A Systematic Analysis of Neural Networks, Fuzzy Logic and Genetic Algorithms in Tumor Classification
Abstract
1. Introduction
- What are the trends in applying ANNs for segmentation tasks, as reflected in the types of ANNs, datasets, and segmentation approaches used in the recent literature?
- How do different training and testing mechanisms and evaluation metrics correlate with performance outcomes (e.g., accuracy, precision) in ANN-based segmentation models across the selected papers?
2. Methodology
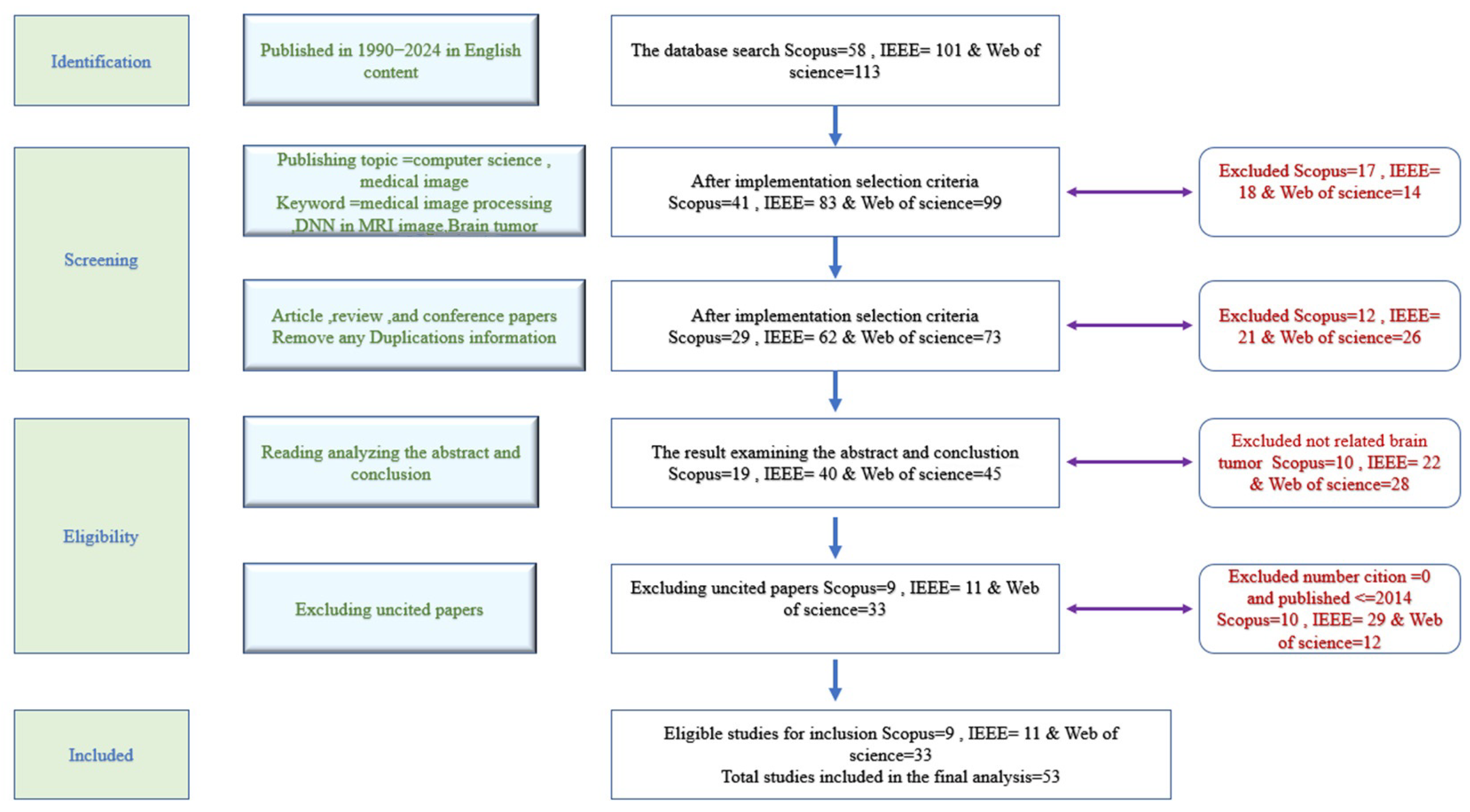
2.1. PRISMA Basic Spets
- Search strategy;
- Selection criteria;
- Ensuring result quality.
- Data Extraction
- Web of Science: in total, 5 papers removed;
- IEEE: in total, 4 papers removed;
- Scopus: in total, 10 papers removed.
