Abstract
The human microbiome plays a critical role in health and disease, with recent innovations in microbiome research offering groundbreaking insights that could reshape the future of healthcare. This study explored emerging methodologies, such as long-read sequencing, culturomics, synthetic biology, machine learning, and AI-driven diagnostics, that are transforming the field of microbiome–host interactions. Unlike traditional broad-spectrum approaches, these tools enable precise interventions, such as detecting foodborne pathogens and remediating polluted soils for safer agriculture. This work highlights the integration of interdisciplinary approaches and non-animal models, such as 3D cultures and organ-on-a-chip technologies, which address the limitations of current research and present ethical, scalable alternatives for microbiome studies. Focusing on food safety and environmental health, we examine how microbial variability impacts pathogen control in food chains and ecosystem resilience, integrating socioeconomic and environmental factors. The study also emphasizes the need to expand beyond bacterial-focused microbiome research, advocating for the inclusion of fungi, viruses, and helminths to deepen our understanding of therapeutic microbial consortia. The combination of high-throughput sequencing, biosensors, bioinformatics, and machine learning drives precision strategies, such as reducing food spoilage and enhancing soil fertility, paving the way for sustainable food systems and environmental management. Hence, this work offers a comprehensive framework for advancing microbiome interventions, providing valuable insights for researchers and professionals navigating this rapidly evolving field.
1. Introduction
Microbiome-based interventions encompass a range of strategies aimed at modulating microbial communities to promote health and environmental sustainability [1,2,3,4]. “Microbiome-based interventions” is an umbrella term encompassing all strategies aimed at altering microbiota for beneficial outcomes across health, food, and environmental contexts. On the other hand, “microbiome-based therapeutics (MBT)” is a specific subset focused on targeted applications to prevent or treat diseases, especially within clinical or medical frameworks. Hence, this review uses the term MBT within the broader context of microbiome interventions applicable to both food safety and environmental health. These interventions include approaches such as fecal microbiota transplantation, administration of probiotics or prebiotics, dietary modifications, and the use of bacterial consortia [1]. The term has sometimes been used to describe microbiome-based solutions for approaches to address new and existing threats to food security, nutrition, health, and agrifood systems’ sustainability [5]. Specifically, MBT is the modification and application of microbial communities with the potential to improve health and prevent diseases [6,7]. Microbiome-based interventions leverage the complexity of symbiotic relationships between diverse microbial communities—bacteria, viruses, fungi, and archaea—to enhance food safety and environmental health, targeting challenges like pathogen contamination in food production and ecosystem degradation [8]. For example, probiotics reduce Salmonella in livestock, ensuring safer meat [9], while bioremediation microbes detoxify oil spills, supporting clean soil and water for agriculture [10]. The ability of microbial communities to interact dynamically with their hosts and habitat provides a strong structural basis for their therapeutic potential [8]. In food systems, MBT minimizes spoilage and foodborne infections, with long-read sequencing pinpointing pathogens like E. coli in beef [11] and synthetic biology engineering microbes to degrade pollutants faster [12].
The role of microbiomes in food safety is critical, as they regulate pathogen growth and food quality. The microbiome is a complex ecosystem of microorganisms, including bacteria, viruses, fungi, and archaea, that inhabit a particular environment, such as the human gut or the soil. Probiotics and prebiotics alter animal gut microbiomes, reducing pathogens and improving meat safety [9], while artificial intelligence (AI) predicts contamination risks across supply chains [13]. In humans, the gut microbiome aids digestion, metabolism, and immune regulation, with dysbiosis linked to health issues like obesity and diabetes [14]. However, this review focuses on microbiome-based interventions’ broader applications, such as preventing foodborne illnesses and restoring ecosystems, rather than human health alone. From an agricultural/environmental perspective, mycorrhizal fungi are essential for plants to grow and stay healthy, and thus microbiome-based techniques such as biofertilization or biocontrol use friendly fungi to increase crop yields while reducing the need for artificial pesticides and fertilizers, as they stimulate the plants towards more environment-friendly farming methods [15].
Microbiome interventions enhance food safety by tackling pathogens at the source. For instance, bacteriophages target resistant bacteria in food production, curbing antimicrobial resistance (AMR) [16], while culturomics uncovers unculturable microbes for novel applications [17]. Microbiomes in the environment are very important for sustaining healthy ecosystems through their roles in the formation of soil structure, nutrient cycling, and bioremediation [10]. For example, the utilization of microbial agents, communities, or their metabolites in bioremediation transform contaminants (such as oil spills or heavy metal pollution) in the environment into less toxic compounds that are more easily degraded or removed from the environment [18,19]. The effective application of microbial communities in the environment has been used in the restoration of contaminated environments, pollution reduction, and improvement in ecosystem resilience to anthropogenic disturbances [20].
Microbiome research has evolved, and it is important to look at the journey of the methodologies that have shaped our understanding over the years. From the early days of culture-based techniques to today’s cutting-edge AI-driven research, these advancements have revolutionized how we study microbiome–pathogen interactions. Figure 1 illustrates briefly how microbiome research has progressed. The urgency to apply MBT strategically has intensified with rapid advancements in high-throughput sequencing and advanced bioinformatics, which enable more efficient and targeted microbial interventions [21,22]. Recent reviews highlight the transformative potential of integrating artificial intelligence (AI) and machine learning into microbiota analysis, offering unprecedented precision in understanding microbial diversity and functions [23]. Similarly, the One Health approach emphasizes the interconnectedness of human, animal, and environmental microbiomes, reinforcing the need for holistic strategies to tackle global challenges like AMR and ecosystem degradation [24]. Advancing microbiome research requires an interdisciplinary approach, integrating expertise from microbiology, genomics, data science, and environmental science. For example, AI-driven models predict microbial shifts in food processing, while synthetic biology crafts consortia for bioremediation, bridging food safety and environmental applications [14].

Figure 1.
Timeline of key advancements in microbiome research methodologies.
Despite these advances, gaps remain in translating these technologies into practical applications for food safety and environmental health, particularly in expanding beyond bacterial-focused research to include fungi, viruses, and helminths [25]. This review addresses these gaps by providing a comprehensive framework for advancing microbiota-based technologies, synthesizing current knowledge, and exploring emerging methodologies such as culturomics, synthetic biology, and AI-driven diagnostics to improve food safety and environmental sustainability. By focusing on actionable interventions—like reducing pathogen loads in food chains and restoring contaminated ecosystems—this work builds on recent insights by Mu et al. [26] to guide researchers and practitioners in this rapidly evolving field.
2. Microbiome-Based Interventions for Ecosystem Sustainability
Microbiomes are emerging as pivotal players in environmental health due to their abilities to enhance nutrient cycling, remediate pollutants, and maintain soil and water quality [6]. Advancements in long-read sequencing and AI are unlocking precise microbial insights, enabling targeted interventions like bioremediation of oil spills [27] and pathogen suppression in agricultural soils [28]. Gulliver et al. [6] asserted how microbial interventions will be a vital step in ecological restoration, showing promising application of microbiomes in the bioremediation of pollutants in contaminated environments. One Health, which is an integrated approach to tackle the health of humans, animals, and their shared environment, reinforces microbiome strategies for sustainable development, as reported by Tomasulo et al. [29]. Microbial biotechnology tackles industrial pollutants, such as heavy metals in farmland, supporting safer food production [30]. According to the above studies, it can be argued that microbiomes are the top priority to enhance environmental resilience [31] and that they should be implemented in the strategy for tackling the global environmental challenge. Figure 2 illustrates microbiome-based interventions aimed at promoting ecosystem sustainability.
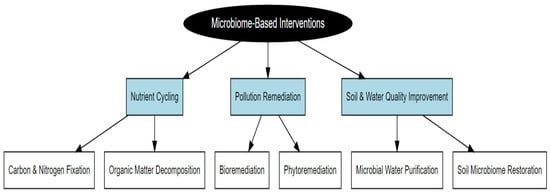
Figure 2.
Microbiome-based interventions for ecosystem sustainability.
To ensure the reliability and effectiveness of microbiome-based interventions, it is essential to evaluate their consistency and reproducibility across varying environmental conditions. Reproducibility is particularly critical for large-scale applications in agriculture, bioremediation, and public health. For instance, in a multi-site field trial, Zhang et al. [32] applied a synthetic microbial consortium to degraded farmland soil and observed a consistent 35% increase in crop yield over three consecutive growing seasons, highlighting the intervention’s reproducibility. Similarly, a study by Elgazali et al. [33] demonstrated the bioremediation potential of hydrocarbon-degrading bacteria, observing a total petroleum hydrocarbon reduction of 61% with bacterial augmentation, which improved to a 73% reduction when combined with nutrient amendments (nitrogen, potassium, and phosphorus). Statistical modeling, such as Bayesian hierarchical models, can further enhance reproducibility assessments by accounting for environmental variability, as shown in the study by Palmero et al. [34]. Their research demonstrated that Bayesian inference provides meaningful uncertainty estimations, especially under limited data conditions. These findings underscore the need for large-scale, multi-environment trials combined with robust statistical frameworks to validate the consistency of microbiome-based interventions.
2.1. Bioremediation for Environmental Health
The focus on environmental pollution and waste management has fostered interest in microbiome-based strategies, which are cheap, sustainable, and generally eco-friendly. This is because microbial communities possess metabolic versatility that enables them to degrade pollutants, transform toxic molecules, and recycle waste.
Bioremediation is the utilization of microbes to promote the biodegradation (or detoxification) of harmful pollutants, such as hydrocarbons and heavy metals, that contaminate the soil and water. Kaur et al. [35] highlight the use of hydrocarbon-degrading microbes to break down polycyclic aromatic hydrocarbons (PAHs). These bacteria play a pivotal role in the bioremediation of oil spills in the marine environment as they have the potential to degrade PAHs and prevent ecological damage to the environment due to the toxicity of this recalcitrant pollutant. AI takes it further, modeling how fast they will work or where to deploy them [36], while synthetically engineered biology consortia chew through PAHs even quicker [37,38]. Long-read sequencing helps identify hydrocarbon-degrading bacteria, such as Pseudomonas, showing exactly which microbes are involved in breaking down pollutants [22]. For instance, in a recent application by Zhou et al. [39], nanopore sequencing technology was used to detect foodborne microorganisms, including Listeria monocytogenes. They combined this sequencing approach with machine learning algorithms to enhance the identification and characterization of pathogens in various food samples. This integration facilitated rapid and accurate detection, demonstrating the potential for real-time monitoring in food safety applications. Similarly, a study by Perdigão et al. [40] demonstrated that bioremediation strategies at a fuel-contaminated port significantly enhanced the abundance of hydrocarbon-degrading bacteria and achieved over 60% petroleum hydrocarbon removal within 15 days.
2.2. Successful Environmental Microbiome Projects
Several successful projects integrating microbiome-based interventions into environmental damage management have demonstrated the significant potential of bioremediation and phytoremediation techniques. Microbial communities, which reside both internally and externally on plants, play diverse and crucial ecological roles, contributing to the effectiveness of these remediation strategies. One example is the microbial response to the Deepwater Horizon oil spill in the Gulf of Mexico, where naturally occurring hydrocarbon-degrading bacteria such as Alcanivorax and Pseudomonas helped to degrade oil components, thereby facilitating faster recovery of the marine ecosystem [27]. The use of these microbes highlights the role of microbial diversity in bioremediation, whereby microbial metabolism of hydrocarbons converts them into less toxic substances that are easier to process by the biogeochemical cycle. A deeper analysis of the Deepwater Horizon case reveals that microbial taxa such as A. borkumensis and Cycloclasticus were enriched in oil-contaminated zones. Metagenomic data indicated the upregulation of genes linked to hydrocarbon degradation (alkB, CYP153). Comparatively, a study by Mason et al. [41] found similar microbial patterns in the Ixtoc I oil spill, suggesting conserved functional adaptations among marine microbiomes. Furthermore, nutrient enrichment strategies, including nitrogen and phosphate amendments, significantly accelerated biodegradation in both cases, underscoring the synergy between microbial consortia and nutrient availability in bioremediation. This case is in line with the findings of Ferguson et al. [27], who highlight the significance of microbial biodiversity in marine ecosystems for maintaining resistance to environmental perturbations, especially in regions directly affected by oil exploration and spills.
Another success story comes from the Pérez Zeledón landfill in Costa Rica, where bioremediation was used for the treatment of leachate, a highly contaminated liquid made by the decomposition of waste [42]. This leachate can reach underground water tables or even the sea, covering large areas with pollutants. In the project, microbial consortia helped reduce the concentration of the pollutants, improving water quality and decreasing environmental impact. This shows how microbial communities can be used in waste management to deal with complex environmental problems such as leachate from landfills, which represents an important threat to water quality. In parallel to the Pérez Zeledón landfill project, Masís-Meléndez et al. [42] published an article on the role of microbial interventions in waste sanitation and environmental management.
