBRCond proposed model presented in Table 7 suggests that the model presents good performance. Therefore, the model explains almost 95% of the observed variance, and an adjusted R2 score value of 0.94 confirms that this performance is obtained without overfitting (it maintains a good generalization). In addition, the difference of 0.01 between R2 Score and Adjusted R2, suggests the fact that the proposed model is well-built, without overfitting on features. The value of the MAE metric (3.74 μS/cm) shows that the obtained predictions present a reduced variation from the real values. At the same time, the RMSE metric value (7.97 μS/cm) suggests a slight presence of larger isolated errors, but within acceptable limits.
In Figure 9, it is presented the model residuals versus predicted values.
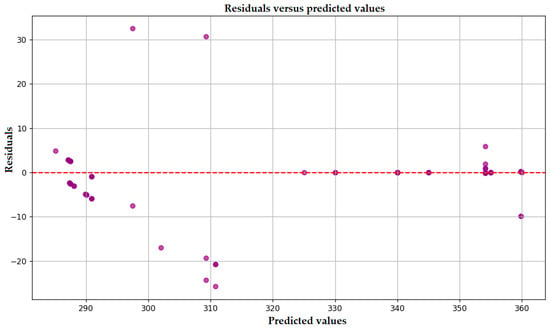
Figure 9.
Residuals versus predicted values (purple dots represent individual residuals, and the red dashed line indicates zero residuals)-HGBRCond model.
Figure 9 demonstrates that this model performs well across for most samples, with deviations limited to isolated outliers. These anomalies arise from inherent experimental variability in biodegradation systems, pilot-scale biodegradation process characteristics (microbial stochasticity, instrumental measurement uncertainty-calibration drift, sensor malfunctions), and the inherently non-linear process behavior (a strict linear relationship between predicted and experimental data is not to be expected.
3.2.1. HGBRCond Model Validation, Sensitivity Analysis and Morris Method Screening
To demonstrate that the HGBRCond model is performant, valid and that it can be used to make good predictions, was used cross-validation (k-fold) ML technique (technique that divides the used data set into k-equal size folds to evaluate model effectiveness). Respectively it is identified the optimal configuration setting and analyzed the model performance on new data, providing a more robust estimation of the model predictive ability. In Table 8 are presented the identified optimal configuration (obtained using GridSearchCV- the tuning process to determine the optimal values) and their associated tested interval.

Table 8.
Optimized hyperparameters.
It should be noted that the used validation methodology (train-validation test stages) followed strict practice in order to prevent data leakage:
- Data splitting: The dataset was initially split intro training (70%, n = 424) and testing (30%, n = 182) sets. The test set was never used during model development, hyperparameters tuning or any other evaluations;
- Parameter optimization: This stage was performed exclusively on the training dataset (70%) using 10-fold cross-validation. Within each fold, the training set was subdivided into 90% for training and 10% for validation. The parameters were optimized by averaging the performance obtained across all 10-folds. Importantly, no information from the test set was used at any stage of the parameters identification;
- Model final evaluation: Once the optimal parameters were identified through 10-fold cross-validation on the training set, the HGBRCond model was trained on the entire training set (70%). The model was then evaluated once again on the unused (reserved) test dataset (30%). The performance metrics reported in Table 9 (R2 = 0.8772 ± 0.0110, RMSE = 10.2353 ± 0.54092388 [μS/cm], and MAE = 4.8599 ± 0.2388 [μS/cm]) demonstrate the model final evaluation on unused data.
 Table 9. R2 Score, RMSE, and MAE performance metrics values per k-fold for HGBRCond model performance evaluation (cross-validation performance-10-fold).
Table 9. R2 Score, RMSE, and MAE performance metrics values per k-fold for HGBRCond model performance evaluation (cross-validation performance-10-fold).
As outputs, the k-fold validation technique supplied, in the case of the proposed HGBRCond model performance evaluation, the R2 Score, RMSE, and MAE performance metrics values (obtained using the best identified values of the hyperparameters from Table 8) are presented in Table 9, for different numbers of k-folds (where k = ).
Table 9 provides detailed cross-validation results for the HGBRCond model across 10 folds cross-validation:
- R2 = 0.8772 ± 0.0110: the model captures 87.72% of the target variable (conductivity C2) explained variance, with low standard deviation (SD = 0.0110) confirming stable performance across folds; an R2 > 0.7 indicates excellent HGBRCond model predictive capability;
- RMSE = 10.2353 ± 0.5409 μS/cm: the root mean absolute error is 10.24 μS/cm; relative to the target variable (conductivity-C2) range (285–360 μS/cm; range = 75 μS/cm), this represents 13.65% of the target scale—a good performance per empirical criteria (RMSE 10–20%: good; <10%: excellent); the low standard deviation (SD = 0.54) demonstrates model consistency and an good model stability across validation sets;
- MAE = 4.8599 ± 0.2388 μS/cm: the mean absolute error is excellent at under 5 μS/cm, with minimal variability across folds.
The initial evaluation on a single test data set shown a R2 Score value of 0.95 (Table 7), but cross-validation supplied a more reliable and robust estimation obtained using different k-folds (R2 value is 0.8772-Table 9) for the proposed model generalization capability. As a result, the validated and trusted value (value that will be taken as reference to report the proposed model performance) for R2 Score value is 0.8772. The difference of 0.07 between the initial R2 Score value (0.95) and R2 Score value obtained through cross-validation (0.8772) suggests the fact that the proposed model is quite stable.
In addition, a sensitivity analysis of the proposed HGBRCond model parameters was conducted in order to demonstrate that the model is well calibrated and robust. In this sense, how each model settings (learning-rate, max_depth, max_iter, min_samples_leaf, L2_regularization and max_bins) influence the proposed model performance (respective the variation in R2 Score, RMSE and MAE performance metrics) was analyzed. In this sense, the parameters optimal values obtained by cross-validation (k-fold) were used, presented in Table 8, due to the fact that through cross-validation was obtained the model settings, which ensures, a robust performance and model generalization (therefore, the sensitivity analysis is more representative for the real behavior of the proposed model).
In Table 10, Table 11 and Table 12 the sensitivity analysis results for the HGBRCond model hyperparameters is presented, respectively, as are the model parameters impact on R2 Score, RMSE, and MAE metrics variation.

Table 10.
Sensitivity Analysis–Hyperparameters impact on R2 Score.

Table 11.
Sensitivity Analysis–Hyperparameters impact on RMSE.

Table 12.
Sensitivity Analysis–Hyperparameters impact on MAE.