- Web of Science: in total, 15 additional papers removed;
- IEEE: in total, 13 additional papers removed.
- A.
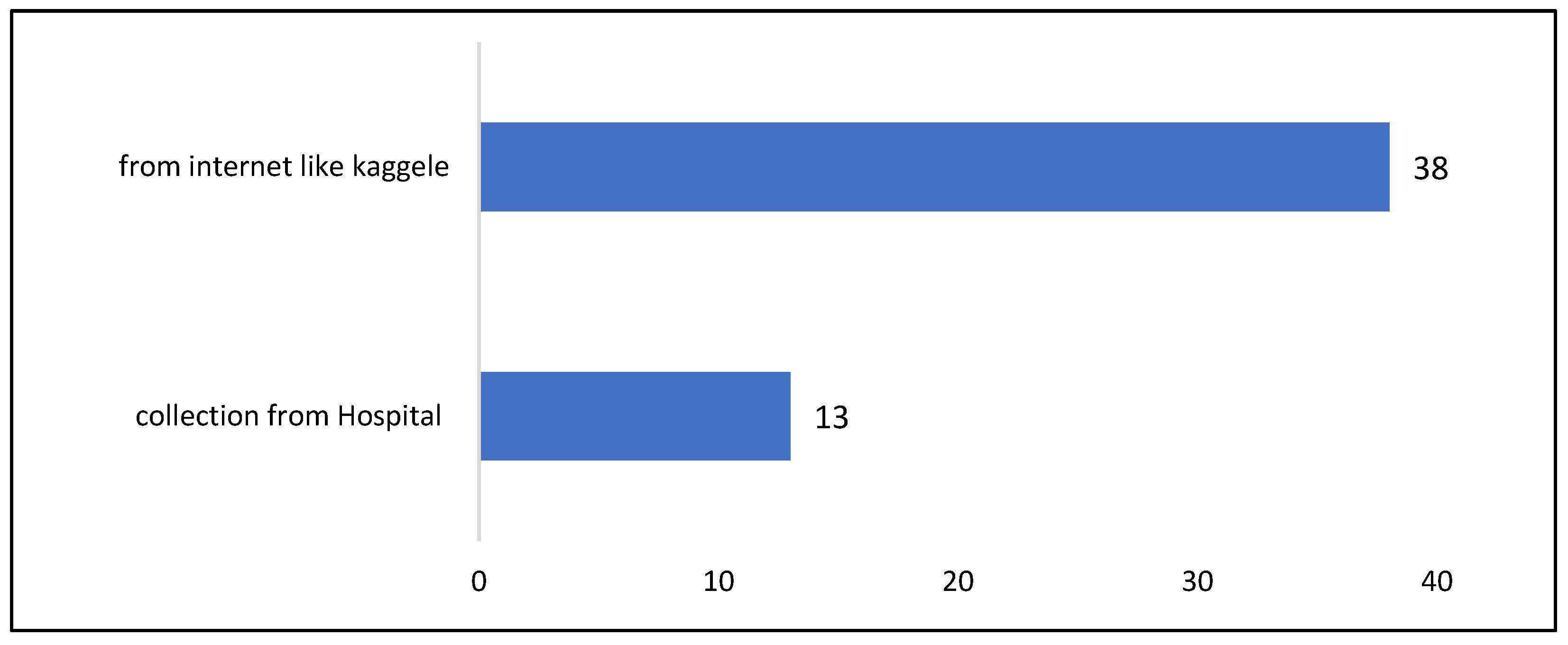
- Extracting articles that employed magnetic resonance imaging (MRI)The abstract summarizes the research work, making it useful for identifying relevant studies. Analyzing and evaluating the abstract content helped isolate research that specifically employed MRI. Any study that did not rely on MRI for conducting experiments was excluded. As a result, 28 articles were excluded from Web of Science, 22 from IEEE, and 10 from the Scopus database.
- B.
- Extracting highly cited articlesThis step involved identifying and selecting articles with high citation counts from the past decade (2014–2024). As a result, 12 papers were excluded from Web of Science, 29 from IEEE, and 10 from the Scopus database.
2.2. Data Analysis
- A conceptual contribution refers to descriptive, comparative, analytical, or review-based research;
- A practical contribution indicates that the research involved designing, developing, implementing a program, or presenting a novel algorithm.
- The “field” perspective examines the specific parameters researchers worked with;
- The “parameter” perspective reflects the targeted application areas.