Lastly, microbiome-assisted phytoremediation in contaminated agricultural soils is another successful example of microbiome-based intervention. Sedum alfredii and its microbiome were used for the remediation of heavy metals (cadmium and zinc) in polluted soils to reduce soil toxicity and produce safe crops [43]. This also demonstrates the emerging trend of combining plants with a microbial community for sustainable remediation efforts. As described by Zhang et al. [43], phytoremediation systems are highly effective and economical. Microbiome-assisted plant remediation can be used as a cost-effective solution for the restoration of contaminated farmlands. While microbiome interventions offer promising solutions for environmental sustainability, their long-term ecological consequences remain underexplored. For example, large-scale soil microbiome engineering could unintentionally disrupt native microbial diversity, potentially altering nutrient cycling and soil fertility over time [27]. Additionally, introducing genetically modified microbial strains for bioremediation may lead to the horizontal gene transfer of antibiotic-resistance genes, as observed in a decade-long monitoring study by Zhao et al. [44]. The authors found that engineered Pseudomonas strains introduced for oil degradation transferred resistance genes to indigenous soil bacteria, raising concerns about unintended ecological impacts. Furthermore, long-term use of microbiome-based probiotics in livestock farming could alter gut microbiota composition, potentially affecting animal health and zoonotic pathogen transmission [45]. These findings underscore the need for longitudinal ecological monitoring to assess the sustainability and safety of microbiome interventions over time.
3. Leveraging Microbiome Profiling and Metagenomics for Advanced Foodborne Pathogen Control
The microbiome critically influences food safety and quality. Complex microbial communities can obscure foodborne pathogen detection, complicating food safety efforts [46]. Contaminated foods impact public health and economics through illnesses, recalls, and reduced consumer trust [47]. Fresh produce, often consumed raw, serves as a reservoir for pathogens, necessitating rigorous screening. Long-read sequencing improves detection of risks in raw vegetables [48], while AI predicts contamination hotspots [49]. Rising outbreaks and antimicrobial-resistance (AMR) gene transfer risks [50] highlight the urgent need for alternative pathogen detection approaches.
3.1. Metagenomic Approaches in Pathogen Detection
Advancements in metagenomic sequencing technologies have revolutionized the detection and characterization of foodborne pathogens within complex microbiomes. Unlike traditional culture-based methods that are laborious and ineffective for non-culturable microbes, metagenomics offers a more culture-independent approach, enabling the identification of a broad spectrum of microorganisms, including those that are viable but non-culturable [51]. Metagenomics therefore allows for a thorough understanding of microbial diversity and possible health hazards by enabling the simultaneous investigation of bacterial, viral, and fungal pathogens in food matrices [51].
Figure 3 illustrates the sequential steps involved in microbiome data analysis, starting from sample collection and DNA extraction to bioinformatic processing and statistical interpretation.
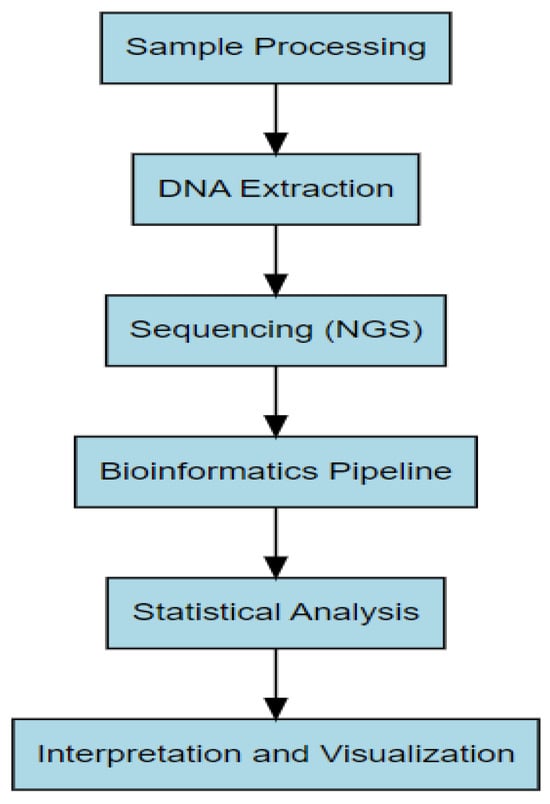
Figure 3.
Workflow for microbiome data analysis, from sample processing to statistical interpretation.
Long-read sequencing identifies all microbes present, whether they can be cultured or not [51,52]. AI analyzes this data, detecting E. coli in beef and identifying emerging threats [53]. For example, a study by Yang et al. [11] demonstrated the efficacy of metagenomic shotgun sequencing in identifying pathogens along the beef production chain, revealing critical insights into the microbial landscape of meat products. This technology enhances pathogen detection by providing a comprehensive view of the entire microbial community, allowing for the identification of both known and novel pathogens that might otherwise be overlooked. It also informs food safety interventions by pinpointing areas of contamination, which can lead to more precise and effective interventions. For example, pathogen loads in meat products have been shown to be reduced through food safety practices like knife trimming and steam vacuuming. These practices are guided by the data provided through metagenomic analysis, which identifies the most critical sources of contamination and informs the development of targeted interventions to reduce pathogen loads effectively.
Furthermore, the application of omics technologies, including metagenomics, as discussed by Cook and Nightingale [54], provides a comprehensive understanding of microbial communities in food processing environments. Omics technologies, which include genomics, transcriptomics, proteomics, and metabolomics, allow researchers to analyze the genetic and functional potential of entire microbial communities. This integrated approach facilitates the identification of contamination routes and supports the implementation of targeted food safety measures. By combining omics data with food safety management systems, we can enhance risk assessments, tailor mitigation strategies to specific pathogens, and ultimately improve food safety outcomes. The integration of omics technologies offers a more holistic and data-driven approach to food safety, enabling more precise identification of hazards and more effective control measures.
Metabolomics typically involves collecting microbial samples, extracting metabolites using GC-MS or LC-MS platforms, and interpreting data through multivariate analysis. In a food safety context, targeted metabolomics revealed aflatoxin-producing pathways in contaminated peanuts [55]. Culturomics, on the other hand, relies on high-throughput culture conditions, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) for identification, and has successfully isolated novel anaerobes from fermented dairy products [56]. Synthetic biology applications require CRISPR or recombination-based pathway engineering, validated through microbial growth curves and pollutant breakdown efficiency. For example, Pseudomonas putida engineered for enhanced biodegradation of polyethylene achieved a 25–30% weight reduction over 60 days under controlled laboratory conditions [57]. These integrative approaches not only advance our understanding of microbial function and diversity but also offer scalable solutions to pressing challenges in health, industry, and environmental sustainability.
3.2. Microbiome Profiling and Sanitation Effectiveness
Microbiome profiling through techniques such as 16S rRNA sequencing has become a standard practice for assessing sanitation effectiveness in food production systems. Dong and Feng [58] emphasize the importance of understanding microbiome shifts in controlled environment farming systems, where sanitation practices are critical for preventing contamination. However, the limitations of 16S rRNA sequencing in distinguishing closely related species highlight the need for more advanced techniques to accurately characterize microbial communities.
The longitudinal consistency and cross-sectional distinctiveness of microbiomes in retail chicken breast, as reported by Li et al. [59], further illustrate the importance of microbiome profiling in food safety. AI monitors changes in chicken breast microbiomes, helping optimize sanitation practices [23]. Understanding the microbial communities associated with food products can inform processing practices and enhance food safety measures.
3.3. Implications for Foodborne Pathogen Surveillance
The surveillance of foodborne pathogens is essential for preventing outbreaks and ensuring public health. Painter et al. [60] provides insights into the attribution of foodborne illnesses to specific food commodities, emphasizing the need for robust surveillance systems to track and manage foodborne pathogens effectively. The integration of microbiome data into these surveillance efforts can enhance our understanding of pathogen transmission dynamics and inform targeted interventions. Long-read sequencing and AI enhance pathogen tracking by mapping their transmission routes [61].
Furthermore, the recognition of foodborne pathogens through innovative technologies, such as terahertz spectroscopy, as discussed by Zeng et al. [62], represents a promising avenue for improving pathogen detection in food products. These advancements can facilitate rapid and accurate identification of pathogens, thereby enhancing food safety. A comprehensive understanding of the diverse microbiomes involved in food safety is essential, as different microbiomes play distinct roles in influencing pathogen behavior and food quality, as outlined in Table S1 (see Supplementary Materials).
4. Current Applications for Microbiome-Based Interventions
Microbiome-based interventions have emerged as an important tool in food safety, environmental health, and treatment of infectious diseases, due to their positive effects on the immune system, overall human health, and the environment. Microbiome-based therapeutics find application in probiotics, prebiotics, and synbiotics, especially in ensuring improved gut health and food safety. They also find applications in fecal microbiota transplantation (FMT) and phage therapy for the treatment of diseases.
4.1. Probiotics in Food Safety
The use of probiotics as health-promoting ingredients in a variety of meals, particularly in fermented goods, pharmaceutical/dietary supplements, or animal diets to support and stabilize animal growth and products, has grown throughout the last two decades [63]. Probiotics are used to improve gut health, boost the immune system, and outcompete pathogenic bacteria [64]. Primarily, probiotics enhance food safety by maintaining a healthy gut microbiota, which is crucial for preventing various disease states and intestinal disorders associated with an unbalanced gut [65]. The idea that keeping the gut microbiota in a healthy state helps guard against digestive problems such as gastrointestinal infections, irritable bowel syndrome, and even cancer is being supported by growing scientific research [63]. Probiotics like Lactobacillus and Bifidobacterium species help to outcompete pathogenic bacteria by adhering to the gut lining, producing antimicrobial substances, and enhancing the host’s immune response [64]. These beneficial bacteria help to enhance immune function and maintain the integrity of the gut lining, thereby reducing the risk of infections and inflammatory conditions. Additionally, a balanced gut microbiota can modulate the body’s inflammatory response and has been linked to lower incidence of colorectal cancer [66].
Probiotics can be introduced into food through various methods to enhance their health benefits. One common approach is fermentation, where live probiotic strains are incorporated into the food product along with traditional starter cultures such as Lactobacillus bulgaricus and Streptococcus thermophilus in food products like yogurt, kefir, and sauerkraut [67,68]. This fermentation process not only enriches the food with live probiotic cultures that confer health benefits but also enhances its flavor and shelf life [67]. Additionally, probiotics can be introduced through encapsulation, an advanced technique that involves coating probiotic bacteria with a protective layer [69]. This layer helps shield the bacteria from harsh conditions during processing, storage, and digestion, improving their viability when consumed [70]. Encapsulation is often used in functional foods and dietary supplements to ensure that probiotics remain active until they reach the gut, protecting them from degradation that might occur during the production process [69].The role of probiotics in food safety has been demonstrated by numerous studies. For instance, ref. [71] investigated the ability of Lactobacillus rhamnosus GG (ATCC 53103) and L. casei (ATCC 393) to inhibit and prevent biofilm formation by pathogens like E. coli O157:H7, Salmonella enterica, and L. monocytogenes on food contact surfaces. Results showed that these probiotics significantly reduced pathogenic adhesion and biofilm formation, particularly on stainless steel surfaces in juice processing, highlighting their potential to improve the safety and quality of fruit-based products. Similarly, ref. [72] encapsulated Lactobacillus paraplantarum FT-259 and Lactococcus lactis QMF in casein/pectin microparticles and tested their ability to inhibit Listeria and Staphylococcus in fresh Minas cheese stored at 8 °C. The encapsulated probiotics maintained high viability and effectively reduced pathogen levels, with the encapsulated Lactobacillus paraplantarum showing enhanced antilisterial activity and Lactococcus lactis significantly inhibiting Listeria in both free and encapsulated forms, indicating that these microparticles are effective carriers for enhancing the safety of the cheese. The inhibitory activity of these lactic acid bacteria is attributable to synthesized lactic acid and associated bacteriocins, which are active against a broad range of pathogenic and spoilage organisms [73].
AI and machine learning are rapidly becoming integral tools in microbiome-based therapeutics, helping to improve outcomes in food safety, medical applications, and environmental monitoring [74]. By leveraging predictive modeling, machine learning, and deep learning techniques, researchers can better understand microbiome dynamics and predict microbial behavior in various environments. Food Safety: AI-driven pathogen detection in food microbiomes has significantly enhanced the ability to identify harmful microorganisms, enabling proactive food safety measures [75]. Medical Applications: Machine learning models are being used to predict microbiome shifts in diseases, offering insights into disease progression and therapeutic interventions [76]. Environmental Monitoring: AI tools are increasingly employed to analyze microbial communities in polluted environments, offering a more efficient means to assess environmental health [22].
4.2. Prebiotics in Food Safety
Unlike probiotics, which introduce live foreign bacteria into the human GIT, prebiotics encourage the growth of certain beneficial species that are already present in the gut, particularly but not limited to lactobacilli and bifidobacteria [77]. Prebiotics are non-digestible food ingredients that beneficially affect the host by selectively stimulating the growth and activity of beneficial microorganisms in the gut [78]. Prebiotic oligosaccharides like fructooligosaccharides are found in a variety of fruits and vegetables, and therefore consuming them through diet may be a more natural way to obtain prebiotics [77]. The oligosaccharides found in human breast milk are thought to be the prototypical prebiotic because they encourage the colons of exclusively breastfed newborns to grow bifidobacteria and lactobacilli preferentially [77]. This phenomenon may be responsible for some of the immunological and other advantages that breastfed infants experience [79]. In food animals, certain natural microflora species (such as Bifidobacteria, Butyrivibrio, and Lactobacillus) that are known to function antagonistically against pathogens benefit from a competitive advantage provided by some prebiotics [80]. Additionally, prebiotics may lower colonic inflammation and colitis [81], supply some limiting nutrients directly to the intestinal mucosa, and provide the substrates necessary for intestinal bacteria to ferment, thereby increasing the synthesis of B vitamins [80].