As indicated in Table 10, the proposed model demonstrates high stability for the majority of tuning parameters. The only exception is min_samples_leaf, which has a moderate influence on model performance (sensitivity between 0.02 and 0.05) and needs precise tuning. Other parameters (learning_rate, max_depth, max_iter, L2_regularization, and max_bins) have low influence (<0.02), causing only minor variations in the model performance.
According to Table 11, learning_rate, max_iter, and L2_regularization have minimal impact on model performance, with RMSE changes ≤ 0.024, indicating reduced model sensitivity. Max_depth (RMSE = 0.2438) and max_bins (RMSE = 0.479) show moderate sensitivity and require more attention during tuning. The most critical parameter is min_samples_leaf (RMSE = 0.8826), which needs precise fine-tuning because its values in the range 9.71–10.58 can seriously affect model performance.
In Table 12, max_bins (MAE range = 1.026) and min_samples_leaf (MAE range = 0.971) have a major influence on MAE (small changes in these parameters produce significant variation). In contrast, L2_regularization, max_iter, max_depth, and learning_rate show low sensitivity, indicating that the model remains stable and MAE exhibits minor changes with their modification.
Therefore, for the majority of hyperparameters, the model remains stable under these parameters changes, the only sensible ones being min_samples_leaf and max_bins, which need to be more carefully tuned than the others.
Next, using the Morris method screening analysis [59,60], the results presented in Table 13 and Figure 10 were obtained.

Table 13.
Sensibility table.
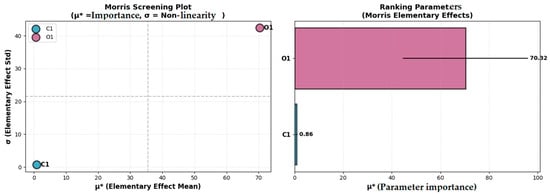
Figure 10.
Morris screening plot and ranking parameters.
According to Table 13 and Figure 10, the most influential parameter is O1 (), with moderate nonlinear effects (ratio = 0.60) and a relative importance of 98.79%.
The C1 parameter has a reduced influence on the model output () and a relative importance of only 1.2%, while presenting significant nonlinear behavior (ratio = 0.78).
In order to strengthen the affirmations about model robustness and stability were included confidence intervals (CI), as seen in Table 14 and Figure 11.

Table 14.
CI lower and upper intervals (selection).
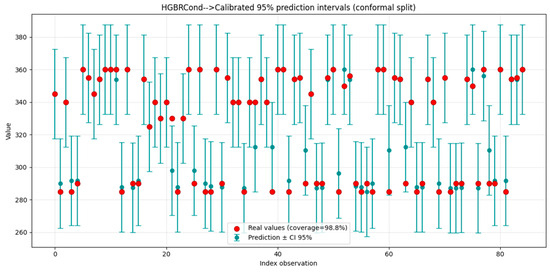
Figure 11.
HGBRCond calibrated 95% prediction intervals.
In Figure 11 are presented the calibrated 95% prediction intervals for HGBRCond model, obtained using split conformal method [59]. These intervals achieve a coverage of approximatively 98.8% of the real values, highlighting the fact that are well calibrated and adequately (are sufficient large enough to capture all the data real variations), providing a reliable basis for evaluating model resilience and consistency.
Regarding the HGBRCond model computational time, in the achieved experiments a time in the order of seconds (respectively, 63 s) was obtained. In the present study, we focused mainly on the HGBRCond model development, validation, robustness, various analysis and a brief comparison of the proposed model with alternative models, while a systematic comparison of the proposed model computational time with other model was considered for future work. HGBRCond model requires approximatively 63 s making all the stages (data loading and preprocessing, cross-validation, final model training, sensitivity analysis, predictions, supplementary analysis, etc.), whereas other studied models (RFR, SVR, KNR, RR, DTR, GBR, LR) have similar computational times, the differences being only of a few seconds. Therefore, these small computational times differences are practical negligible and do not affect the conclusions regarding the predictive performance and robustness of the proposed model.
Beyond its predictive performance, the proposed HGBRCond model is efficient and robust from computationally point of view because it uses histogram-based gradient boosting implementation, with a total computational time of 63 s (with a training time of 7.99 s and an efficient batch inference of 0.0735 s for test set predictions, showing stable cross-validation errors with well-calibrated 95% prediction intervals. Although the present study is limited to a representative experimental dataset (obtained from a pilot wastewater treatment plant), respectively, it used a pilot-scale dataset, a thorough model validation on real industrial data (including computational times obtained under production type conditions), representing a natural continuation of the present work (future work).
3.2.2. HGBRCond Model Feature Importance and SHAP Analysis
In Section 3.2.1 of the present paper was demonstrated that HGBRCond model is a valid one through cross-validation (is a good model—it works well on new data sets) and robust to parameters variations through sensitivity analysis (for the majority of the configuration settings).
In this section, feature importance in order to explain what makes the model to work well (model interpretability) was used, respectively, to determine which features has the greatest contribution to the model predictions.

Table 15.
HGBRCond model feature importance.
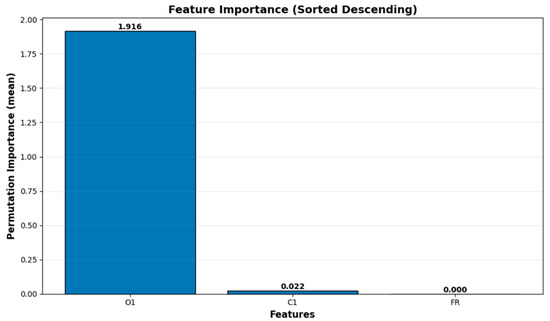
Figure 12.
HBGRCond model feature importance (Sorted descending).
As shown in Table 15 and Figure 12, the O1 variable has the highest contribution (importance = 1.9164; SD = 0.2545; relative importance = 98%) to HGBRCond model predictions, while the C1 feature presents a smaller but statistically significant contribution (importance = 0.0022; SD = 0.0148; relative importance = 1.13%). In addition, as it can be observed in the same table, the flowrate (FR) feature has zero variation and zero importance (importance = 0; SD = 0; relative importance = 0%), highlighting the fact that FR was kept constant in the experimental design. This does not mean that FR is physically irrelevant, rather, it does not contribute to the model due to the controlled experimental design. The variable importance analysis confirms the internal logic of the model, with O1 as the main predictor, C1 as a secondary predictor, while FR has no contribution to the model predictions (it is constant).
Feature importance analysis identified O1 as the dominant predictor and C1 as secondary one. In order to strengthen feature importance analysis results, SHAP analysis was also achieved, of which the results are presented in Table 16 and Figure 13, Figure 14, Figure 15 and Figure 16.

Table 16.