3. Results
3.1. Descriptive Analysis
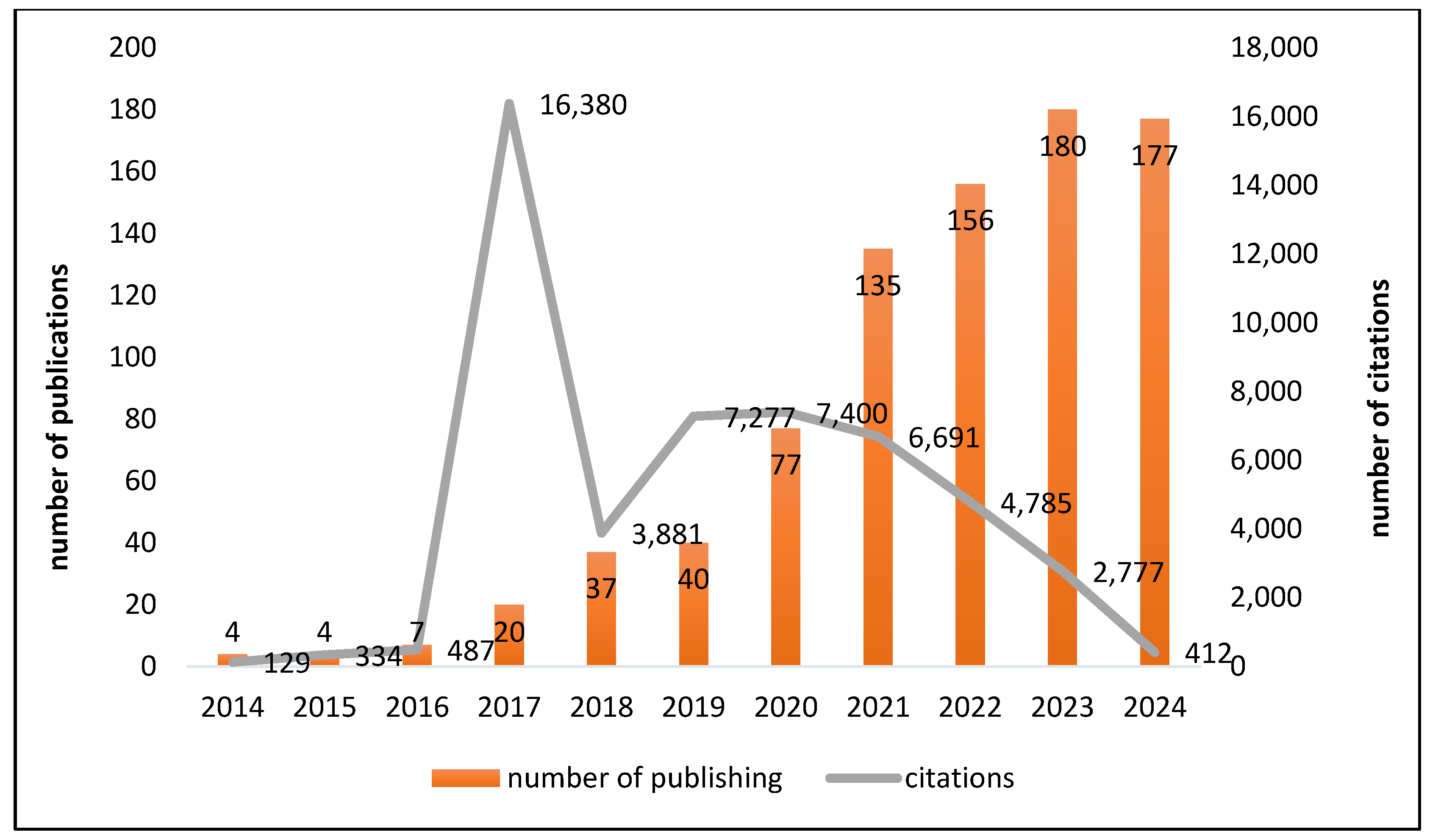
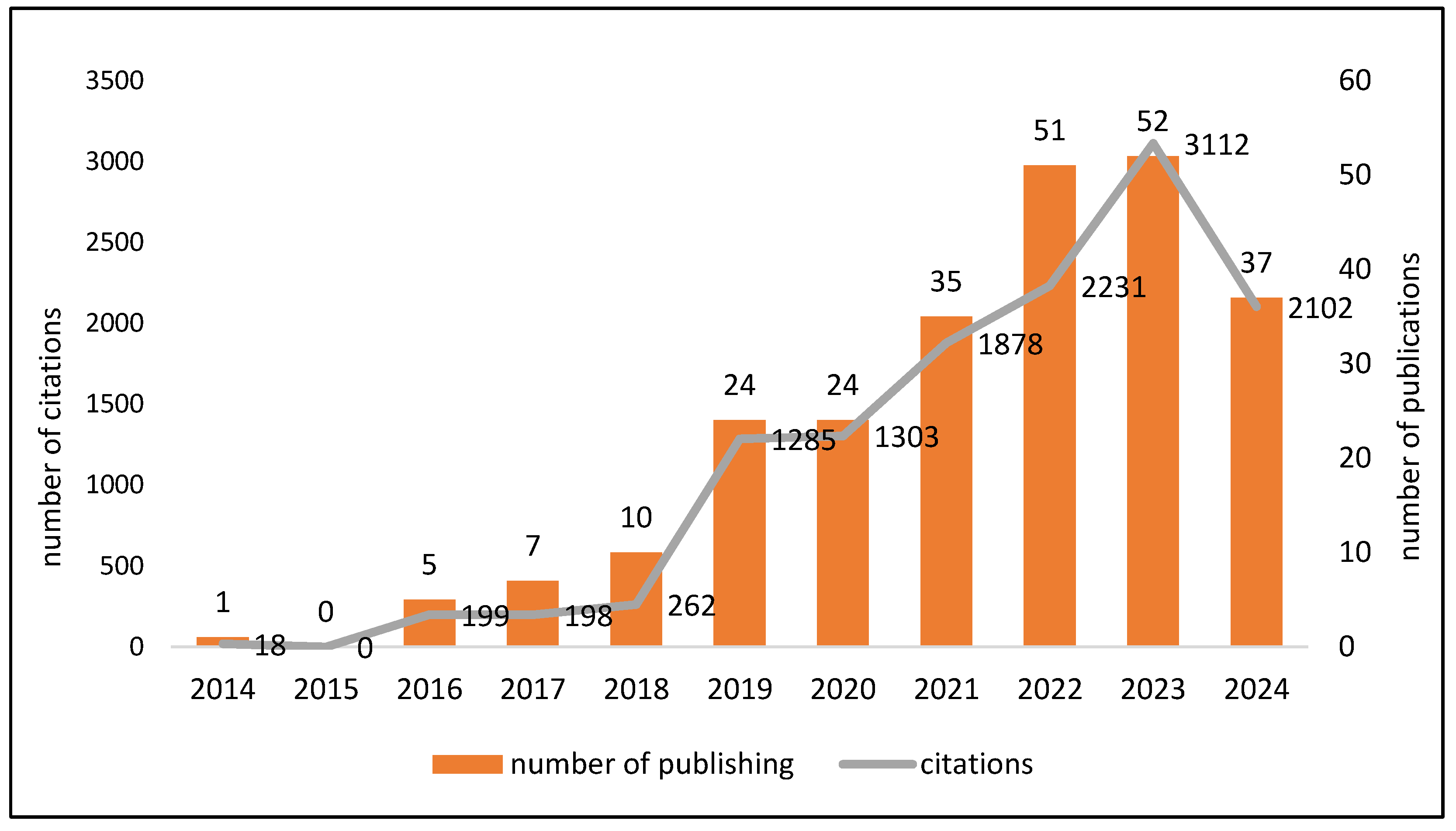
3.2. Trends in Citations and Publications over Time
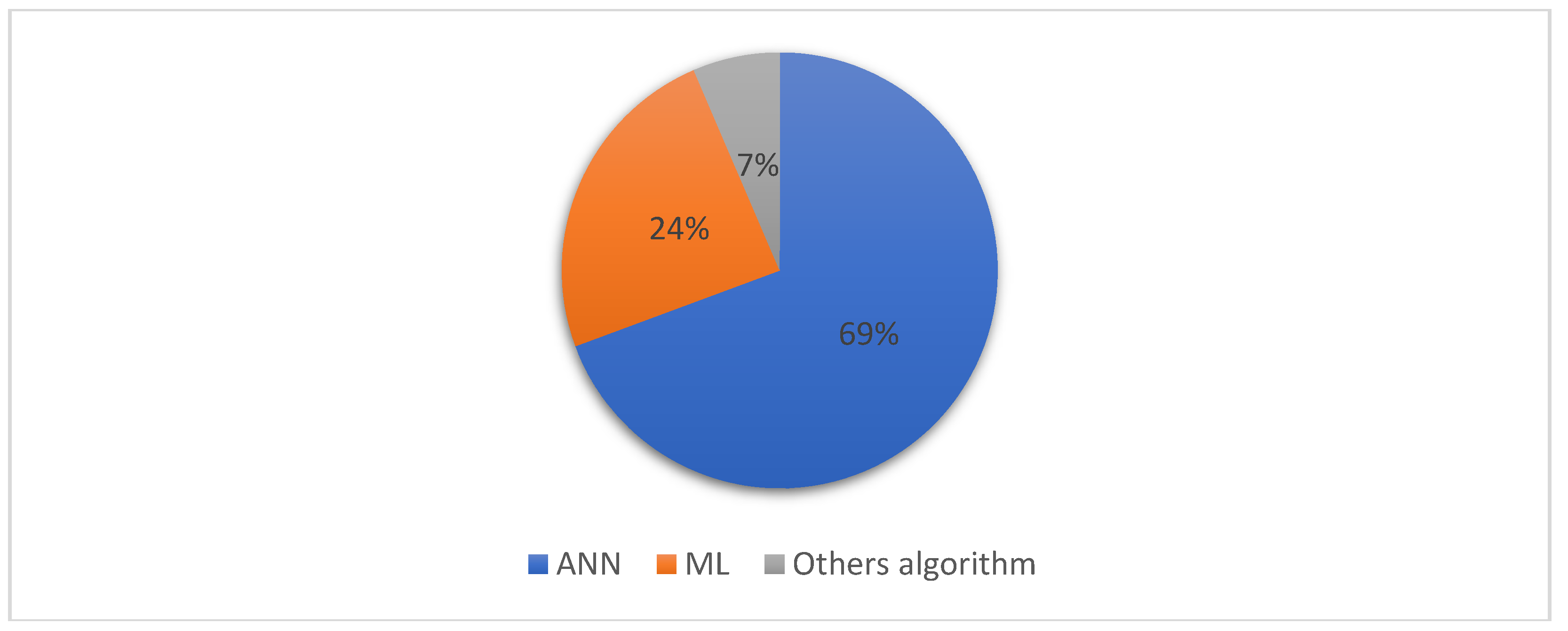
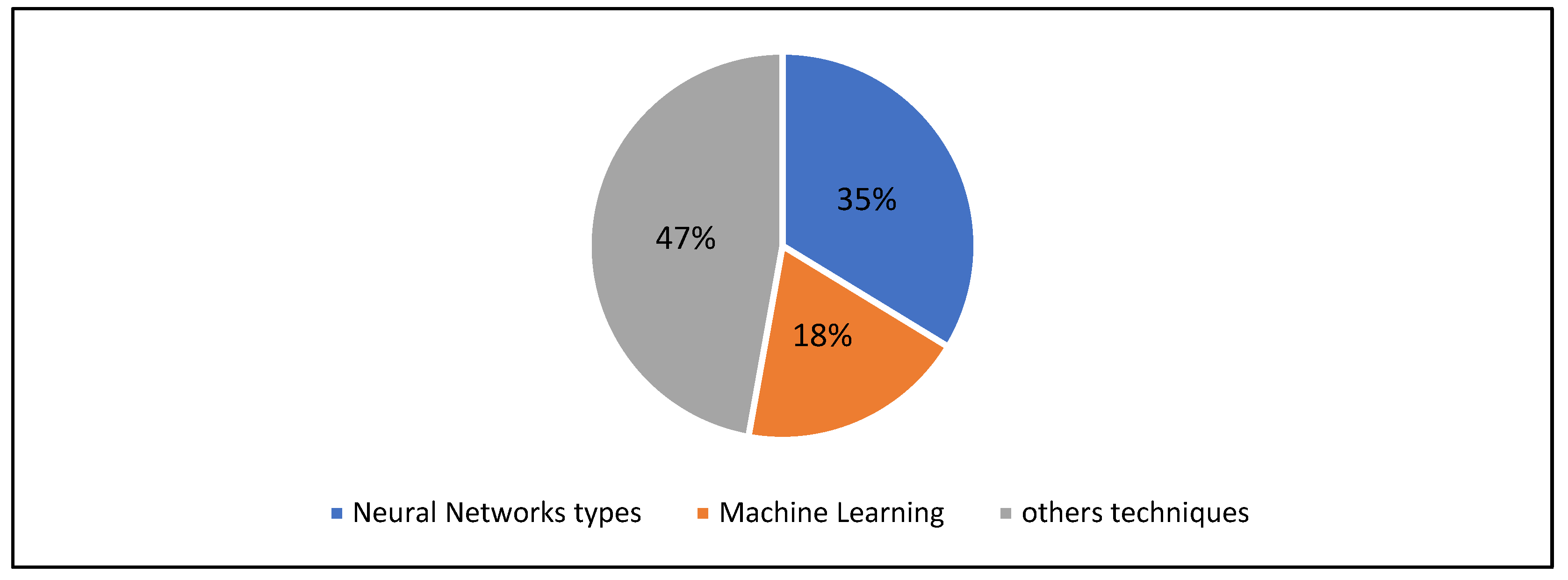