Different studies have demonstrated the role of prebiotics in food safety, such as a reduction in the presence of foodborne pathogens by promoting the growth of beneficial bacteria, serving as a substrate for probiotic growth, enhancing gastrointestinal functions, the immune system, calcium and magnesium absorption, blood glucose levels, and plasma lipids [82]. For instance, ref. [83] examined the effects of probiotics and prebiotics on controlling Salmonella spp. in chickens, revealing that B. subtilis probiotic and Levoxyl prebiotic treatments significantly improved survival rates and reduced the growth of Salmonella enteritidis and Salmonella typhimurium. While the B. subtilis probiotic was found to have antimicrobial properties that inhibited the growth of these harmful bacteria, Levoxyl, a prebiotic, supported the growth of beneficial gut microbes that competed with Salmonella for nutrients and space. Together, these treatments improved the survival rates of chickens by enhancing the balance of gut microbiota and reducing the prevalence of Salmonella infections, indicating that the oral application of these treatments in infected broiler chickens could effectively enhance their resistance against nalidixic acid-resistant Salmonella strains. Similarly, ref. [84] investigated the effect of Salmonella Typhimurium infection on the gut microbiota in laying chickens and evaluated how an in-feed probiotic supplement can restore microbial balance and reduce Salmonella load. They found that Salmonella infection caused significant dysbiosis in the gut by reducing the abundance of beneficial microbial genera such as Lactobacillus, Bacteroides, and Alistipes and increasing the presence of genera such as Eisenbergiella and Oscillibacter, which favor Salmonella growth. Probiotic supplementation helped restore gut microbiota balance by increasing the abundance of previously displaced beneficial genera and enhancing levels of propionate and butyrate, which were not significantly affected by Salmonella challenge. Continuous probiotic use notably reduced the overall Salmonella load in feces and internal organs, highlighting its effectiveness in controlling Salmonella infections. Similarly, prebiotics may also have a beneficial effect on the proliferation of probiotic bacteria, as shown in a study by [85], where they found that the prebiotic effect of rice increased the number of Bifidobacterium cells in yogurt mixed with rice as opposed to plain yogurt.
4.3. Synbiotics in Food Safety
Synbiotics are a blend of probiotics and prebiotics that have a positive effect on the host by enhancing the implantation and survival of live microbial strains in the gastrointestinal system [86]. They can work in synergy or complement each other to perform a variety of tasks, such as antioxidant activities, which can improve the health of the host [87]. The antioxidant effects of synbiotics are attributed to their activation of nuclear factors that enhance the expression of antioxidant enzymes, generate key antioxidant molecules, and neutralize harmful singlet oxygen and free radicals [87,88]. When probiotic bacteria and prebiotic compounds are combined, antibacterial compounds like bacteriocin are released, which can stop the growth of harmful bacteria [82]. Due to their synergistic effects, synbiotics are expected to confer more benefits than probiotics or prebiotics alone [89]. Synbiotics have been shown to help prevent the aberrant growth of harmful bacteria in the intestine in animals. For instance, it has been discovered that Campylobacter jejuni, the causative agent of foodborne gastroenteritis in humans with poultry and poultry products serving as the primary source of infection, can be moderated using synbiotics. Galactooligosaccharides and the microencapsulated Bifidobacterium longum subsp. longum PCB133 strain were used in an in vivo experiment by [90] to evaluate the effectiveness of the synbiotic in grill chickens. Comparing the treated group to the control group that did not get any special treatment, the results showed a substantial drop in C. jejuni quantification and a significant increase in total bifidobacteria after two weeks of treatment. In an observational study, ref. [91] investigated the prevalence of infectious diseases in infants transitioning from exclusive breastfeeding to symbiotic-supplemented formula, finding reduced infection rates. Results from the study showed that compared to babies who received normal formula, the incidence of infectious disease and growth were found to be positively impacted by supplementation with synbiotic-enriched formula.
4.4. Phage Therapy
Bacteriophages (phages), which are viruses that can specifically target and replicate within bacterial cells, have been utilized as a form of antibacterial monotherapy for over 100 years to treat bacterial infections [6]. Phages replicate via two main life cycles that impact their therapeutic use: virulent phages rapidly kill their bacterial hosts, while temperate phages can integrate into the host genome or switch to a lytic cycle [92]. Temperate phages can also prevent reinfection in their host and alter bacterial traits through lysogenic conversion [93]. Phage therapy involves administering pathogenic phages directly to a patient with the aim of lysing the bacterial pathogen causing a reduction or elimination of the pathogenic bacteria [92]. The primary feature of phages is their high specificity of infection: they usually identify only a small subset of bacterial strains, minimizing the harm to the host’s regular microbial community [94]. Furthermore, because phages reproduce exclusively at the site of infection, they are self-limiting and self-dosing, and they vanish when their particular bacterial pathogen is eliminated [95]. Phages have shown promise in effectively killing bacteria resistant to broad-spectrum antibiotics, as their mechanisms of action differ from those of antibiotics, and resistance can often be overcome using different phage isolates, cocktails, or modified phages [96]. This is demonstrated in a study by [97], where a personalized bacteriophage therapy successfully treated a 68-year-old diabetic patient with necrotizing pancreatitis and a multidrug-resistant A. baumannii infection, which had persisted despite multiple-antibiotic therapy. The intravenous and percutaneous administration of nine different lytic bacteriophages into the abscess cavities of the patient led to the resolution of the infection and a return to health, highlighting the potential of tailored phage therapies for managing resistant infections. Phage therapy is also gaining attention for controlling bacterial biofilms, especially as resistance to antibiotics increases. A study by [98] demonstrated that combining bacteriophage T4 with the antibiotic cefotaxime enhanced the eradication of biofilms formed by E. coli, showing that phage-antibiotic synergy can significantly improve treatment outcomes. This synergy, where sublethal antibiotic concentrations boost phage activity, resulted in better biofilm control and reduced antibiotic concentrations needed for effective treatment.
4.5. Fecal Microbiota Transplantation
Gut microbiota imbalances, known as dysbiosis, have been linked to various diseases, driving increased interest in FMT over the past decade [99]. FMT involves transferring screened manipulated donor stool into a patient’s gastrointestinal tract to restore microbiota diversity and functionality, aiming to correct the dysbiotic state [99,100]. FMT has been integral in restoring microbial diversity and structure in the colon, leading to high cure rates for Clostridium difficile infection (CDI) [101]. CDI is primarily caused by the suppression and disruption of colon microbiota, often due to antibiotic use, and is significantly with high morbidity and mortality [102]. FMT restores microbial diversity in the colon by inhibiting C. difficile and preventing reinfection through several mechanisms such as bile acid-mediated inhibition of spore germination and vegetative growth, competition for nutrients, direct suppression by antimicrobial peptides, and activation of immune-mediated colonization resistance [101].
Several studies have reported the effectiveness of FMT in the treatment of recurrent and refractory CDI, with a recent meta-analysis by [102] concluding that FMT successfully resolved symptoms of CDI in 92% of the cases treated. Similarly, a study by [103] of 111 patients treated with FMT for recurrent or refractory CDI in Israel from 2013 to 2017 reported an overall success rate of 87.4%, with no significant difference in success between different routes of administration (lower gastrointestinal route by colonoscopy (45%), capsules (33%), and upper gastrointestinal route (22%)). Since it has proven effective in treating CDI, FMT is currently being investigated for its potential to treat other GI disorders such as inflammatory bowel disease (IBD). A systematic review of 17 studies by [104] involving 41 patients showed that most IBD patients treated with FMT experienced symptom reduction (19/25), disease remission (15/24), cessation of IBD medications (13/17), and remission of C. difficile infection in every patient receiving such treatment (15/15), suggesting that FMT may be an effective treatment option, particularly when standard therapies have failed.
5. Challenges and Limitations Associated with MBT
5.1. Regulatory and Safety Concerns
Despite significant advancements in MBT using various techniques like probiotics, prebiotics, synbiotics, and FMT to counteract dysbiosis and restore gut microbiota, this research area still faces challenges, such as the evolving regulatory landscape, and safety concerns arising from their unclear mechanism of action and the inherent risks of introducing live microorganisms into the human body [105,106]. While the stable colonization of therapeutic organisms in the gut can enhance their effectiveness, it raises concerns about pharmacological control and the potential spread of genetically modified DNA to native microbiota due to natural horizontal gene transfer [107]. Also, although most genetically modified organisms (GMOs) developed in laboratories tend to be less competitive than their natural counterparts, there is a risk that these engineered organisms could escape into the environment and inadvertently colonize other areas such as natural ecosystems or agricultural environments [108]. This presents a challenge in MBT, as the unintended spread of GMOs could potentially disrupt natural ecosystems and interactions.
Recent studies have shown the vast potential of MBT, as evidenced by scientific literature and media, with practices like FMT showing high efficacy in treating recurrent CDI [109], and probiotics, especially strains of Lactobacillus rhamnosus and Saccharomyces boulardii, helping in reducing the duration and severity of diarrhea, including antibiotic-associated and infectious diarrhea [110]. However, FMT products pose safety risks due to the potential presence of uncharacterized pathogens that might pose threats to immunocompromised individuals, prompting the U.S. Food and Drug Administration (FDA) to issue multiple safety alerts regarding serious adverse events associated with their use [111]. Similarly, a study of Lactobacillus bacteremia cases in ICU patients at Boston Children’s Hospital by [112] found that those receiving Lactobacillus rhamnosus strain GG (LGG) probiotics had a significantly higher risk of developing Lactobacillus bacteremia compared to those who did not receive probiotics. Out of 22,174 ICU patients, 522 were given LGG probiotics (usually through a feeding tube), and 6 of these patients developed Lactobacillus bacteremia, while only 2 of the 21,652 patients who did not receive LGG probiotics experienced the same condition.
Until 2022, FMT and other microbiota-based therapies in Europe faced regulatory uncertainty because they were not classified as drugs. This lack of classification stemmed from the highly variable composition of gut microbiota samples, which made it challenging to standardize and regulate these therapies under existing drug frameworks [113]. The diverse nature of microbiota means that each FMT or microbiota-based product can differ significantly in its microbial content [114,115], complicating efforts to apply conventional drug regulations and approval processes. As a result, these therapies existed in a regulatory gray area, lacking clear guidelines and standards for their use and oversight [113].
In the United States, the classification of MBT such as the use of stool in FMT as a drug and a biological product under traditional statutory declarations by regulatory bodies affects the required clinical trial rigor, safety evaluations, and manufacturing standards, creating significant regulatory challenges [116,117]. This is exemplified by the FDA’s 2013 decision to regulate stool for FMT as a biologic drug, necessitating an investigational new drug (IND) application for clinical trials [118]. Since clinical experience and preliminary data for FMT already showed its efficacy as treatment for recurrent or refractory CDI [119,120], investigators and clinicians pushed back against the policy, arguing that the regulatory burden of maintaining an IND may prohibit many clinicians from offering a potentially life-saving therapy [121]. In consideration of this input from major stakeholders, the FDA eventually allowed “enforcement discretion” for FMT to treat CDI that was not responding to standard antibiotic therapies without requiring them to submit an IND application, resulting in most physicians and stool banks not obtaining IND approval for such treatments [116,117]. The agency, however, insisted that an IND application would be necessary if FMT were to be used for research purposes or to treat ailments other than CDI [122].
The FDA classifies products based on the claims made by the manufacturer, not their ingredients or properties [116]. Ethical and socioeconomic factors significantly influence the design, implementation, and outcomes of microbiome research. For instance, in FMT trials, stringent donor screening is required to minimize the risk of transmitting infectious agents. However, inconsistent regulatory frameworks across countries raise ethical concerns regarding patient safety and data privacy [113]. Socioeconomic factors also impact the accessibility of microbiome-based interventions [123]. Additionally, ethical considerations extend to data privacy in large-scale microbiome studies. For example, in human gut microbiome sequencing projects, ensuring participant consent and data security is paramount. To prevent privacy breaches while maintaining data transparency and reproducibility, it is essential to de-identify microbiome data. For instance, Gaikwad and Naoghare [124] discuss the privacy risks associated with microbiome data and suggest that adopting well-known anonymization techniques can help protect participant privacy in microbiome studies. Additionally, Wagner et al. [125] propose the use of secure computation methods to enable comparative analysis over combined microbiome datasets without revealing individual sample information, thereby addressing privacy concerns in microbiome research.
If a product, including a probiotic, is claimed to cure, mitigate, treat, or prevent disease, it is classified as a drug, thereby requiring a costly IND application. Similarly, human studies investigating these claims also require an IND application, which poses barriers for researchers due to the need to provide proprietary manufacturing information [126]. The “locked-in” rule, which forbids marketing a substance as a food product if it is first examined as an IND, even if the study is intended to encourage usage of that product as a food rather than a medication, increases limitations to high-quality clinical research on probiotics [126]. Considering that these substances can be marketed as a dietary supplement or food before conducting an IND application, the FDA inadvertently provides a loophole that allows marketing a substance as a dietary supplement or food before seeking an IND approval, preserving the option to continue marketing it in those forms even if substantial clinical investigations show no benefit [126]. This has discouraged manufacturers and researchers from rigorously evaluating probiotic claims, contributing to the FDA not approving any probiotic products as live biotherapeutic agents [116,126].