SHAP feature importance.
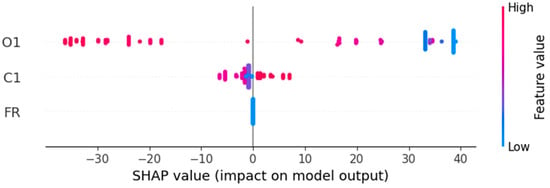
Figure 13.
SHAP summary plot.
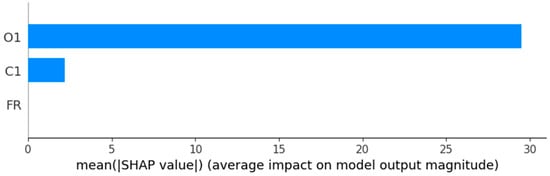
Figure 14.
Feature importance bar chart.
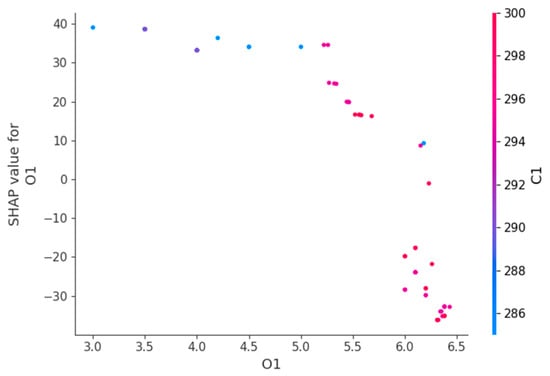
Figure 15.
Feature dependence plot for O1.
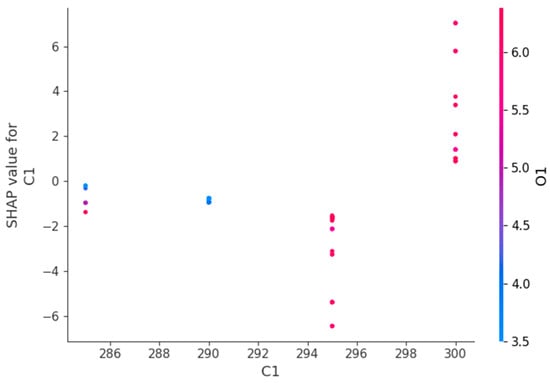
Figure 16.
Feature dependence plot for C1.
As seen in Table 16, Figure 13 and Figure 14, the results confirms the ones obtained by feature importance analysis.
Thus, O1 emerges as the dominant predictor, exhibiting the highest influence on the HGBRCond model output, with a mean absolute SHAP value of 29.49 μS/cm. Its contribution drives the largest share of both positive and negative prediction impact, accounting for 93.11% of the total variability in model predictions, O1 small variations producing significant chances in model predictions. The SHAP values for O1 varies between −36.20 and 39.04, having a mean contribution of 5.29 and a standard deviation of 30.38, a fact that highlights its impact substantial variability on different samples. This highlights that O1 feature has a critical role in target variable (C2) prediction, having both positive and negative influence depending on the concentration levels. The C1 feature has a model secondary impact (mean absolute SHAP value of 2.18 μS/cm), displaying a minor role (helpful but not essential), explaining only 6.88% of the predictions with a mean absolute SHAP value of 2.18 μS/cm, while the SHAP distribution is between −6.44 to 7.03 (negative influence on predictions). The flowrate (FR) feature has zero contribution (across all metrics) to the HGBRCond model predictions (can be eliminated, without performance loss, as it explains 0% of the predictions).
Figure 15 displays the O1 influence on the HGBRCond model predictions. Therefore, it has a positive contribution for low O1 values, meaning that O1 significantly increases the predicted results. Its positive effects diminishes as O1 increases, while at higher values of O1, the effect is negative, suggesting that O1 reduces the model prediction. The color gradient illustrates that the O1 impact on the predicted results depends on C1 level.
Figure 16 reports that C1 has a non-linear effect on the HGBRCond model predictions. Therefore, C1 has a negative or reduced impact at low values, while at high values it has a positive contribution to the model predictions. The color gradient indicates the fact that C1 influence depends on O1 level.
The comprehensive quantitative analyses provide rigorous statistical evidence of HGBRCond model robustness as shown in Table 17, consolidating all robustness results obtained through cross-validation, sensitivity analysis, Morris screening, SHAP analysis and confidence intervals, in one location.

Table 17.
Comprehensive quantitative model robustness and validation summary.
3.2.3. HGBRCond Model Predictions for Water Conductivity
In Section 3.2.1 of the present paper, it was demonstrated using validation procedure, that HGBRCond model is a valid and performant one. In addition, for the same model, a sensitivity analysis of the model parameters was conducted in order to demonstrate that it achieves robust performance. Taking into consideration all the elements presented in Section 3.2.1 and Section 3.2.2, it can be concluded that HGBRCond model can be used to achieve reliable predictions for various water physical–chemical essential parameters, such as conductivity (an essential indicator for water quality assessment and monitoring).
The predictions obtained for conductivity parameter (), using HGBRCond model, are those highlighted in Table 18.

Table 18.
Conductivity (C2) prediction using HGBRCond model.
According to Table 18, the predicted values for the target variable (conductivity-C2) using the input water flowrate (FR), the initial conductivity C1 (initial conductivity) and O1 (oxygen), are confirming the results obtained through chemical experiments. As can be observed, FR parameter is constant and it not contributing with information to the proposed model.
The correlation of statistical interpretations obtained using ML models and feature importance, SHAP analysis can have potential applicability in industrial processes because a conductivity prediction can optimize the purification process of contaminated waters by streamlining the consumption of reagents and energy. It could also be useful for the timely detection of failures in a WWTP, could identify accidental chemical contamination, or could generate chemical signals in the event of corrosion, chemical deposits or fluctuations of chemicals used in a plant for the control of chemical indicators in a water or steam treatment facility.
The application of ML in the control of industrial wastewater pollution effectively reduces the gap between the prediction of contamination by analyzing the concentration of pollutants and making decisions based on engineering information. The evolution of industrial technology can lead to the formation of new chemical compounds that may be difficult to detect by classical methods or their detection requires a long time and additional costs for the digitalization of the infrastructure, other pollutant monitoring solutions being presented in [13]. As shown in this study, an effective alternative for the analysis of pollutants would be the research methods using intelligent methods, as ML regression algorithms. Biodegradation of chemical pollutants can be achieved through bioremediation based on predictions made using ML methods.