3.3. AI Techniques Used in Brain Tumor Classification
3.4. Dataset Analysis
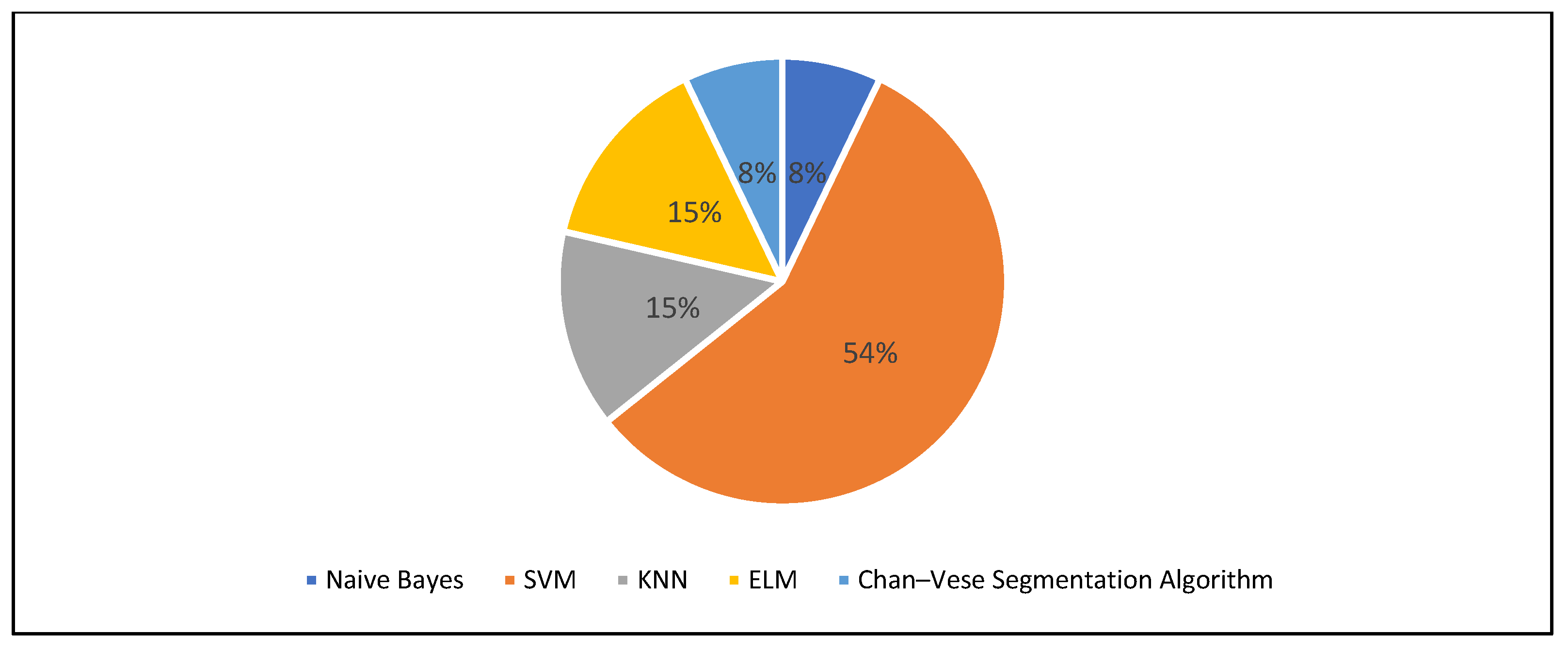
3.5. Segmentation Approaches
3.6. Literature Classification
3.6.1. Classification of Articles into “Conceptual Contribution” and “Practical Contribution” Groups
- A conceptual contribution refers to research that involves description, comparison, analysis, or review-based studies on the topic;
- A practical contribution indicates that the research involved designing, developing, or implementing a program or presenting a novel algorithm.
3.6.2. Separating Articles That Address Segmentation
3.6.3. Studying the Train/Test Mechanism, Evaluation Metrics, and Segmentation Approaches
4. Discussion
- Support vector machines (SVM), noted in five papers;
- MATLAB, Keras, and Z-score normalization.