5.2. Technical and Logistical Challenges in Implementation
One of the foremost technical challenges in the development of effective MBT is the isolation and cultivation of effective microbial strains, which is challenging due to the complexity of the human microbiome and the specific conditions required for their growth, as many beneficial microbes thrive only in the unique environment of the human body [127]. The human microbiome is incredibly diverse, with thousands of bacterial species coexisting in various body sites. Determining which specific strains have therapeutic potential involves extensive screening and validation, which is challenging because the effectiveness of probiotics varies by strain and is often specific to different body environments and diseases [128]. For example, Bacteroides sp. colonizes the colon and cecum, Lactobacillus sp. and E. coli Nissle 1917 thrive in the small intestine, while Lactobacillus lactis does not colonize the intestine, making disease biogeography crucial in determining the suitable probiotic for treatment [129].
Identifying the mechanistic elements of host–microbe interactions and microbe–microbe interactions of potential probiotic strains has proven challenging, as they are usually influenced by many internal and external factors such as diet, antimicrobials, genotype, and environment [130]. For example, the critical role that vaginal delivery plays in transferring beneficial microbes from mother to newborn that colonize the baby’s skin and gut highlights a technical and logistical challenge in implementing MBT in ensuring the appropriate seeding and maintenance of beneficial microbial communities in individuals, especially those born via cesarean section [130]. Cesarean births prevent this beneficial microbial transfer, leading to the acquisition of environmental microbes, which increases the risk of chronic diseases in adulthood, such as diabetes, obesity, asthma, and neurological disorders [131]. This can however be mitigated through methods such as vaginal microbial transfer (also known as “vaginal seeding”), where beneficial bacteria from the mother’s vaginal canal are applied to the newborn’s skin or mouth immediately after birth, helping to introduce beneficial microbes that are typically acquired during a vaginal delivery [132].
There are practical challenges associated with the preparation of probiotics, especially for commercial purposes. Probiotics face destabilization during manufacturing and storage due to stressors such as heat, oxygen, mechanical force, osmotic shock, and changes in pH [127]. The commonly used process of spray-drying often induces thermal stress, with elevated temperatures potentially denaturing proteins and causing cell damage in probiotics, which presents significant challenges in the implementation of MBT, as maintaining the stability and viability of probiotics becomes difficult [133]. Osmotic shock can also result during the drying process because it raises intracellular osmolarity, which modifies the cellular membrane, thereby reducing the viability of microbiome therapies [133]. Similarly, the ability of probiotics to survive during production and storage is impacted by oxidative stress, as their proteins, lipids, and DNA can be harmed by reactive oxygen species, which are produced when oxygen is partially reduced to water [134]. Spray-dried cells may be more susceptible to oxidative stress because dehydration causes cellular damage [127].
5.3. Microbial Resistance and Stability Issues
The promise of MBT in treating various conditions is tempered by significant challenges, including microbial resistance and stability issues, which can impact the efficacy and safety of these therapies and pose hurdles for their widespread adoption [135]. The use of probiotics or engineered microbes in microbiome-based therapies could potentially introduce antibiotic-resistance genes into the gut microbiota, which could lead to the failure of antibiotic treatments [135]. In an in vivo study by Jacobsen et al. [136] using gnotobiotic rats, two wild-type strains of Lactobacillus plantarum from fermented sausages were found to transfer tet(M) and erm(B) resistance gene–encoding plasmids to Enterococcus faecalis within the gastrointestinal tract, with the first transconjugants detected in fecal samples just two days after donor introduction. This study was one of the first to demonstrate the in vivo transfer of antibiotic-resistant plasmids from wild-type L. plantarum to E. faecalis, highlighting the potential for resistance genes to be transferred within the gut environment. While probiotics are generally considered safe, concerns exist about their potential risks, including causing diseases like bacteremia or endocarditis, toxic or metabolic effects on the gastrointestinal tract, and the transfer of antibiotic resistance within gut flora [137], remain. Also, while intrinsic or mutation-based antibiotic resistance in probiotic bacteria is not inherently a safety concern, as they can even help restore gut microbiota post-antibiotic treatment, the presence of resistance genes on mobile genetic elements in them poses a risk of transferring resistance to potential gut pathogens [137,138]. These bacteria can develop resistance through spontaneous gene mutations or horizontal gene transfer, which poses a high risk in non-pathogenic bacteria found in the gut [137]. Lactobacilli have shown high spontaneous mutation rates to antibiotics like nitrofurazone, kanamycin, and streptomycin (10−4 to 10−5 frequency/rate), indicating significant variability in their antibiotic-resistance profiles even within the same genus and species [135]. Molecular research suggests that the efficiency and safety of microbiome therapeutics may be compromised by human interactions with bacteria due to constant exposure to various bacterial strains through diet, environment, and medical treatments, which can increase the uptake of antibiotic-resistant genes already present in the environment and gut microbiota [138,139].
Although lactobacilli are generally recognized as safe and are widely used in various fermented foods and beverages, recent studies have shown that some strains carry antibiotic-resistance genes and can transfer these genes to other bacteria, posing a threat to human health [140]. An analysis of 33 Lactobacillus strains from fermented milk in China [140] revealed resistance to vancomycin and varying susceptibilities to other antibiotics, with certain Lactobacillus strains being found to carry specific resistance genes: erm(B) for erythromycin, tet(W) for tetracycline (in three L. helveticus strains), gyrA for ciprofloxacin (in all tested L. helveticus strains and two L. casei strains), and vanX for vancomycin (in most Lactobacillus strains). The kanamycin-resistance gene aph(3″)-III was detected in only one L. helveticus strain, highlighting the challenge of ensuring the safety of MBT due to the potential for antibiotic-resistance gene transfer [140]. Similarly, a study by [141] found that some commercially available probiotics contained lactic acid bacteria strains with antibiotic-resistance genes. Although the overall incidence was low, these resistance genes were found in mobile genetic elements like plasmids and transposons, conferring resistance to a range of antibiotics including aminoglycosides, β-lactams, and macrolides. Specific resistance genes, such as msrC, vanX, and dfrA, were detected in strains like Enterococcus faecium, Lactobacillus plantarum, Streptococcus thermophilus, and Lactococcus lactis, which poses a significant threat to food safety [141].
The increasing incorporation of probiotic bacteria into food products to enhance health benefits has been laden with stability challenges, which frequently call into question their efficacy in commercial products [142]. The primary challenge in developing functional foods is maintaining high levels of viable probiotic bacteria during processing, with an additional difficulty in ensuring their safe delivery to the gut, where they can provide health benefits [143]. Due to instability, the viability of these probiotics can diminish significantly during preparation, shelf storage, in vivo application, and when passing through the human gastrointestinal tract, thereby compromising their intended therapeutic effects [142]. Following oral delivery, probiotics face severe conditions in the gastrointestinal tract, such as the highly acidic gastric fluids (pH 1–3) and a two-hour gastric emptying period, which significantly reduce their viability in the stomach [144]. The cytoplasmic pH and glycolytic enzyme activity of probiotics are reduced by the extremely acidic pH of the stomach, which has an impact on the F1Fo-ATPase proton pump that is necessary for probiotic survival in acidic environments [145]. This presents a critical limitation for MBT, as maintaining the stability and viability of probiotic strains is essential to ensure their effectiveness in promoting health and preventing diseases [143]. Additionally, ensuring that probiotics remain active and potent throughout their shelf life and after ingestion involves overcoming significant technical and formulation hurdles, which adds to the complexity and cost of developing effective microbiome-based therapeutic products [127].
6. Future Directions and Research Needs
New technologies and innovation in the field of MBT are rapidly expanding and will in the near future reduce the continuous reliance on conventional therapeutics. This is based on the breadth of its potential use and market value. The advent of new sequencing technologies and culture-independent methods are promising for understanding microbiome–host interactions. Currently new technologies and innovations are being introduced to overcome current challenges, design large-scale clinical trials, and generate robust objective research aimed at improving patient outcomes. Next-generation sequencing metagenomics, specifically amplicon and shotgun sequencing, are widely adopted in microbiome research; however, both methods are limited by their short-read sequencing technologies. Emerging data supports the adoption of newer sequencing tools such as Pacific Biosciences (PacBio) single-molecule real-time sequencing and Oxford Nanopore Technologies’ nanopore sequencing, both long-read sequencing, to manage the limitations mentioned [52]. Currently, there is ongoing debate regarding the cost-effectiveness and accuracy of long-read sequencing compared to short-read sequencing, which limits its widespread adoption [146]. Additionally, the application of the 16S ribosomal RNA gene as a phylogenetic marker has proven to be a relatively efficient and cost-effective microbiome analytic tool. High-throughput technology is currently being adopted to provide insight into the microbial determinants of health and disease [147]. Advances have been made using culturomics (a computational method for analyzing cultural trends and a culturing technique for identifying bacterial species) to address the limitations of traditional culture methods that are labor-intensive and limit the identification of gut microbiome, with about 80% gut microbiota previously being unculturable [148]. The statistics of culturable microbes culturomics have heightened and extended our understanding of bacterial diversity [17]. A significant evolution in metatranscriptomics, metabolomics, culturomics, and synthetic biology and their application in microbiome research is regarded as a potential tool to advance research. A multi-omics approach was reported in a recent landmark study on microbial communities present on human fetal organs during the second trimester of gestation. Mishra et al. [146] profiled microbes on fetal organs, which play certain roles in microbial exposure in fetal immune-priming, using 16S rRNA gene sequencing; however, subject-specific transit times and selection bias pose a limit to this advancement [147]. Recent years have seen a dramatic rise in gut microbiome studies, which has been enabled by the rapidly evolving high-throughput sequencing methods. Microbial-based interventions are gradually exceeding the limits of feces-based transplants. They are used sparingly in the treatment of various diseases, with the mainstay in the treatment of recurrent CDI, the largest area of focus in microbiome clinical development. Recently IBD, including ulcerative colitis and Crohn’s disease, which is thought to be affected by the gut microbiome, is being explored with a rise in ongoing clinical trials [149]. Again, therapeutic interventions are starting to gain attention around microbiota–gut–brain bidirectional pathway in neurodegenerative diseases with compelling evidence that an alteration in gut microbiome drives neurodegenerative disease pathogenesis [150,151,152,153]. Immune-checkpoint inhibitors, which are immunotherapies that boost anticancer immune responses, are being explored in the field of microbial therapeutics [154]. Finally, AI is gaining attention for its potential in shaping clinical diagnostics, prognosis of diseases, and treatment options to bridge the gap between present knowledge of the human microbiome and targeted health interventions [13].
The collaboration among stakeholders and partnerships between industry, learning institutions, governments, and individuals can aid progression in specialist knowledge and public awareness about microbiome-based therapies. Interdisciplinary approaches require collaboration across different disciplines including bioinformatics, microbiology, mathematics, biochemistry, ecology, social sciences, and humanities, amongst others, due to the relevance of microorganisms in all forms of life. Presently, a large fraction of microbiome field research has focused on human populations in clinical contexts, with dietary, social, economic, and environmental factors representing major variability and limitations in these studies. One such limitation relies on recent evidence that suggests that the environment rather than genetics predominantly shapes human gut microbiome [155]. More exploration in microbiome research outside clinical contexts targets more diverse human populations that focus on human behavior, environment and physiology, although humanities research has explored microbiomes, particularly in areas of medical ethics and history [156,157,158]. Nevertheless, research on the intricate entanglements between microbiomes and the environment is advancing amidst challenges, although these research areas are still in their infancy.
New studies are beginning to highlight certain variabilities posed by early-life socioeconomic resources [159], social relationships [160,161], and socioeconomic conditions in microbiome interventions [162]. Mathematical techniques are now available for research to determine the population dynamics of gut–microbiome, causal interactions, predict stable microcommunities, and identify crucial microbes in varying contexts [163]. Ethical and social disciplines are equally involved in human microbiome research to explore ethical, legal, and social aspects of human microbiome research as well as clinical applications due to the potential risk associated with some interventions [112,164].
Despite the advances in MBT research, there remains a lack of standardized set of methodology for sample collection of microbiomes, storage, data analysis, and dosage calculation [165,166,167]. Additionally, there has been a partial tilt towards the use of animal models in recent studies in microbiome therapeutics and their generalizability to humans, that is, preclinical testing and safety profiling are skipped for most microbiome mimetic/drug products based on prior approval of the therapeutic with reported variable treatment regulation globally. Disease-specific testing of therapeutic efficacy is offered little or no attention in most countries [168,169]. However interspecies limitations with the use of animal models in anatomy, metabolism, physiology, and genetics are too significant to overlook and limit holistic test patterns due to the notorious variability in responses [105,170]. In line with this, a significant gap and potential research area exists concerning the use of non-animal models such as three-dimensional (3D) cultures, which are currently used to suffice for the limitations of animal models. Development of non-animal alternatives has led to groundbreaking research that overcomes the limitations of animal models and ex vivo tissues. However, these alternatives are still constrained by the inability to model microbiome–host immune interactions in certain target locations, and they can potentially harm the host, as seen with organoids derived from tissue-specific adult stem cells or pluripotent stem cells [171]. The generation of mini-human organs on a chip addressed the limitations of organoids, and although development has been reportedly successful, this may not represent adaptive host–microbe responses, which are equally relevant for microbiome studies [172]. Furthermore, since a microbial consortium can be used in the production of therapeutic molecules or antitoxins by manipulating individual microbes or a microbial consortia genome [12], advances in microbiome therapeutics demand a well-characterized library of biosensors integrated with AI to identify relevant candidate disease biomarkers, utilize microbes as a class of diagnostics, and trigger therapeutic responses [105]. Presently, the escape of engineered organisms into the environment is limited by advancement in technologies. Nevertheless, the possibility of escape of engineered organisms triggers concerns of environmental perturbations. Again, the adoption of vast machine learning could bridge the existing gaps in the design of new therapeutics that are specifically targeted to microbiomes of interest. This would expand its adoption in microbiome precision medicine and microbiome profiling [173,174,175] and requires continuous research. Importantly, most microbiome studies are heavily bacteria-biased, and more research is required to analyze potential therapeutic benefits from overlooked microbial interactions among viruses, fungi, intestinal helminths, and protozoan parasites [176].