4. Discussion
A sample whose BOD/COD ratio is higher than 0.3 is considered biodegradable. In the case of the present experiment, it can be stated that the ratio was between 0.68 and 0.48 which means that the tested water with mineral contamination was biodegradable. Considering that the used flowrate was constant, it must be taken into account that the described ML model can have potential application in industry, in systems with controlled feed flow.
In order to demonstrate that the proposed model HGBRCond is performant, valid, and that it can be used to make reliable predictions, a cross-validation (k-fold) technique was used. The model achieved R2 = 0.877 (95% CI: [0.855, 0.899]), indicating that approximately 88% of conductivity variance is explained. The prediction errors (RMSE = 10.23 μS/cm corresponding to 3.2% relative error; MAE = 4.85 μS/cm corresponding to 1.5% relative error) across the operational conductivity range (285–360 μS/cm), demonstrating sufficient precision for potential industrial applications.
Sensitivity analysis was conducted to evaluate HGBRCond model robustness across tuning parameters variations. The analysis revealed that HGBRCond has stable performance (R2 Score, RMSE, MAE) across most hyperparameters (learning_rate, max_depth, max_iter, L2_regularization), but presents critical sensitivity to min_samples_leaf and max_bins (requires careful tuning of these two parameters). This sensitivity is particularly important given the modest dataset size (n = 424) because suboptimal min_samples_leaf leads to model overfitting, while poor max_bins compromises model’s ability to capture nonlinear oxygen-conductivity dynamics.
Although cross-validation demonstrated the overall performance of the HGBRCond model and sensitivity analysis highlighted prediction robustness, these methods do not provide information about individual feature contributions. Feature importance analysis revealed O1 as main predictor, reflecting its direct physical role in oxygen-conductivity dynamics. The negligible contribution of C1 (1.13%) suggests that this feature supplies minimal additional predictive information in the experimental context. Because of its lack of importance due to its constant value with the experimental design, FR feature can be eliminated from the model (it must be mentioned, that this conclusion about FR cannot be generalized).
The HGBRCond model performs well from a chemical-biological perspective due to its histogram binning and gradient boosting based architecture that captures the conductivity behavior in biodegradation process, respectively, O1-driven mineralization (SHAP > 0.45) corresponds to aerobic kinetics and histogram efficiency (max_bins = 128, min_samples_leaf = 20) handles nonlinear interactions on used dataset (n = 424). These aspects lead to an R2 value of 0.8772 (SD ± 0.0110) with stable 95% CI: [0.855, 0.899] and 63 s total runtime.
The authors acknowledge several important limitations of the HGBRCond model. The model shows strong dependence on O1 concentration measurements, making it susceptible to oxygen sensor malfunctions. It also presents high influence to critical parameters (max_bins and min_samples_leaf), requiring careful tuning to avoid overfitting. Furthermore, the model performance is constrained by the limited dataset size (n = 424) and narrow conductivity range (285–360 μS/cm), which limit its applicability under diverse operational conditions. The constant flowrate (FR) maintained during controlled experiments prevents the model from learning flow-dependent dynamics, restricting its use in variable flowrate scenarios. Finally, the model requires recalibration and validation before potential industrial implementation, as it was optimized exclusively for controlled biodegradation settings.
All these model limitations are reflecting the controlled nature of the experimental design and the evident need for model validation under various operating conditions. Overall, the model could fail if O1 sensor failure occurs, FR regime shift, parameters overfitting and industrial scaling without feature recalibrations.
Next, it was strategically enhanced the discussion regarding a more structured, comparative discussion (data type, experimental vs. industrial context, predictor variables and error scales) in a concise comparative table (Table 19) focusing on different key metrics.

Table 19.
HGBRCond model comparative analysis.
Table 19 addresses the following aspects: data type (pilot vs. industrial), context, predictors, error scales, and HGBRCond model advantages. The comparative analysis from Table 19 suggests that the HGBRCond model has multiple advantages for possible practical industrial applications such as
- While LSTM (R2 = 0.88), XGBoost (R2 = 0.82), and ANN combined with PCA (R2 = 0.88) supplies comparable or superior performance, they are operating in batch context, full-scale or using historical data, limiting their applicability in continuous monitoring; on the other hand, HGBRCond model with R2 = 0.877 uses pilot data, that allows its testing and validation before possible implementation at industrial scale;
- The HGBRCond model’s main advantage is stability, highlighted through cross-validation (SD = 0.011) and sensitivity analysis;
- Unlike other complex models such as Hybrid CNN-LSM (RMSE = 53.83 µS/cm) or LSTM which are require high computational resources and have high computational complexity, HGBRCond model uses simple and measurable predictors (such as O1 and C1), eliminating the necessity of complex laboratory analysis or multiple parameters (such as DO, pH, BOD, COD or NH4.).
So, this targeted comparison demonstrates HGBRCond superior interpretability (SHAP analysis, Morris screening, feature importance) and real-time feasibility (efficient batch inference of 0.0735 s for test set predictions) versus literature XGBoost/GBR, despite smaller pilot dataset. Also, HGBRCond distinguishes itself through an optimal combination of competitive accuracy (R2 = 0.877; SD = 0.011), superior computational efficiency, stable interpretability guaranteed by SHAP, native handling of missing values, high-speed training and real-time inference, eliminating the need for complex preprocessing techniques (like PCA) and offering superior scalability compared to LSTM, GPR, and traditional ensemble methods, at reduced operational costs.
While XGBoost [13] and GBR [12] achieve an R2 = 0.82 on industrial data, HGBRCond model provides a comparable accuracy (R2 = 0.877) presenting several advantages, such as it operates only with two predictors (O1-main predictor and C1-secondary predictor); therefore, it minimized the costs with sensors infrastructure and maintenance complexity, while XGBoost requires multiple parameters (DO, pH, BOD) for full-scale operations. Compared with traditional GBR, HGBRCond histogram-based splitting algorithm provide faster training times (63 s total runtime versus typical GBR training times that are approximatively between 150 and 300 s for a comparable dataset size) and improved handling of missing values without requiring additional preprocessing. Also, HGBRCond ensures improved interpretability through consistent performance (95% CI: [0.855, 0.899]), while XGBoost and GBR models are usually operating as black-boxes models.
In addition, a comparative analysis of HGBRCond model with XGBoost and traditional GBR model, using as criteria accuracy, limitations, training efficiency, parameter complexity, stability (SD), missing value handling, interpretability, 95% CI and scalability was achieved in Table 20.

Table 20.
Comparative Analysis of XGBoost, traditional GBR, and HGBRCond Models.
It must be mentioned that the training efficiency and stability values for XGBoost [13] and traditional GBR [12] were approximated based on typical performance with sklearn implementations on comparable dataset sizes (n ≈ 400–500), while HGBRCond training efficiency and stability values were measured in the current study.