5. Conclusions
Ethical Considerations
Funding
Conflicts of Interest
References
- Kumar, K.A.; Prasad, A.; Metan, J. A hybrid deep CNN-Cov-19-Res-Net Transfer learning architype for an enhanced Brain tumor Detection and Classification scheme in medical image processing. Biomed. Signal Process. Control 2022, 76, 103631. [Google Scholar]
- Abdou, M.A. Literature review: Efficient deep neural networks techniques for medical image analysis. Neural Comput. Appl. 2022, 34, 5791–5812. [Google Scholar] [CrossRef]
- Muhammad, K.; Khan, S.; Del Ser, J.; De Albuquerque, V.H.C. Deep learning for multigrade brain tumor classification in smart healthcare systems: A prospective survey. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Choudhary, S.; Jain, A.; Singh, K.; Ahmadian, A.; Bajuri, M.Y. Brain tumor classification using deep neural network and transfer learning. Brain Topogr. 2023, 36, 305–318. [Google Scholar] [CrossRef]
- Vankdothu, R.; Hameed, M.A. Brain tumor MRI images identification and classification based on the recurrent convolutional neural network. Meas. Sens. 2022, 24, 100412. [Google Scholar] [CrossRef]
- Ghorbian, M.; Ghorbian, S.; Ghobaei-arani, M. A comprehensive review on machine learning in brain tumor classification: Taxonomy, challenges, and future trends. Biomed. Signal Process. Control 2024, 98, 106774. [Google Scholar] [CrossRef]
- Torshabi, A.E. Investigation the efficacy of fuzzy logic implementation at image-guided radiotherapy. J. Med. Signals Sens. 2022, 12, 163–170. [Google Scholar] [CrossRef]
- Zadeh, L.A. Fuzzy logic. Computer 1988, 21, 83–93. [Google Scholar] [CrossRef]
- Sun, Y.; Xue, B.; Zhang, M.; Yen, G.G.; Lv, J. Automatically designing CNN architectures using the genetic algorithm for image classification. IEEE Trans. Cybern. 2020, 50, 3840–3854. [Google Scholar] [CrossRef]
- Galeazzi, C.; Sacchetti, A.; Cisbani, A.; Babini, G. The PRISMA program. In Proceedings of the IGARSS 2008—2008 IEEE International Geoscience and Remote Sensing Symposium, Boston, MA, USA, 7–11 July 2008; p. IV-105. [Google Scholar]
- Abdullah, S.A.; Al Ashoor, A.A. Ipv6 security issues: A systematic review following prisma guidelines. Baghdad Sci. J. 2022, 19, 1430. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Zuluaga, M.A.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.; Ourselin, S. Interactive medical image segmentation using deep learning with image-specific fine tuning. IEEE Trans. Med. Imaging 2018, 37, 1562–1573. [Google Scholar] [CrossRef] [PubMed]
- Mallick, P.K.; Ryu, S.H.; Satapathy, S.K.; Mishra, S.; Nguyen, G.N.; Tiwari, P. Brain MRI image classification for cancer detection using deep wavelet autoencoder-based deep neural network. IEEE Access 2019, 7, 46278–46287. [Google Scholar] [CrossRef]
- Hossain, T.; Shishir, F.S.; Ashraf, M.; Al Nasim, M.A.; Shah, F.M. Brain tumor detection using convolutional neural network. In Proceedings of the 1st international conference on advances in science, engineering and robotics technology (ICASERT), Dhaka, Bangladesh, 3–5 May 2019; pp. 1–6. [Google Scholar]
- Hemanth, G.; Janardhan, M.; Sujihelen, L. Design and implementing brain tumor detection using machine learning approach. In Proceedings of the 3rd international conference on trends in electronics and informatics (ICOEI), Tirunelveli, India, 23–25 April 2019; pp. 1289–1294. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Pernas, F.J.; Martínez-Zarzuela, M.; Antón-Rodríguez, M.; González-Ortega, D. A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. Healthcare 2021, 9, 153. [Google Scholar] [CrossRef]
- Ghafourian, E.; Samadifam, F.; Fadavian, H.; Jerfi Canatalay, P.; Tajally, A.; Channumsin, S. An ensemble model for the diagnosis of brain tumors through MRIs. Diagnostics 2023, 13, 561. [Google Scholar] [CrossRef]