7. Conclusions
This review highlights the significant advances and emerging directions in MBT, underscoring the novelty and originality of approaches that promise to reshape the landscape of healthcare. This comprehensive assessment brings together diverse technologies and methodologies—from next-generation long-read sequencing to AI-driven diagnostics and therapeutic design—that collectively offer unprecedented insights into the human microbiome. Unlike traditional approaches, which often relied on generalized microbial interventions, recent innovations enable highly specific, targeted manipulations of microbial communities, marking a departure from conventional therapeutics. This work integrates new evidence on microbiome–host interactions with advanced methodologies such as culturomics, synthetic biology, and machine learning, offering a more holistic understanding of the microbiome’s impact on health and disease.
A key contribution of this review is its focus on interdisciplinary collaborations and non-animal model systems, which represent novel responses to the limitations of existing microbiome studies. By advocating for robust, non-animal alternatives like 3D cultures and organ-on-a-chip technologies, this review not only addresses current gaps but also paves the way for more ethical, scalable, and reproducible microbiome research. Moreover, the review’s emphasis on integrating socioeconomic, environmental, and behavioral factors in microbiome research adds a unique dimension to the field, positioning the human microbiome within a broader ecological and social context. This approach is still emerging, yet it has the potential to transform how we understand microbiome variability across populations and individual life stages.
Finally, this review highlights the potential of overlooked microbial relationships—such as those involving fungi, viruses, and helminths—in advancing therapeutic interventions. It calls for a more comprehensive exploration of microbial consortia and their roles in diverse physiological pathways, an area that remains largely uncharted, yet promises substantial therapeutic value. This work synthesizes current knowledge while identifying critical gaps and proposing directions that push beyond traditional research boundaries. By emphasizing the integration of machine learning, biosensors, and bioinformatics with high-throughput sequencing, it highlights the revolutionary potential of precision microbiome medicine and establishes a foundation for future breakthroughs in personalized healthcare. The perspectives presented here serve as a valuable resource for researchers and clinicians navigating the rapidly evolving field of microbiome therapeutics.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/app15095219/s1. References [11,12,13,16,22,23,28,32,46,47,48,49,50,52,54,58,60,62,75,156,170,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200] are cited in the supplementary materials.
Author Contributions
B.O.S.: conceptualization, writing—original draft and editing, N.D.N.: conceptualization, writing—original draft, reviewing, editing, and supervision, C.K.A.: conceptualization, writing—original draft, reviewing, editing, and supervision, J.C.A.: writing—original draft, C.T.E.: writing—original draft, reviewing, and editing, C.C.U.: writing—original draft, reviewing, and editing, F.C.I.: writing—original draft, reviewing, and editing, P.O.: writing—review and editing, O.V.I.: writing—review and editing, P.S.E.: writing—review and editing, H.O.: conceptualization, writing—original draft, editing, and supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
We declare that we do not have any conflicts of interest in publishing this manuscript.
References
- Chervin, C.S.; Gajewski, T. Microbiome-based interventions: Therapeutic strategies in cancer immunotherapy. Immuno-Oncol. Technol. 2020, 8, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Hitch, T.C.; Hall, L.J.; Walsh, S.K.; Leventhal, G.E.; Slack, E.; de Wouters, T.; Walter, J.; Clavel, T. Microbiome-based interventions to modulate gut ecology and the immune system. Mucosal Immunol. 2022, 15, 1095–1113. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.S.; Voolstra, C.R. The baseline is already shifted: Marine microbiome restoration and rehabilitation as essential tools to mitigate ecosystem decline. Front. Mar. Sci. 2023, 10, 1218531. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Voolstra, C.R.; Sweet, M.; Duarte, C.M.; Carvalho, S.; Villela, H.; Lunshof, J.E.; Gram, L.; Woodhams, D.C.; Walter, J. Harnessing the microbiome to prevent global biodiversity loss. Nat. Microbiol. 2022, 7, 1726–1735. [Google Scholar] [CrossRef]
- Callens, K.; Fontaine, F.; Sanz, Y.; Bogdanski, A.; D‘Hondt, K.; Lange, L.; Smidt, H.; Overbeek, L.V.; Kostic, T.; Maguin, E.; et al. Microbiome-based solutions to address new and existing threats to food security, nutrition, health and agrifood systems’ sustainability. Front. Sustain. Food Syst. 2023, 7, 1143808. [Google Scholar] [CrossRef]
- Gulliver, E.L.; Young, R.B.; Chonwerawong, M.; D’Adamo, G.L.; Thomason, T.; Widdop, J.T.; Rutten, E.L.; Rossetto Marcelino, V.; Bryant, R.V.; Costello, S.P. The future of microbiome-based therapeutics. Aliment. Pharmacol. Ther. 2022, 56, 192–208. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Pamer, E.G. Microbiome-based therapeutics. Nat. Rev. Microbiol. 2022, 20, 365–380. [Google Scholar] [CrossRef]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Fathima, S.; Shanmugasundaram, R.; Adams, D.; Selvaraj, R.K. Gastrointestinal microbiota and their manipulation for improved growth and performance in chickens. Foods 2022, 11, 1401. [Google Scholar] [CrossRef]
- Kour, D.; Kaur, T.; Devi, R.; Yadav, A.; Singh, M.; Joshi, D.; Singh, J.; Suyal, D.C.; Kumar, A.; Rajput, V.D. Beneficial microbiomes for bioremediation of diverse contaminated environments for environmental sustainability: Present status and future challenges. Environ. Sci. Pollut. Res. 2021, 28, 24917–24939. [Google Scholar] [CrossRef]
- Yang, X.; Noyes, N.R.; Doster, E.; Martin, J.N.; Linke, L.M.; Magnuson, R.J.; Yang, H.; Geornaras, I.; Woerner, D.R.; Jones, K.L. Use of metagenomic shotgun sequencing technology to detect foodborne pathogens within the microbiome of the beef production chain. Appl. Environ. Microbiol. 2016, 82, 2433–2443. [Google Scholar] [CrossRef] [PubMed]
- McCarty, N.S.; Ledesma-Amaro, R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Abavisani, M.; Foroushan, S.K.; Ebadpour, N.; Sahebkar, A. Deciphering the gut microbiome: The revolution of artificial intelligence in microbiota analysis and intervention. Curr. Res. Biotechnol. 2024, 7, 100211. [Google Scholar] [CrossRef]
- Lin, D.; Medeiros, D.M. The microbiome as a major function of the gastrointestinal tract and its implication in micronutrient metabolism and chronic diseases. Nutr. Res. 2023, 112, 30–45. [Google Scholar] [CrossRef]
- Arıkan, Ş.; Karakoyun, M. Nutrient availability in temperate fruit species: New approaches in bacteria and mycorrhizae. In Sustainable Horticulture; Elsevier: Amsterdam, The Netherlands, 2022; pp. 39–54. [Google Scholar]
- Mayne, J.; Zhang, X.; Butcher, J.; Walker, K.; Ning, Z.; Wójcik, E.; Dastych, J.; Stintzi, A.; Figeys, D. Examining the effects of an anti-Salmonella bacteriophage preparation, BAFASAL®, on ex-vivo human gut microbiome composition and function using a multi-omics approach. Viruses 2021, 13, 1734. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef]
- Nnaji, N.D.; Onyeaka, H.; Miri, T.; Ugwa, C. Bioaccumulation for heavy metal removal: A review. SN Appl. Sci. 2023, 5, 125. [Google Scholar] [CrossRef]
- Nnaji, N.D.; Ughamba, K.T.; Onyeaka, H.; Anyanwu, C.U.; Al-Sharify, Z.; Miri, T. (Eds.) Biostimulatory potentials of plantain skin on soils polluted with used motor oil. In AIP Conference Proceedings; AIP Publishing: Melville, NY, USA, 2023. [Google Scholar]
- Malik, A.; Garg, V.K. Bioremediation for Sustainable Environmental Cleanup; CRC Press: Boca Raton, FL, USA, 2024. [Google Scholar]
- Sharma, A.; Vashistt, J.; Shrivastava, R. Response surface modeling integrated microtiter plate assay for Mycobacterium fortuitum biofilm quantification. Biofouling 2021, 37, 830–843. [Google Scholar] [CrossRef]
- Zhao, B.; Richardson, R.E.; You, F. Advancing microplastic analysis in the era of artificial intelligence: From current applications to the promise of generative AI. Nexus 2024, 1, 100043. [Google Scholar] [CrossRef]
- D’Urso, F.; Broccolo, F. Applications of Artificial Intelligence in Microbiome Analysis and Probiotic Interventions—An Overview and Perspective Based on the Current State of the Art. Appl. Sci. 2024, 14, 8627. [Google Scholar] [CrossRef]
- Alkorta, I.; Garbisu, C. Expanding the focus of the One Health concept: Links between the Earth-system processes of the planetary boundaries framework and antibiotic resistance. Rev. Environ. Health 2024, 40, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Nath, T.C.; Eom, K.S.; Choe, S.; Islam, S.; Sabuj, S.S.; Saha, E.; Tuhin, R.H.; Ndosi, B.A.; Kang, Y.; Kim, S. Insights to helminth infections in food and companion animals in Bangladesh: Occurrence and risk profiling. Parasite Epidemiol. Control 2022, 17, e00245. [Google Scholar] [CrossRef]
- Mu, W.; Kleter, G.A.; Bouzembrak, Y.; Dupouy, E.; Frewer, L.J.; Radwan Al Natour, F.N.; Marvin, H. Making food systems more resilient to food safety risks by including artificial intelligence, big data, and internet of things into food safety early warning and emerging risk identification tools. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13296. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.K.; Li, C.; Chakraborty, A.; Gittins, D.A.; Fowler, M.; Webb, J.; Campbell, C.; Morrison, N.; MacDonald, A.; Hubert, C.R. Multi-year seabed environmental baseline in deep-sea offshore oil prospective areas established using microbial biodiversity. Mar. Pollut. Bull. 2023, 194, 115308. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Singh, B.K.; He, J.-Z.; Han, Y.-L.; Li, P.-P.; Wan, L.-H.; Meng, G.-Z.; Liu, S.-Y.; Wang, J.-T.; Wu, C.-F. Plant developmental stage drives the differentiation in ecological role of the maize microbiome. Microbiome 2021, 9, 171. [Google Scholar] [CrossRef]
- Tomasulo, A.; Simionati, B.; Facchin, S. Microbiome One Health model for a healthy ecosystem. Sci. One Health 2024, 3, 100065. [Google Scholar] [CrossRef]
- Naghavi, N.S.; Samieipour, F. Microbiome therapies: Role of microbial biotechnology in sustainable development. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2020; pp. 163–172. [Google Scholar]
- Ma, L.-c.; Zhao, H.-q.; Wu, L.B.; Cheng, Z.-l.; Liu, C. Impact of the microbiome on human, animal, and environmental health from a One Health perspective. Sci. One Health 2023, 2, 100037. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Zhang, X.; Zhang, X. Artificial intelligence applications in the diagnosis and treatment of bacterial infections. Front. Microbiol. 2024, 15, 1449844. [Google Scholar] [CrossRef]
- Elgazali, A.; Althalb, H.; Elmusrati, I.; Ahmed, H.M.; Banat, I.M. Remediation Approaches to Reduce Hydrocarbon Contamination in Petroleum-Polluted Soil. Microorganisms 2023, 11, 2577. [Google Scholar] [CrossRef]
- Palmero, F.; Hefley, T.J.; Lacasa, J.; Almeida, L.F.; Haro, R.J.; Garcia, F.O.; Salvagiotti, F.; Ciampitti, I.A. A Bayesian approach for estimating the uncertainty on the contribution of nitrogen fixation and calculation of nutrient balances in grain legumes. Plant Methods 2024, 20, 134. [Google Scholar] [CrossRef]