Therefore, analyzing the results presented in Table 20, HGBRCond model has several advantages over XGBoost and traditional GBR methods, such as
- Faster training efficiency (63 s vs. 150–400 s for comparable dataset sizes) ensuring rapid model development and iterative optimization;
- It operates with 67% fewer parameters, requiring only two measurable predictors (O1 and C1) compared to 4–6 features (DO, pH, BOD, COD, NH4) typically needed by XGBoost and GBR, substantially reducing sensor infrastructure costs and system complexity;
- It has 50–78% more stable predictions (SD = 0.011 vs. 0.02–0.04 for ensemble methods), demonstrating superior robustness;
- Provides native missing value handling;
- Superior interpretability through SHAP screening, detecting O1 dominance (98% relative importance);
- It achieves 6.8% higher accuracy (R2 = 0.877 vs. 0.82) despite being trained on pilot-scale data rather than full industrial datasets, demonstrating a good performance.
Unlike many existing ML applications in the water domain, which focus on general water parameters such as groundwater levels, consumption, or potability, this study addresses water conductivity prediction, highlighting the specific challenges and novelty of the task [27,28,29,30].
Overall, the HGBRCond model efficiently predicts water conductivity using only two key features (O1 and C1), with O1 as dominant predictor. It supplies stable predictions, handles missing values, and requires fewer parameters than traditional GBR or XGBoost models. These results highlight that careful feature selection and computationally efficient algorithms can provide interpretable and robust predictions, while performance may vary under different datasets sizes or operational industrial conditions.
5. Conclusions
The limitations of the existing state of the art consists of the lack of comprehensive interpretability frameworks and systematic validation protocols for controlled biodegradation contexts (XGBoost and GBR models with R2 = 0.82 [12,13]), high computational complexity and reduced interpretability (LSTM models [13]). Also, the lack of multi-level interpretability integration (SHAP, Morris screening, confidence intervals) necessary for process understanding (Hybrid CNN-LSM [12]), limitation to specific industrial contexts without robust cross-validation or sensitivity analysis frameworks (GPR [12]) and, limited features interpretability (ANN + PCA [32]).
The current study methodological limitation refers to the lack of explicit integration of chemical-biological knowledge. The chemical parameters were treated as standard numerical inputs without mechanistic constraints, limiting explicit chemical–biological knowledge integration. Because of the fact that HGBRCond model relies on data-driven patters, future work will focus explicitly on the integration of chemical–biological mechanisms (stoichiometry, Michaelis–Menten kinetics) to enhance the model interpretability and extrapolation.
This study contributes to the state of the art by developing a robust, hybrid Histogram-based Boosting Regression model, referred to as HGBRCond, for water conductivity prediction in controlled biodegradation processes, integrating a rigorous statistical validation framework (10-fold cross-validation, sensitivity analysis, CI) that is not applied in the analyzed studies [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35]. In addition, it integrates multi-level interpretability (combines feature importance, Morris screening, SHAP analysis), addressing the interpretability gap from previous approaches (LSTM, CNN-LSM, XGBoost, ANN + PCA) [13,14,32]. Also, HGBRCond model achieves competitive accuracy (R2 = 0.877) through hybrid ensemble (that combines the computational efficiency of histogram-based gradient boosting with systematic hyperparameter optimization), overcoming the existing models performance (XGBoost and GBR models with R2 = 0.82) and limitations [12,13].
The proposed HGBRCond model achieves approximately 6.8% improvement in R2 compared to XGBoost/GBR industrial models while providing explicit interpretability mechanisms absent in the analyzed approaches [12,13]. Compared to LSTM [13], it has a comparable accuracy with significantly reduced computational complexity and improved process interpretability through SHAP and Morris analysis.
Although the HGBRCond model has good predictive performance (R2 = 0.8772 ± 0.0110, RMSE = 10.24 ± 0.54) under controlled conditions, its performance depends on certain factors, such as the model’s strong sensitivity to O1 variable (sensor failures can lead to prediction errors). Also, a small data set (n = 424), constant flowrate (lack of contribution to the model because of controlled experimental design) and, sensitivity to hyperparameters (such as min_samples_leaf and max_bins) that requires careful tuning to avoid overfitting. In addition, the model accuracy can be reduced by variation in flow condition or by the direct industrial scaling without feature recalibration. Overall, the HGBRCond model is robust within its validated domain but it requires careful tuning and monitoring when applied in industrial conditions.
Synthesizing, the HGBRCond model has several limitations, arising from the controlled experimental conditions and dataset characteristics. The model has a strong dependence on O1, so the oxygen sensor failures can cause prediction errors. In addition, it has a strong sensitivity to min_samples_leaf and max_bins critical hyperparameters that requires careful hyperparameter tuning in order to avoid overfitting. Its performance is constrained by a limited dataset (n = 424) and a limited conductivity range (285–360 μS/cm), limiting model applicability under diverse operational conditions. The controlled experimental design resulted in a constant flowrate (FR) makes the model incapable of learning from flow-dependent dynamics, limiting its applicability to variable flowrates conditions. All these limitations indicate that HGBRCond model is optimized for controlled biodegradation setting, requiring recalibration and validation for industrial conditions application.
Future work will first prioritize full-scale validation of the HGBRCond model in operational treatment plants to assess performance under real conditions, across multiple flowrates and conductivity ranges using industrial wastewater samples. In addition, it will achieve model extension in order to predict key water quality parameters (COD, BOD, pH—parameters measured and evaluated experimentally within the current study—Section 3.1.2). A comparative performance analysis will be made in order to compare the model computational efficiency against alternative models, under real operational constraints. Systematic ablation studies will measure model performance degradation under suboptimal hyperparameters choices, providing practical guidance for model tuning and potential industrial implementation settings with larger datasets.
Future work strategy include an experimental validations and model extension plan that contains:
- Model validation across multiple flowrates and extended conductivity ranges using controlled experimental conditions; model testing on real industrial wastewater data to evaluate its performance under real conditions; full-scale validation in operational industrial plants; model computational performance evaluation under real production conditions (experimental validation plan);
- Model extension in order to predict additional wastewater parameters (such as COD, BOD, pH) for a more detailed analysis of wastewater treatment efficiency; the development of dedicated ML-based models for specific toxic pollutants (pesticides, phenols, cyanides, petroleum tars) biodegradation prediction; the computational time comparison with alternative models under industrial operating conditions (model extension strategy).
In conclusion, the paper proposes a validated modeling framework for water conductivity prediction under controlled conditions, respectively, a valid, robust, and well-calibrated model referred as HGBRCond. In addition, the obtained results are providing evidence for the potential of this methodology and establishes a foundation for expanded future studies, that are necessary to rigorously assess scalability and model potential industrial applicability.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app16020694/s1.