- Kaur, R.; Gupta, S.; Tripathi, V.; Chauhan, A.; Parashar, D.; Shankar, P.; Kashyap, V. Microbiome based approaches for the degradation of polycyclic aromatic hydrocarbons (PAHs): A current perception. Chemosphere 2023, 341, 139951. [Google Scholar] [CrossRef] [PubMed]
- Akintola, A.A. AI-driven monitoring systems for bioremediation: Real-time data analysis and predictive modelling. World J. Adv. Res. Rev. 2024, 24, 788–803. [Google Scholar] [CrossRef]
- Davoodi, S.M.; Miri, S.; Brar, S.K.; Martel, R. Formulation of synthetic bacteria consortia for enzymatic biodegradation of polyaromatic hydrocarbons contaminated soil: Soil column study. Environ. Sci. Pollut. Res. 2023, 30, 72793–72806. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Li, G.; Wen, L.; Su, C.; Liu, Y.; Tang, H.; Dai, J. Biodegradation of aromatic pollutants meets synthetic biology. Synth. Syst. Biotechnol. 2021, 6, 153–162. [Google Scholar] [CrossRef]
- Zhou, Y.; Ren, M.; Zhang, P.; Jiang, D.; Yao, X.; Luo, Y.; Yang, Z.; Wang, Y. Application of Nanopore Sequencing in the Detection of Foodborne Microorganisms. Nanomaterials 2022, 12, 1534. [Google Scholar] [CrossRef]
- Perdigão, R.; Tomasino, M.P.; Magalhães, C.; Carvalho, M.F.; Almeida, C.M.R.; Mucha, A.P. Microbial response to a port fuel spill: Community dynamics and potential for bioremediation. Mar. Pollut. Bull. 2024, 203, 116434. [Google Scholar] [CrossRef]
- Mason, O.U.; Hazen, T.C.; Borglin, S.; Chain, P.S.; Dubinsky, E.A.; Fortney, J.L.; Han, J.; Holman, H.-Y.N.; Hultman, J.; Lamendella, R. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012, 6, 1715–1727. [Google Scholar] [CrossRef]
- Masís-Meléndez, F.; Segura-Montero, F.; Quesada-González, A. Control of septage sanitization by limes and lactic acid fermentation. J. Environ. Manag. 2021, 287, 112203. [Google Scholar] [CrossRef]
- Zhang, J.; Cao, X.; Yao, Z.; Lin, Q.; Yan, B.; Cui, X.; He, Z.; Yang, X.; Wang, C.-H.; Chen, G. Phytoremediation of Cd-contaminated farmland soil via various Sedum alfredii-oilseed rape cropping systems: Efficiency comparison and cost-benefit analysis. J. Hazard. Mater. 2021, 419, 126489. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, B.; Cui, Q.; Wu, Y. Genetically modified indigenous Pseudomonas aeruginosa drove bacterial community to change positively toward microbial enhanced oil recovery applications. J. Appl. Microbiol. 2024, 135, lxae168. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, J.; Jiang, L.; Zhang, L.; Raghavan, V.; Wang, J. A comprehensive review on novel synthetic foods: Potential risk factors, detection strategies, and processing technologies. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13371. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, K.G.; Daquigan, N.; White, J.R.; Morin, P.M.; Howard, L.M.; Manetas, J.E.; Ottesen, A.; Ramachandran, P.; Grim, C.J. Microbiomes associated with foods from plant and animal sources. Front. Microbiol. 2018, 9, 2540. [Google Scholar] [CrossRef] [PubMed]
- Anumudu, C.; Hart, A.; Miri, T.; Onyeaka, H. Recent advances in the application of the antimicrobial peptide nisin in the inactivation of spore-forming bacteria in foods. Molecules 2021, 26, 5552. [Google Scholar] [CrossRef] [PubMed]
- Maguire, M.; Ramachandran, P.; Tallent, S.; Mammel, M.K.; Brown, E.W.; Allard, M.W.; Musser, S.M.; González-Escalona, N. Precision metagenomics sequencing for food safety: Hybrid assembly of Shiga toxin-producing Escherichia coli in enriched agricultural water. Front. Microbiol. 2023, 14, 1221668. [Google Scholar] [CrossRef]
- Zehnder, C.; Béen, F.; Vojinovic, Z.; Savic, D.; Torres, A.S.; Mark, O.; Zlatanovic, L.; Abebe, Y.A. Machine learning for detecting virus infection hotspots via wastewater-based epidemiology: The case of SARS-CoV-2 RNA. GeoHealth 2023, 7, e2023GH000866. [Google Scholar] [CrossRef]
- Opara, C.N.; Anumudu, C.K. DIVERSITY, ANTIBIOGRAM AND PLASMID PROFILE OF MICROBIAL CONTAMINANTS OF SOME SELECTED VEGETABLES SOLD IN BAYELSA NIGERIA. Food Environ. Saf. J. 2022, 21, 129–142. [Google Scholar]
- Forbes, J.D.; Knox, N.C.; Ronholm, J.; Pagotto, F.; Reimer, A. Metagenomics: The next culture-independent game changer. Front. Microbiol. 2017, 8, 1069. [Google Scholar] [CrossRef]
- Moss, E.L.; Maghini, D.G.; Bhatt, A.S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat. Biotechnol. 2020, 38, 701–707. [Google Scholar] [CrossRef]
- Ma, L.; Yi, J.; Wisuthiphaet, N.; Earles, M.; Nitin, N. Accelerating the detection of bacteria in food using artificial intelligence and optical imaging. Appl. Environ. Microbiol. 2023, 89, e01828-22. [Google Scholar] [CrossRef]
- Cook, P.W.; Nightingale, K.K. Use of omics methods for the advancement of food quality and food safety. Anim. Front. 2018, 8, 33–41. [Google Scholar] [CrossRef]
- Commey, L.; Mechref, Y.; Burow, M.; Mendu, V. Identification and Characterization of Peanut Seed Coat Secondary Metabolites Inhibiting Aspergillus flavus Growth and Reducing Aflatoxin Contamination. J. Agric. Food Chem. 2024, 72, 23844–23858. [Google Scholar] [CrossRef] [PubMed]
- Gantzias, C.; Lappa, I.K.; Aerts, M.; Georgalaki, M.; Manolopoulou, E.; Papadimitriou, K.; De Brandt, E.; Tsakalidou, E.; Vandamme, P. MALDI-TOF MS profiling of non-starter lactic acid bacteria from artisanal cheeses of the Greek island of Naxos. Int. J. Food Microbiol. 2020, 323, 108586. [Google Scholar] [CrossRef] [PubMed]
- Wirth, N.T.; Kozaeva, E.; Nikel, P.I. Accelerated genome engineering of Pseudomonas putida by I-SceI―mediated recombination and CRISPR-Cas9 counterselection. Microb. Biotechnol. 2020, 13, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Feng, H. Microbial community analysis and food safety practice survey-based hazard identification and risk assessment for controlled environment hydroponic/aquaponic farming systems. Front. Microbiol. 2022, 13, 879260. [Google Scholar] [CrossRef]
- Li, S.; Mann, D.A.; Zhang, S.; Qi, Y.; Meinersmann, R.J.; Deng, X. Microbiome-informed food safety and quality: Longitudinal consistency and cross-sectional distinctiveness of retail chicken breast microbiomes. mSystems 2020, 5, e00589-20. [Google Scholar] [CrossRef]
- Painter, J.A.; Hoekstra, R.M.; Ayers, T.; Tauxe, R.V.; Braden, C.R.; Angulo, F.J.; Griffin, P.M. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 407. [Google Scholar] [CrossRef]
- Sundermann, A.J.; Chen, J.; Miller, J.K.; Martin, E.M.; Snyder, G.M.; Van Tyne, D.; Marsh, J.W.; Dubrawski, A.; Harrison, L.H. Whole-genome sequencing surveillance and machine learning for healthcare outbreak detection and investigation: A systematic review and summary. Antimicrob. Steward. Healthc. Epidemiol. 2022, 2, e91. [Google Scholar] [CrossRef]
- Zeng, W.; Wang, Q.; Wu, C.; Hang, M.; Huang, Z.; Huang, J.; Xia, Z. Recognition of foodborne pathogens terahertz spectrum based on convolutional neural network. J. Phys. 2022, 2226, 012012. [Google Scholar] [CrossRef]
- Peivasteh-Roudsari, L.; Pirhadi, M.; Karami, H.; Tajdar-Oranj, B.; Molaee-Aghaee, E.; Sadighara, P. Probiotics and food safety: An evidence-based review. J. Food Saf. Hyg. 2019, 5, 1–9. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics regulate gut microbiota: An effective method to improve immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Kato, I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016, 3, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Kok, C.R.; Hutkins, R. Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr. Rev. 2018, 76 (Suppl. S1), 4–15. [Google Scholar] [CrossRef]
- Rakib, M.R.H.; Kabir, A.; Amanullah, S.M. Starter cultures used in the production of probiotic dairy products and their potential applications: A Review. Chem. Biomol. Eng. 2017, 2, 83–89. [Google Scholar]
- Frakolaki, G.; Giannou, V.; Kekos, D.; Tzia, C. A review of the microencapsulation techniques for the incorporation of probiotic bacteria in functional foods. Crit. Rev. Food Sci. Nutr. 2021, 61, 1515–1536. [Google Scholar] [CrossRef]
- Anselmo, A.C.; McHugh, K.J.; Webster, J.; Langer, R.; Jaklenec, A. Layer-by-layer encapsulation of probiotics for delivery to the microbiome. Adv. Mater. 2016, 28, 9486. [Google Scholar] [CrossRef]
- Tarifa, M.C.; del Rosario Agustín, M.; Brugnoni, L.I. Biological control of foodborne pathogens by lactic acid bacteria: A focus on juice processing industries. Rev. Argent. De Microbiol. 2023, 55, 378–386. [Google Scholar] [CrossRef]
- Ribeiro, L.L.S.M.; Araújo, G.P.; de Oliveira Ribeiro, K.; Torres, I.M.S.; De Martinis, E.C.P.; Marreto, R.N.; Alves, V.F. Use of encapsulated lactic acid bacteria as bioprotective cultures in fresh Brazilian cheese. Braz. J. Microbiol. 2021, 52, 2247–2256. [Google Scholar] [CrossRef]
- Han, Y.; Akinsemolu, A.A.; Anumudu, C.K.; Miri, T.; Onyeaka, H. Antimicrobial Activity of Diffusible Substances Produced by Lactococcus lactis Against Bacillus cereus in a Non-Contact Co-Culture Model. Hygiene 2024, 4, 469–482. [Google Scholar] [CrossRef]
- Khullar, D. How A.I. Teaches Machines to Discover Drugs. In The New Yorker; Advance Publications, Inc.: Staten Island, NY, USA, 2024. [Google Scholar]
- Yu, W.; Ouyang, Z.; Zhang, Y.; Lu, Y.; Wei, C.; Tu, Y.; He, B. Research progress on the artificial intelligence applications in food safety and quality management. Trends Food Sci. Technol. 2024, 156, 104855. [Google Scholar] [CrossRef]
- Patil, A.; Singh, N.; Patwekar, M.; Patwekar, F.; Patil, A.; Gupta, J.K.; Elumalai, S.; Priya, N.S. AI-driven insights into the microbiota: Figuring out the mysterious world of the gut. Intell. Pharm. 2024, 3, 46–52. [Google Scholar] [CrossRef]
- Tadesse, S. Probiotics, prebiotics and synbiotics as functional food ingredients: Production, health benefits and safety. J. Biol. Act. Prod. Nat. 2012, 2, 124–134. [Google Scholar] [CrossRef]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M. Prebiotics and probiotics; modifying and mining the microbiota. Pharmacol. Res. 2010, 61, 213–218. [Google Scholar] [CrossRef]
- Callaway, T.R.; Edrington, T.; Harvey, R.B.; Anderson, R.C.; Nisbet, D.J. Prebiotics in food animals, a potential to reduce foodborne pathogens and disease. Rom. Biotechnol. Lett. 2012, 17, 7808–7816. [Google Scholar]
- Pouillart, P.R.; Dépeint, F.; Abdelnour, A.; Deremaux, L.; Vincent, O.; Mazière, J.-C.; Madec, J.-Y.; Chatelain, D.; Younes, H.; Wils, D. Nutriose, a prebiotic low-digestible carbohydrate, stimulates gut mucosal immunity and prevents TNBS-induced colitis in piglets. Inflamm. Bowel Dis. 2010, 16, 783–794. [Google Scholar] [CrossRef]
- Fazilah, N.F.; Ariff, A.B.; Khayat, M.E.; Rios-Solis, L.; Halim, M. Influence of probiotics, prebiotics, synbiotics and bioactive phytochemicals on the formulation of functional yogurt. J. Funct. Foods 2018, 48, 387–399. [Google Scholar] [CrossRef]
- Enan, G.; Amen, S.; Abd El-badiea, A.; Abd El-Hack, M.E.; Abdel-Shafi, S. The pathogen inhibition effects of probiotics and prebiotics against spp. in chicken. Ann. Anim. Sci. 2023, 23, 537–544. [Google Scholar] [CrossRef]
- Khan, S.; Chousalkar, K.K. Salmonella Typhimurium infection disrupts but continuous feeding of Bacillus based probiotic restores gut microbiota in infected hens. J. Anim. Sci. Biotechnol. 2020, 11, 29. [Google Scholar] [CrossRef]
- Kumari, A.; Ranadheera, C.; Prasanna, P.; Senevirathne, N.; Vidanarachchi, J. Development of a rice incorporated synbiotic yogurt with low retrogradation properties. Int. Food Res. J. 2015, 22, 2032. [Google Scholar]
- Khurana, H.; Kanawjia, S. Recent trends in development of fermented milks. Curr. Nutr. Food Sci. 2007, 3, 91–108. [Google Scholar] [CrossRef]