Author Contributions
Conceptualization, M.C. and C.G.G.; methodology M.C. and C.G.G.; software, M.C.; validation, M.C.; formal analysis, M.C. and C.G.G.; investigation, M.C. and C.G.G.; resources, M.C. and C.G.G.; data curation, M.C.; writing—original draft preparation, M.C. and C.G.G.; writing—review and editing, M.C.; visualization, M.C.; supervision, M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by internal funding from Petroleum-Gas University of Ploiesti, Romania.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ANN | Artificial Neural Network |
| BOD | Biochemical oxygen demand |
| COD | Chemical oxygen demand |
| CNN-LSTM | Convolutional Neural Network-Long Short-Term Memory |
| CI | Confidence Intervals |
| CV | Coefficient of variation |
| DTR | Decision Tree Regression |
| DO | Dissolved oxygen |
| EVS | Explained variance score |
| FR | Water flowrate |
| GB | Gradient Boosting model |
| GBR | Gradient Boosting Regression |
| CFU/mL | Total bacterial colony count |
| GPR | Gaussian process regression |
| HGBR | Histogram-based Gradient Boosting Regression |
| HGBRCond | Histogram-based Gradient Boosting Regression proposed mathematical model for water conductivity prediction |
| IDS | Intelligent Digital Sensor |
| KNR | KNeighbors Regression |
| KNN | k-nearest neighbors |
| LR | Linear Regression |
| LSTM | Long Short-Term Memory |
| MAE | Mean absolute error |
| MAPE | Mean absolute percentage error |
| MedAE | Median absolute error |
| ML | Machine Learning |
| MLP | Multilayer perceptron |
| MM | Mineral medium |
| OECD | Guidelines for the testing of chemicals of the Organization for Economic Cooperation and Development |
| PCA | Principal Component Analysis |
| R2 Score | Coefficient of determination |
| RFR | Random Forest Regression |
| RMSE | Root Mean Square Error |
| RR | Ridge Regression |
| SD | Standard Deviation |
| SVR | Support Vector Regression |
| SVM | Support Vector Machine |
| SHAP | SHapley Additive exPLanations |
| TOC | Total organic carbon |
| TEM | Transmission electron microscopy |
| TDS | Total Dissolved Solids |
| WWTP | Wastewater treatment plants |
| WQI | Water quality index |
References
- Chen, M.; Li, Y.; Jiang, X.; Zhao, D.; Liu, X.; Zhou, J.; He, Z.; Zheng, C.; Pan, X. Study on soil physical structure after the bioremediation of Pb pollution using microbial-induced carbonate precipitation methodology. J. Hazard. Mater. 2021, 411, 125103. [Google Scholar] [CrossRef]
- Chang, Y.C.; Peng, Y.-P.; Chen, K.-F.; Chen, T.-Y.; Tang, C.-T. The effect of different in situ chemical oxidation (ISCO) technologies on the survival of indigenous microbes and the remediation of petroleum hydrocarbon-contaminated soil. Process Saf. Environ. Prot. 2022, 163, 105–115. [Google Scholar] [CrossRef]
- OECD. Guideline for Testing of Chemicals-301, Adopted by Council on 17 July 1992. Available online: https://www.google.ro/books/edition/OECD_Guidelines_for_the_Tesing_of_Chemi/7s5yoSa3vykC?hl=en&gbpv=1&printsec=frontcover (accessed on 15 October 2025).
- Su, Y.; Cheng, Z.; Hou, Y.; Lin, S.; Gao, L.; Wang, Z.; Bao, R.; Peng, L. Biodegradable and conventional microplastics posed similar toxicity to marine algae Chlorella vulgaris. Aquat. Toxicol. 2022, 244, 106097. [Google Scholar] [CrossRef]
- Murdock, J.N.; Wetzel, D. FT-IR Microspectroscopy Enhances Biological and Ecological Analysis of Algae. Appl. Spectrosc. Rev. 2009, 44, 335–361. [Google Scholar] [CrossRef]
- Traverso-Soto, J.M.; Figueredo, M.; Punta-Sánchez, I.; Campana, O.; Ciufegni, E.; Hampel, M.; Buoninsegni, J.; Quiñones, M.A.M.; Anfuso, G. Assessment of Organic Pollutants Desorbed from Plastic Litter Items Stranded on Cadiz Beaches (SW Spain). Toxics 2025, 13, 673. [Google Scholar] [CrossRef]
- Davis, A.B.; Evans, M.; McKindles, K.; Lee, J. Co-Occurrence of Toxic Bloom-Forming Cyanobacteria Planktothrix, Cyanophage, and Symbiotic Bacteria in Ohio Water Treatment Waste: Implications for Harmful Algal Bloom Management. Toxins 2025, 17, 450. [Google Scholar] [CrossRef]
- Renganathan, P.; Gaysina, L.A.; Gutiérrez, C.G.; Puente, E.O.R.; Sainz-Hernández, J.C. Harnessing Engineered Microbial Consortia for Xenobiotic Bioremediation: Integrating Multi-Omics and AI for Next-Generation Wastewater Treatment. J. Xenobiot. 2025, 15, 133. [Google Scholar] [CrossRef] [PubMed]
- Wolff, D.; Krah, D.; Dötsch, A.; Ghattas, A.; Wick, A.; Ternes, T. Insights into the variability of microbial community composition and micropollutant degradation in diverse biological wastewater treatment systems. Water Res. 2018, 143, 313–324. [Google Scholar] [CrossRef]
- Saini, S.; Tewari, S.; Dwivedi, J.; Sharma, V. Biofilm-mediated wastewater treatment: A comprehensive review. Mater. Adv. 2023, 4, 1415–1443. [Google Scholar] [CrossRef]
- Xiong, H.; Zhou, X.; Cao, Z.; Xu, A.; Dong, W.; Jiang, M. Microbial biofilms as a platform for diverse biocatalytic applications. Bioresour. Technol. 2024, 386, 129396. [Google Scholar] [CrossRef]
- Negri, F.; Galeazzi, A.; Gallo, F.; Manenti, F. Reshaping Industrial Maintenance with Machine Learning: Fouling Control Using Optimized Gaussian Process Regression. Ind. Eng. Chem. Res. 2025, 64, 6633–6654. [Google Scholar] [CrossRef]
- Li, Y.; Xu, J.; Anastasiu, D.C. An Extreme-Adaptive Time Series Model Based on Probability-Enhanced LSTM Neural Networks. Proc. AAAI Conf. Artif. Intell. 2023, 37, 8684–8691. [Google Scholar] [CrossRef]