- Mounir, M.; Ibijbijen, A.; Farih, K.; Rabetafika, H.N.; Razafindralambo, H.L. Synbiotics and their antioxidant properties, mechanisms, and benefits on human and animal health: A narrative review. Biomolecules 2022, 12, 1443. [Google Scholar] [CrossRef] [PubMed]
- Cukkemane, A.; Kumar, P.; Sathyamoorthy, B. A metabolomics footprint approach to understanding the benefits of synbiotics in functional foods and dietary therapeutics for health, communicable and non-communicable diseases. Food Res. Int. 2020, 128, 108679. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; de Moreno de LeBlanc, A.; de Souza Oliveira, R.P.; Todorov, S.D. Use of Synbiotics (Probiotics and Prebiotics) to Improve the Safety of Foods. In Practical Food Safety: Contemporary Issues and Future Directions; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; pp. 497–516. [Google Scholar]
- Baffoni, L.; Gaggìa, F.; Di Gioia, D.; Santini, C.; Mogna, L.; Biavati, B. A Bifidobacterium-based synbiotic product to reduce the transmission of C. jejuni along the poultry food chain. Int. J. Food Microbiol. 2012, 157, 156–161. [Google Scholar] [CrossRef]
- Picaud, J.C.; Chapalain, V.; Paineau, D.; Zourabichvili, O.; Bornet, F.R.; Duhamel, J.F. Incidence of infectious diseases in infants fed follow-on formula containing synbiotics: An observational study. Acta Paediatr. 2010, 99, 1695–1700. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B.; Delattre, A.-S.; Lavigne, R. Learning from bacteriophages-advantages and limitations of phage and phage-encoded protein applications. Curr. Protein Pept. Sci. 2012, 13, 699–722. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Costa, A.R.; Kluskens, L.D.; Azeredo, J. Revisiting phage therapy: New applications for old resources. Trends Microbiol. 2015, 23, 185–191. [Google Scholar] [CrossRef]
- Kutter, E.; De Vos, D.; Gvasalia, G.; Alavidze, Z.; Gogokhia, L.; Kuhl, S.; Abedon, S.T. Phage therapy in clinical practice: Treatment of human infections. Curr. Pharm. Biotechnol. 2010, 11, 69–86. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.M.; Alkawareek, M.Y.; Donnelly, R.F.; Gilmore, B.F. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol. Med. Microbiol. 2012, 65, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Allegretti, J.R.; Mullish, B.H.; Kelly, C.; Fischer, M. The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. Lancet 2019, 394, 420–431. [Google Scholar] [CrossRef]
- Almeida, C.; Oliveira, R.; Baylina, P.; Fernandes, R.; Teixeira, F.G.; Barata, P. Current trends and challenges of fecal microbiota transplantation—An easy method that works for all? Biomedicines 2022, 10, 2742. [Google Scholar] [CrossRef] [PubMed]
- Khoruts, A.; Sadowsky, M.J. Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Quraishi, M.N.; Widlak, M.; Bhala, N.A.; Moore, D.; Price, M.; Sharma, N.; Iqbal, T. Systematic review with meta-analysis: The efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment. Pharmacol. Ther. 2017, 46, 479–493. [Google Scholar] [CrossRef]
- Greenberg, S.A.; Youngster, I.; Cohen, N.A.; Livovsky, D.M.; Strahilevitz, J.; Israeli, E.; Melzer, E.; Paz, K.; Fliss-Isakov, N.; Maharshak, N. Five years of fecal microbiota transplantation-an update of the Israeli experience. World J. Gastroenterol. 2018, 24, 5403. [Google Scholar] [CrossRef]
- Anderson, J.; Edney, R.; Whelan, K. Systematic review: Faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment. Pharmacol. Ther. 2012, 36, 503–516. [Google Scholar] [CrossRef]
- Mimee, M.; Citorik, R.J.; Lu, T.K. Microbiome therapeutics—Advances and challenges. Adv. Drug Deliv. Rev. 2016, 105, 44–54. [Google Scholar] [CrossRef]
- Thanush, D.; Basavaraj, H.; Gowrav, M. Current regulation and initial considerations for successful development and commercialization of microbiome therapies. Adv. Gut Microbiome Res. 2023, 2023, 6657515. [Google Scholar] [CrossRef]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.A.; Alm, E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Ceroni, F.; Algar, R.; Stan, G.-B.; Ellis, T. Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat. Methods 2015, 12, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Fischer, M. Expert opinion on fecal microbiota transplantation for the treatment of Clostridioides difficile infection and beyond. Expert Opin. Biol. Ther. 2020, 20, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Mantegazza, C.; Molinari, P.; D’Auria, E.; Sonnino, M.; Morelli, L.; Zuccotti, G.V. Probiotics and antibiotic-associated diarrhea in children: A review and new evidence on Lactobacillus rhamnosus GG during and after antibiotic treatment. Pharmacol. Res. 2018, 128, 63–72. [Google Scholar] [CrossRef]
- USFDA. Fecal Microbiota for Transplantation: Safety Alert—Risk of Serious Adverse Events Likely Due to Transmission of Pathogenic Organisms 2020. Available online: https://www.fda.gov/safety/medical-product-safety-information/fecal-microbiota-transplantation-safety-alert-risk-serious-adverse-events-likely-due-transmission#:~:text=Product%20Safety%20Information-,Fecal%20Microbiota%20for%20Transplantation%3A%20Safety%20Alert%20%2D%20Risk%20of%20Serious%20Adverse,to%20Transmission%20of%20Pathogenic%20Organisms&text=ISSUE%3A%20FDA%20is%20informing%20health,microbiota%20for%20transplantation%20(FMT) (accessed on 14 August 2024).
- Yelin, I.; Flett, K.B.; Merakou, C.; Mehrotra, P.; Stam, J.; Snesrud, E.; Hinkle, M.; Lesho, E.; McGann, P.; McAdam, A.J. Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat. Med. 2019, 25, 1728–1732. [Google Scholar] [CrossRef]
- Manrique, P.; Montero, I.; Fernandez-Gosende, M.; Martinez, N.; Cantabrana, C.H.; Rios-Covian, D. Past, present, and future of microbiome-based therapies. Microbiome Res. Rep. 2024, 3, 23. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Wang, Z.-K.; Yang, Y.-S.; Chen, Y.; Yuan, J.; Sun, G.; Peng, L.-H. Intestinal microbiota pathogenesis and fecal microbiota transplantation for inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 14805. [Google Scholar] [CrossRef]
- Hoffmann, D.E. Introduction: The promise and challenges of microbiome-based therapies. J. Law Med. Ethics 2019, 47, 476–481. [Google Scholar] [CrossRef]
- Khoruts, A.; Hoffmann, D.E.; Palumbo, F.B. The impact of regulatory policies on the future of fecal microbiota transplantation. J. Law Med. Ethics 2019, 47, 482–504. [Google Scholar] [CrossRef]
- USFDA. Enforcement Policy Regarding Investigational New Drug Requirements for Use of Fecal Microbiota for Transplantation to Treat Clostridium Difficile Infection Not Responsive to Standard Therapies. 2022. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/enforcement-policy-regarding-investigational-new-drug-requirements-use-fecal-microbiota (accessed on 14 August 2024).
- Cammarota, G.; Masucci, L.; Ianiro, G.; Bibbò, S.; Dinoi, G.; Costamagna, G.; Sanguinetti, M.; Gasbarrini, A. Randomised clinical trial: Faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment. Pharmacol. Ther. 2015, 41, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Hvas, C.L.; Jørgensen, S.M.D.; Jørgensen, S.P.; Storgaard, M.; Lemming, L.; Hansen, M.M.; Erikstrup, C.; Dahlerup, J.F. Fecal microbiota transplantation is superior to fidaxomicin for treatment of recurrent Clostridium difficile infection. Gastroenterology 2019, 156, 1324–1332.e3. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, D.; Palumbo, F.; Ravel, J.; Roghmann, M.-C.; Rowthorn, V.; von Rosenvinge, E. Improving regulation of microbiota transplants. Science 2017, 358, 1390–1391. [Google Scholar] [CrossRef]
- Grigoryan, Z.; Shen, M.J.; Twardus, S.W.; Beuttler, M.M.; Chen, L.A.; Bateman-House, A. Fecal microbiota transplantation: Uses, questions, and ethics. Med. Microecol. 2020, 6, 100027. [Google Scholar] [CrossRef]
- Fanfan, D.; Mulligan, C.J.; Groer, M.; Mai, V.; Weaver, M.; Huffman, F.; Lyon, D.E. The intersection of social determinants of health, the microbiome, and health outcomes in immigrants: A scoping review. Am. J. Biol. Anthropol. 2024, 183, 3–19. [Google Scholar] [CrossRef]
- Gaikwad, P.L.; Naoghare, M. Data Anonymization Approach for Data Privacy. Int. J. Sci. Res. 2015, 4, 1534–1539. Available online: https://www.ijsr.net/getabstract.php?paperid=12121502 (accessed on 5 May 2025).
- Wagner, J.; Paulson, J.N.; Wang, X.; Bhattacharjee, B.; Corrada Bravo, H. Privacy-preserving microbiome analysis using secure computation. Bioinformatics 2016, 32, 1873–1879. [Google Scholar] [CrossRef]
- Freedman, S.B.; Schnadower, D.; Tarr, P.I. The probiotic conundrum: Regulatory confusion, conflicting studies, and safety concerns. JAMA 2020, 323, 823–824. [Google Scholar] [CrossRef]
- Baral, K.C.; Bajracharya, R.; Lee, S.H.; Han, H.-K. Advancements in the pharmaceutical applications of probiotics: Dosage forms and formulation technology. Int. J. Nanomed. 2021, 16, 7535–7556. [Google Scholar] [CrossRef]
- McFarland, L.V.; Evans, C.T.; Goldstein, E.J. Strain-specificity and disease-specificity of probiotic efficacy: A systematic review and meta-analysis. Front. Med. 2018, 5, 124. [Google Scholar] [CrossRef]
- Yadav, M.; Chauhan, N.S. Microbiome therapeutics: Exploring the present scenario and challenges. Gastroenterol. Rep. 2022, 10, goab046. [Google Scholar] [CrossRef] [PubMed]
- Cruz, N.; Abernathy, G.A.; Dichosa, A.E.; Kumar, A. The age of next-generation therapeutic-microbe discovery: Exploiting microbe-microbe and host-microbe interactions for disease prevention. Infect. Immun. 2022, 90, e00589-21. [Google Scholar] [CrossRef] [PubMed]
- Blustein, J.; Liu, J. Time to consider the risks of caesarean delivery for long term child health. BMJ 2015, 350, h2410. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.T.; Hourigan, S.K.; Hoffmann, D.E.; Levy, L.; von Rosenvinge, E.C.; Chou, B.; Dominguez-Bello, M.-G. Bacterial baptism: Scientific, medical, and regulatory issues raised by vaginal seeding of C-section-born babies. J. Law Med. Ethics 2019, 47, 568–578. [Google Scholar] [CrossRef]
- Kim, J.; Muhammad, N.; Jhun, B.H.; Yoo, J.-W. Probiotic delivery systems: A brief overview. J. Pharm. Investig. 2016, 46, 377–386. [Google Scholar] [CrossRef]
- Mills, S.; Stanton, C.; Fitzgerald, G.F.; Ross, R. Enhancing the stress responses of probiotics for a lifestyle from gut to product and back again. Microb. Cell Factories 2011, 10, S19. [Google Scholar] [CrossRef]
- Jose, N.M.; Bunt, C.R.; Hussain, M.A. Implications of antibiotic resistance in probiotics. Food Rev. Int. 2015, 31, 52–62. [Google Scholar] [CrossRef]
- Jacobsen, L.; Wilcks, A.; Hammer, K.; Huys, G.; Gevers, D.; Andersen, S.R. Horizontal transfer of tet (M) and erm (B) resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiol. Ecol. 2007, 59, 158–166. [Google Scholar] [CrossRef]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic resistance among commercially available probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Imperial, I.C.; Ibana, J.A. Addressing the antibiotic resistance problem with probiotics: Reducing the risk of its double-edged sword effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef]
- Zheng, M.; Zhang, R.; Tian, X.; Zhou, X.; Pan, X.; Wong, A. Assessing the risk of probiotic dietary supplements in the context of antibiotic resistance. Front. Microbiol. 2017, 8, 908. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Pan, L.; Li, L.; Lu, J.; Kwok, L.; Menghe, B.; Zhang, H.; Zhang, W. Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food Sci. 2017, 82, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhang, Z.-Y.; Ke, D.; Jian-Ping, Y.; Xiao-Kui, G. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed. Environ. Sci. 2009, 22, 401–412. [Google Scholar]
- Chen, J.; Wang, Q.; Liu, C.-M.; Gong, J. Issues deserve attention in encapsulating probiotics: Critical review of existing literature. Crit. Rev. Food Sci. Nutr. 2017, 57, 1228–1238. [Google Scholar] [CrossRef]