- Karbasi, M.; Ali, M.; Bateni, S.M.; Jun, C.; Jamei, M.; Farooque, A.A.; Yaseen, Z.M. Multi-step ahead forecasting of electrical conductivity in rivers by using a hybrid Convolutional Neural Network-Long Short-Term Memory (CNN-LSTM) model enhanced by Boruta-XGBoost feature selection algorithm. Dent. Sci. Rep. 2024, 14, 1991. [Google Scholar] [CrossRef]
- Hridoy, A.M.; Shawkat, A.I.; Bordin, C.; Acharjee, M.R.; Masood, A.; Baki, A.O.; Al Mamun, A. Advanced machine learning models for accurate water quality classification and WQI prediction: Implications for aquatic disease risk management. Sci. Total Environ. 2025, 1008, 180965. [Google Scholar] [CrossRef]
- Cechinel, M.A.P.; Neves, J.; Fuck, J.V.R.; de Andrade, R.C.; Spogis, N.; Riella, H.G.; Padoin, N.; Soares, C. Enhancing wastewater treatment efficiency through machine learning-driven effluent quality prediction: A plant-level analysis. J. Water Process Eng. 2024, 58, 104758. [Google Scholar] [CrossRef]
- Dikmen, F.; Demir, A.; Özkaya, B.; Raza, M.O.; Rasheed, J.; Asuroglu, T.; Alsubai, S. AI-driven wastewater management through comparative analysis of feature selection techniques and predictive models. Sci. Rep. 2025, 15, 25347. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Wang, J.; Ye, G.; Wang, Y. Data-driven prediction of effluent quality in wastewater treatment processes: Model performance optimization and missing-data handling. J. Water Process Eng. 2025, 71, 107352. [Google Scholar] [CrossRef]
- Lv, J.; Du, L.; Lin, H.; Wang, B.; Yin, W.; Song, Y.; Chen, J.; Yang, J.; Wang, A.; Wang, H. Enhancing effluent quality prediction in wastewater treatment plants through the integration of factor analysis and machine learning. Bioresour. Technol. 2024, 393, 130008. [Google Scholar] [CrossRef]
- Yin, H.; Chen, Y.; Zhou, J.; Xie, Y.; Wei, Q.; Xu, Z. A probabilistic deep learning approach to enhance the prediction of wastewater treatment plant effluent quality under shocking load events. Water Res. X 2025, 26, 100291. [Google Scholar] [CrossRef] [PubMed]
- Fitriyani, N.; Syafrudin, M.; Chamidah, N.; Rifada, M.; Susilo, H.; Aydin, D.; Qolbiyani, S.L.; Lee, S.W. A Novel Approach Utilizing Bagging, Histogram Gradient Boosting, and Advanced Feature Selection for Predicting the Onset of Cardiovascular Diseases. Mathematics 2025, 13, 2194. [Google Scholar] [CrossRef]
- Zamfir, F.-S.; Carbureanu, M.; Mihalache, S.F. Application of Machine Learning Models in Optimizing Wastewater Treatment Processes: A Review. Appl. Sci. 2025, 15, 8360. [Google Scholar] [CrossRef]
- Grbčić, L.; Druzeta, S.; Kranjčević, L. Water distribution network leak localization with histogram-based gradient boosting histogram-based gradient boosting water network leak localization. J. Hydroinform. 2023, 25, 663–684. [Google Scholar] [CrossRef]
- Makumbura, R.K.; Mampitiya, L.; Rathnayake, N.; Meddage, D.; Henna, S.; Dang, T.L.; Hoshino, Y.; Rathnayake, U. Advancing Water Quality Assessment and Prediction Using Machine Learning Models, Coupled with Explainable Artificial Intelligence (XAI) Techniques Like Shapley Additive Explanations (SHAP) For Interpreting the Black-Box Nature. Results Eng. 2024, 23, 102831. [Google Scholar] [CrossRef]
- Bhuria, R.; Gill, K.S.; Upadhyay, D.; Devliyal, S. Predicting Water Purity by Riding the Ensemble Waves with Gradient Boosting Classification Technique. In Proceedings of the 2024 2nd International Conference on Sustainable Computing and Smart Systems (ICSCSS), Coimbatore, India, 10–12 July 2024; pp. 1365–1368. [Google Scholar] [CrossRef]
- Nagarajan, G.; Reddy, N.K.; Kumar, Y.V.; Reddy, A.; Thota, C. Water Quality Classification Using XG Boost. In Proceedings of the 2024 4th International Conference on Trends in Quantum Computing and Emerging Business Technologies (TQCEBT), Pune, India, 22–23 March 2024; Volume 190, pp. 1–3. [Google Scholar] [CrossRef]
- Sharma, J.; Gill, K.S.; Kumar, M. Innovating Water Purity Analysis with Gradient Boosting Classification Techniques. In Applied Intelligence and Computing; SCRS: Delhi, India, 2023; pp. 159–168. [Google Scholar] [CrossRef]
- Sattari, M.T.; Mirabbasi, R.; Shamsi Sushab, R.; Abraham, J. Prediction of Groundwater Level in Ardebil Plain Using Support Vector Regression and M5 Tree Model. Ground Water 2018, 56, 636–646. [Google Scholar] [CrossRef]
- Ainapure, B.; Baheti, N.; Buch, J.; Appasani, B.; Jha, A.V.; Srinivasulu, A. Drinking water potability prediction using machine learning approaches: A case study of Indian rivers. Water Pract. Technol. 2023, 18, 3004–3020. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Le, H.T.T. Water Level Prediction at TICH-BUI River in Vietnam Using Support Vector Regression. In Proceedings of the 2019 International Conference on Machine Learning and Cybernetics (ICMLC), Kobe, Japan, 7–10 July 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Sarkar, H.; Goriwale, S.S.; Ghosh, J.K.; Ojha, C.S.P.; Ghosh, S.K. Potential of machine learning algorithms in groundwater level prediction using temporal gravity data. Groundw. Sustain. Dev. 2024, 25, 101114. [Google Scholar] [CrossRef]
- Oliveira-Esquerre, K.P.; Mori, M.; Bruns, R. Simulation of an industrial wastewater treatment plant using artificial neural networks and principal components analysis. Braz. J. Chem. Eng. 2002, 19, 365–372. [Google Scholar] [CrossRef]
- Tchobanoglous, G.; Burton, F.L.; Stensel, H.D. Wastewater Engineering: Treatment and Reuse, 4th ed.; McGraw-Hill: New York, NY, USA, 2003. [Google Scholar]
- Prabu, P.; Alluhaidan, A.S.; Aziz, R.; Basheer, S. AquaFlowNet a machine learning based framework for real time wastewater flow management and optimization. Sci. Rep. 2025, 15, 19182. [Google Scholar] [CrossRef]