- Cassani, L.; Gomez-Zavaglia, A.; Simal-Gandara, J. Technological strategies ensuring the safe arrival of beneficial microorganisms to the gut: From food processing and storage to their passage through the gastrointestinal tract. Food Res. Int. 2020, 129, 108852. [Google Scholar] [CrossRef]
- Lee, S.H.; Bajracharya, R.; Min, J.Y.; Han, J.-W.; Park, B.J.; Han, H.-K. Strategic approaches for colon targeted drug delivery: An overview of recent advancements. Pharmaceutics 2020, 12, 68. [Google Scholar] [CrossRef]
- Yao, M.; Xie, J.; Du, H.; McClements, D.J.; Xiao, H.; Li, L. Progress in microencapsulation of probiotics: A review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 857–874. [Google Scholar] [CrossRef]
- Mishra, A.; Lai, G.C.; Yao, L.J.; Aung, T.T.; Shental, N.; Rotter-Maskowitz, A.; Shepherdson, E.; Singh, G.S.N.; Pai, R.; Shanti, A. Microbial exposure during early human development primes fetal immune cells. Cell 2021, 184, 3394–3409.e20. [Google Scholar] [CrossRef]
- Kwa, W.T.; Sundarajoo, S.; Toh, K.Y.; Lee, J. Application of emerging technologies for gut microbiome research. Singap. Med. J. 2023, 64, 45–52. [Google Scholar] [CrossRef]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ‘unculturable’human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef]
- Hu, K.A.; Gubatan, J. Gut microbiome–based therapeutics in inflammatory bowel disease. Clin. Transl. Discov. 2023, 3, e182. [Google Scholar] [CrossRef]
- Dodiya, H.B.; Lutz, H.L.; Weigle, I.Q.; Patel, P.; Michalkiewicz, J.; Roman-Santiago, C.J.; Zhang, C.M.; Liang, Y.; Srinath, A.; Zhang, X. Gut microbiota–driven brain Aβ amyloidosis in mice requires microglia. J. Exp. Med. 2021, 219, e20200895. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Chau, S.W.; Liu, Y.; Chan, J.W.; Wang, J.; Ma, S.L.; Zhang, J.; Chan, P.K.; Yeoh, Y.K.; Chen, Z. Gut microbiome dysbiosis across early Parkinson’s disease, REM sleep behavior disorder and their first-degree relatives. Nat. Commun. 2023, 14, 2501. [Google Scholar] [CrossRef] [PubMed]
- Palacios, N.; Wilkinson, J.; Bjornevik, K.; Schwarzschild, M.A.; McIver, L.; Ascherio, A.; Huttenhower, C. Metagenomics of the gut microbiome in Parkinson’s disease: Prodromal changes. Ann. Neurol. 2023, 94, 486–501. [Google Scholar] [CrossRef]
- Seo, D.-o.; O’Donnell, D.; Jain, N.; Ulrich, J.D.; Herz, J.; Li, Y.; Lemieux, M.; Cheng, J.; Hu, H.; Serrano, J.R. ApoE isoform–and microbiota-dependent progression of neurodegeneration in a mouse model of tauopathy. Science 2023, 379, eadd1236. [Google Scholar] [CrossRef]
- Lee, K.A.; Thomas, A.M.; Bolte, L.A.; Björk, J.R.; de Ruijter, L.K.; Armanini, F.; Asnicar, F.; Blanco-Miguez, A.; Board, R.; Calbet-Llopart, N. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. 2022, 28, 535–544. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar]
- Formosinho, J.; Bencard, A.; Whiteley, L. Environmentality in biomedicine: Microbiome research and the perspectival body. Stud. Hist. Philos. Sci. 2022, 91, 148–158. [Google Scholar] [CrossRef]
- Sariola, S.; Butcher, A. In critique of anthropocentrism: A more-than-human ethical framework for antimicrobial resistance. Med. Humanit. 2022, 48, e16. [Google Scholar]
- Blanton, L.V.; Charbonneau, M.R.; Salih, T.; Barratt, M.J.; Venkatesh, S.; Ilkaveya, O.; Subramanian, S.; Manary, M.J.; Trehan, I.; Jorgensen, J.M. MICROBIOME gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 2016, 351. [Google Scholar] [CrossRef] [PubMed]
- Amaral, W.Z.; Lubach, G.R.; Proctor, A.; Lyte, M.; Phillips, G.J.; Coe, C.L. Social influences on Prevotella and the gut microbiome of young monkeys. Biopsychosoc. Sci. Med. 2017, 79, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Bennett, G.; Malone, M.; Sauther, M.L.; Cuozzo, F.P.; White, B.; Nelson, K.E.; Stumpf, R.M.; Knight, R.; Leigh, S.R.; Amato, K.R. Host age, social group, and habitat type influence the gut microbiota of wild ring-tailed lemurs (Lemur catta). Am. J. Primatol. 2016, 78, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.V.-A.; Foster, K.R. Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 2018, 16, 647–655. [Google Scholar] [CrossRef]
- Narayana, J.K.; Mac Aogáin, M.; Goh, W.W.B.; Xia, K.; Tsaneva-Atanasova, K.; Chotirmall, S.H. Mathematical-based microbiome analytics for clinical translation. Comput. Struct. Biotechnol. J. 2021, 19, 6272–6281. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, H.; Lan, C.; Ren, J. Help, hope and hype: Ethical considerations of human microbiome research and applications. Protein Cell 2018, 9, 404–415. [Google Scholar] [CrossRef]
- Adetunji, J.B.; Michael, O.S.; Adetunji, C.O.; Ajayi, O.O.; Ogundolie, F.A. Procedures for sampling of small and larger samples of microbiome. In An Introduction to the Microbiome Health and Diseases; Elsevier: Amsterdam, The Netherlands, 2024; pp. 33–47. [Google Scholar]
- Carney, S.M.; Clemente, J.C.; Cox, M.J.; Dickson, R.P.; Huang, Y.J.; Kitsios, G.D.; Kloepfer, K.M.; Leung, J.M.; LeVan, T.D.; Molyneaux, P.L. Methods in lung microbiome research. Am. J. Respir. Cell Mol. Biol. 2020, 62, 283–299. [Google Scholar] [CrossRef]
- Vandeputte, D.; Tito, R.Y.; Vanleeuwen, R.; Falony, G.; Raes, J. Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 2017, 41 (Suppl. S1), S154–S167. [Google Scholar] [CrossRef]
- FitzGerald, M.J.; Spek, E.J. Microbiome therapeutics and patent protection. Nat. Biotechnol. 2020, 38, 806–810. [Google Scholar] [CrossRef]
- Merrick, B.; Allen, L.; Zain, N.M.M.; Forbes, B.; Shawcross, D.L.; Goldenberg, S.D. Regulation, risk and safety of faecal microbiota transplant. Infect. Prev. Pract. 2020, 2, 100069. [Google Scholar] [CrossRef]
- Kim, T.-W.; Che, J.-H.; Yun, J.-W. Use of stem cells as alternative methods to animal experimentation in predictive toxicology. Regul. Toxicol. Pharmacol. 2019, 105, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, C.; Alves da Silva, M.; Saraiva, M.; Neyazi, M.; Olsson, I.A.S.; Bartfeld, S. Organoids as host models for infection biology—A review of methods. Exp. Mol. Med. 2021, 53, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Tovaglieri, A.; Sontheimer-Phelps, A.; Geirnaert, A.; Prantil-Baun, R.; Camacho, D.M.; Chou, D.B.; Jalili-Firoozinezhad, S.; de Wouters, T.; Kasendra, M.; Super, M. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 2019, 7, 43. [Google Scholar] [CrossRef]
- Gupta, V.K.; Kim, M.; Bakshi, U.; Cunningham, K.Y.; Davis, J.M., III; Lazaridis, K.N.; Nelson, H.; Chia, N.; Sung, J. A predictive index for health status using species-level gut microbiome profiling. Nat. Commun. 2020, 11, 4635. [Google Scholar] [CrossRef]
- Lian, X.; Yang, S.; Li, H.; Fu, C.; Zhang, Z. Machine-learning-based predictor of human–bacteria protein–protein interactions by incorporating comprehensive host-network properties. J. Proteome Res. 2019, 18, 2195–2205. [Google Scholar] [CrossRef]
- Veiga, P.; Suez, J.; Derrien, M.; Elinav, E. Moving from probiotics to precision probiotics. Nat. Microbiol. 2020, 5, 878–880. [Google Scholar] [CrossRef]
- Amato, K.R.; Maurice, C.F.; Guillemin, K.; Giles-Vernick, T. Multidisciplinarity in microbiome research: A challenge and opportunity to rethink causation, variability, and scale. BioEssays 2019, 41, 1900007. [Google Scholar] [CrossRef]
- Islam, W.; Noman, A.; Naveed, H.; Huang, Z.; Chen, H.Y.H. Role of environmental factors in shaping the soil microbiome. Environ. Sci. Pollut. Res. 2020, 27, 41225–41247. [Google Scholar] [CrossRef]
- Bilal, H.; Khan, M.N.; Khan, S.; Shafiq, M.; Fang, W.; Khan, R.U.; Rahman, M.U.; Li, X.; Lv, Q.-L.; Xu, B. The role of artificial intelligence and machine learning in predicting and combating antimicrobial resistance. Comput. Struct. Biotechnol. J. 2025, 27, 423–439. [Google Scholar] [CrossRef]
- Noyes, N.R.; Yang, X.; Linke, L.M.; Magnuson, R.J.; Dettenwanger, A.; Cook, S.; Geornaras, I.; Woerner, D.E.; Gow, S.P.; McAllister, T.A.; et al. Resistome diversity in cattle and the environment decreases during beef production. eLife 2016, 5, e13195. [Google Scholar] [CrossRef]
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529. [Google Scholar] [CrossRef] [PubMed]
- UCD Centre for Food Safety, Dublin, Ireland; Van Hoorde, K.; Butler, F. Use of next-generation sequencing in microbial risk assessment. EFSA J. 2018, 16, e16086. [Google Scholar]
- Danchin, A. Artificial intelligence-based prediction of pathogen emergence and evolution in the world of synthetic biology. Microb. Biotechnol. 2024, 17, e70014. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Wang, H.; Shi, T.; Wang, B. The genus Cladosporium: A prospective producer of natural products. International Int. J. Mol. Sci. 2024, 25, 1652. [Google Scholar] [CrossRef] [PubMed]
- Ashaolu, T.J.; Greff, B.; Varga, L. Action and immunomodulatory mechanisms, formulations, and safety concerns of probiotics. Biosci. Microbiota Food Health 2025, 44, 4–15. [Google Scholar] [CrossRef]
- FAO/WHO. Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; World Health Organization, Food and Agriculture Organization of the United Nations: London, ON, Canada, 2002; Volume 30, pp. 16–22. [Google Scholar]
- Khodaei, D.; Hamidi-Esfahani, Z. Influence of bioactive edible coatings loaded with Lactobacillus plantarum on physicochemical properties of fresh strawberries. Postharvest Biol. Technol. 2019, 156, 110944. [Google Scholar] [CrossRef]
- Silva, D.R.; Sardi, J.d.C.O.; Pitangui, N.d.S.; Roque, S.M.; da Silva, A.C.B.; Rosalen, P.L. Probiotics as an alternative antimicrobial therapy: Current reality and future directions. J. Funct. Foods 2020, 73, 104080. [Google Scholar] [CrossRef]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef]
- Wang, X.; Cui, Y.; Sang, C.; Wang, B.; Yuan, Y.; Liu, L.; Yuan, Y.; Yue, T. Fungi with potential probiotic properties isolated from Fuzhuan brick tea. Food Sci. Hum. Wellness 2022, 11, 686–696. [Google Scholar] [CrossRef]
- Guryanova, S.V. Immunomodulation, bioavailability and safety of bacteriocins. Life 2023, 13, 1521. [Google Scholar] [CrossRef] [PubMed]
- Khaneghah, A.M.; Abhari, K.; Eş, I.; Soares, M.B.; Oliveira, R.B.; Hosseini, H.; Rezaei, M.; Balthazar, C.F.; Silva, R.; Cruz, A.G.; et al. Interactions between probiotics and pathogenic microorganisms in hosts and foods: A review. Trends Food Sci. Technol. 2020, 95, 205–218. [Google Scholar] [CrossRef]
- Corsetti, A.; Settanni, L.; Valmorri, S.; Mastrangelo, M.; Suzzi, G. Identification of subdominant sourdough lactic acid bacteria and their evolution during laboratory-scale fermentations. Food Microbiol. 2007, 24, 592–600. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Van Kerrebroeck, S.; Harth, H.; Huys, G.; Daniel, H.M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef]
- Bali, V.; Panesar, P.S.; Bera, M.B.; Kennedy, J.F. Bacteriocins: Recent trends and potential applications. Crit. Rev. Food Sci. Nutr. 2016, 56, 817–834. [Google Scholar] [CrossRef]
- Werning, M.L.; Hernández-Alcántara, A.M.; Ruiz, M.J.; Soto, L.P.; Dueñas, M.T.; López, P.; Frizzo, L.S. Biological functions of exopolysaccharides from lactic acid bacteria and their potential benefits for humans and farmed animals. Foods 2022, 11, 1284. [Google Scholar] [CrossRef]
- Van Zyl, W.F.; Deane, S.M.; Dicks, L.M. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef]
- Stupar, J.; Holøymoen, I.G.; Hoel, S.; Lerfall, J.; Rustad, T.; Jakobsen, A.N. Diversity and antimicrobial activity towards Listeria spp. and Escherichia coli among lactic acid bacteria isolated from ready-to-eat seafood. Foods 2021, 10, 271. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).