- Rasool, J.M.; Somashekar, J.A. A Comprehensive Review of Machine Learning Applications in Wastewater Treatment: Current State, Comparative Analysis, and Future Directions. J. Innov. Technol. 2025, 2025, 1–14. [Google Scholar] [CrossRef]
- Hossen, A.M.; Salam, T. Advancing Water Quality Assessment: Leveraging XGBoost for Precise Predictive Modeling; A Machine Learning Technique. In Proceedings of the 2024 IEEE International Conference on Computing, Applications and Systems (COMPAS), Chattogram, Bangladesh, 5–26 September 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Gheorghe, C.G.; Dusescu, C.; Carbureanu, M. Asphaltenes biodegradation in biosystems adapted on selective media. Rev. Chim. 2016, 67, 2106–2110. [Google Scholar]
- Popovici, D.R.; Gheorghe, C.G.; Dusescu Vasile, C.M. Assessment of the Active Sludge Microorganisms Population During Wastewater Treatment in a Micro-Pilot Plant. Bioengineering 2024, 11, 1306. [Google Scholar] [CrossRef]
- Eshamuddin, M.; Zuccaro, G.; Nourrit, G.; Albasi, C. The influence of process operating conditions on the microbial community structure in the moving bed biofilm reactor at phylum and class level: A review. J. Environ. Chem. Eng. 2024, 12, 113266. [Google Scholar] [CrossRef]
- Gheorghe, C.G.; Dusescu-Vasile, C.M.; Popovici, D.R.; Bombos, D.; Dragomir, R.E.; Dima, F.M.; Bajan, M.; Vasilievici, G. Monitoring the Biodegradation Progress of Naphthenic Acids in the Presence of Spirulina platensis Algae. Toxics 2025, 13, 368. [Google Scholar] [CrossRef]
- Manga, M.; Boutikos, P.; Semiyaga, S.; Olabinjo, O.; Muoghalu, C.C. Biochar and its potential application for the improvement of the anaerobic digestion process: A critical review. Energies 2022, 16, 4051. [Google Scholar] [CrossRef]
- Hassan, A.; Hamid, F.; Pariatamby, A.; Suhaimi, N.; Razali, N.; Ling, K.; Mohan, P. Bioaugmentation-assisted bioremediation and biodegradation mechanisms for PCB in contaminated environments: A review on sustainable clean-up technologies. J. Environ. Chem. Eng. 2023, 11, 110055. [Google Scholar] [CrossRef]
- Chakraborty, S.; Talukdar, A.; Dey, S.; Bhattacharya, S. Role of fungi, bacteria and microalgae in bioremediation of emerging pollutants with special reference to pesticides, heavy metals and pharmaceuticals. Discov. Environ. 2025, 3, 91. [Google Scholar] [CrossRef]
- Yang, Z.; Peng, C.; Cao, H.; Song, J.; Gong, B.; Li, L.; Wang, L.; He, Y.; Liang, M.; Lin, J.; et al. Microbial functional assemblages predicted by the FAPROTAX analysis are impacted by physicochemical properties, but C, N and S cycling genes are not in mangrove soil in the Beibu Gulf, China. Ecol. Indic. 2022, 139, 108887. [Google Scholar] [CrossRef]
- Tyagi, I.; Tyagi, K.; Ahamad, F.; Bhutiani, R.; Kumar, V. Assessment of bacterial community structure, associated functional role, and water health in full-scale municipal wastewater treatment plants. Toxics 2024, 13, 3. [Google Scholar] [CrossRef]
- La Cognata, R.; Piazza, S.; Freni, G. Pollutant Monitoring Solutions in Water and Sewerage Networks: A Scoping Review. Water 2025, 17, 1423. [Google Scholar] [CrossRef]
- Carbureanu, M.; Roșca, C.-M. Evaluating Wastewater pH Prediction Solutions in Custom Python and C# Models. In Proceedings of the 5th International Conference on Emerging Trends and Technologies on Intelligent Systems, Noida, India, 27–28 March 2025; pp. 19–21. [Google Scholar]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random Forest: A Classification and Regression Tool for Compound Classification and QSAR Modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef]
- Srisuradetchai, P.; Suksrikran, K. Random kernel k-nearest neighbors’ regression. Front. Big Data 2024, 7, 1402384. [Google Scholar] [CrossRef]
- Schreiber-Gregory, D.N. Ridge Regression and Multicollinearity: An In-Depth Review. Model. Assist. Stat. Appl. 2018, 13, 359–365. [Google Scholar] [CrossRef]
- Kassim, N.M.; Santhiran, S.; Alkahtani, A.A.; Islam, M.A.; Tiong, S.K.; Mohd Yusof, M.Y.; Amin, N. An Adaptive Decision Tree Regression Modeling for the Output Power of Large-Scale Solar (LSS) Farm Forecasting. Sustainability 2023, 15, 13521. [Google Scholar] [CrossRef]
- Singh, U.; Rizwan, M.; Alaraj, M.; Alsaidan, I. A Machine Learning-Based Gradient Boosting Regression Approach for Wind Power Production Forecasting: A Step towards Smart Grid Environments. Energies 2021, 14, 5196. [Google Scholar] [CrossRef]
- Chicco, D.; Warrens, M.J.; Jurman, G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Comput. Sci. 2021, 7, e623. [Google Scholar] [CrossRef]
- Cap. 2.6.12. Biological tests. Microbial examination of nonsterile products Total viable aerobic count Plate count methods. In European Pharmacopoeia 5.0; Council of Europe: Strasbourg, France, 2004; p. 154.
- Validation of microbial recovery from pharmacopeia articles cap 1227 Estimating the number of colony forming units. In USP Pharmacopoeia 29; The United States Pharmacopeia Convention: Frederick, MD, USA, 2021.
- SR EN ISO 5667-15:2010; Calitatea apei. Prelevare. Partea 15: Ghid General Pentru Conservarea şi Tratarea Probelor de Nămol şi Sediment. Asociația Română de Standardizare ASRO: București, Romania, 2010.
- Friedman, J. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3147. [Google Scholar]
- Oliveira, R.I.; Orenstein, P.; Ramos, T.; Romano, J.V. Split conformal prediction and non-exchangeable data. J. Mach. Learn. Res. 2024, 25, 1–38. [Google Scholar]
- Morris, M.D. Factorial Sampling Plans for Preliminary Computational Experiments. Technometrics 1991, 33, 161–174. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.