- Menze, B.; Isensee, F.; Wiest, R.; Wiestler, B.; Maier-Hein, K.; Reyes, M.; Bakas, S. Analyzing magnetic resonance imaging data from glioma patients using deep learning. Comput. Med. Imaging Graph. 2021, 88, 101828. [Google Scholar] [CrossRef]
- Kurdi, S.Z.; Ali, M.H.; Jaber, M.M.; Saba, T.; Rehman, A.; Damaševičius, R. Brain tumor classification using meta-heuristic optimized convolutional neural networks. J. Pers. Med. 2023, 13, 181. [Google Scholar] [CrossRef]
- Daimary, D.; Bora, M.B.; Amitab, K.; Kandar, D. Brain tumor segmentation from MRI images using hybrid convolutional neural networks. Procedia Comput. Sci. 2020, 167, 2419–2428. [Google Scholar] [CrossRef]
- Reddy, D.; Bhavana, V.; Krishnappa, H. Brain tumor detection using image segmentation techniques. In Proceedings of the 2018 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 3–5 April 2018; pp. 18–22. [Google Scholar]
- Ramalakshmi, K.; Rajagopal, S.; Kulkarni, M.B.; Poddar, H. A hyperdimensional framework: Unveiling the interplay of RBP and GSN within CNNs for ultra-precise brain tumor classification. Biomed. Signal Process. Control. 2024, 96, 106565. [Google Scholar]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef]
- Balamurugan, T.; Gnanamanoharan, E. Brain tumor segmentation and classification using hybrid deep CNN with LuNetClassifier. Neural Comput. Appl. 2023, 35, 4739–4753. [Google Scholar] [CrossRef]
- Liu, Z.; Tong, L.; Chen, L.; Zhou, F.; Jiang, Z.; Zhang, Q.; Wang, Y.; Shan, C.; Li, L.; Zhou, H. Canet: Context aware network for brain glioma segmentation. IEEE Trans. Med. Imaging 2021, 40, 1763–1777. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Lin, J.; Lu, C.; Chen, H.; Lin, H.; Zhao, B.; Shi, Z.; Qiu, B.; Pan, X.; Xu, Z. CKD-TransBTS: Clinical knowledge-driven hybrid transformer with modality-correlated cross-attention for brain tumor segmentation. IEEE Trans. Med. Imaging 2023, 42, 2451–2461. [Google Scholar] [CrossRef] [PubMed]
- Al-Masni, M.A.; Kim, D.-H. CMM-Net: Contextual multi-scale multi-level network for efficient biomedical image segmentation. Sci. Rep. 2021, 11, 10191. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, G.; Zhang, Q.; Han, J.; Han, J.; Yu, Y. Cross-modality deep feature learning for brain tumor segmentation. Pattern Recognit. 2021, 110, 107562. [Google Scholar] [CrossRef]
- Xu, Q.; Ma, Z.; Na, H.; Duan, W. DCSAU-Net: A deeper and more compact split-attention U-Net for medical image segmentation. Comput. Biol. Med. 2023, 154, 106626. [Google Scholar] [CrossRef]
- Peng, J.; Kim, D.D.; Patel, J.B.; Zeng, X.; Huang, J.; Chang, K.; Xun, X.; Zhang, C.; Sollee, J.; Wu, J. Deep learning-based automatic tumor burden assessment of pediatric high-grade gliomas, medulloblastomas, and other leptomeningeal seeding tumors. Neuro-Oncology 2022, 24, 289–299. [Google Scholar] [CrossRef]
- Ladkat, A.S.; Bangare, S.L.; Jagota, V.; Sanober, S.; Beram, S.M.; Rane, K.; Singh, B.K. Deep Neural Network-Based Novel Mathematical Model for 3D Brain Tumor Segmentation. Comput. Intell. Neurosci. 2022, 2022, 4271711. [Google Scholar] [CrossRef]
- Preetha, C.J.; Meredig, H.; Brugnara, G.; Mahmutoglu, M.A.; Foltyn, M.; Isensee, F.; Kessler, T.; Pflüger, I.; Schell, M.; Neuberger, U. Deep-learning-based synthesis of post-contrast T1-weighted MRI for tumour response assessment in neuro-oncology: A multicentre, retrospective cohort study. Lancet Digit. Health 2021, 3, e784–e794. [Google Scholar] [CrossRef]
- Xu, Y.; He, X.; Xu, G.; Qi, G.; Yu, K.; Yin, L.; Yang, P.; Yin, Y.; Chen, H. A medical image segmentation method based on multi-dimensional statistical features. Front. Neurosci. 2022, 16, 1009581. [Google Scholar] [CrossRef]
- Telrandhe, S.R.; Pimpalkar, A.; Kendhe, A. Detection of brain tumor from MRI images by using segmentation & SVM. In Proceedings of the 2016 World Conference on Futuristic Trends in Research and Innovation for Social Welfare (Startup Conclave), Coimbatore, India, 29 February–1 March 2016; pp. 1–6. [Google Scholar]
- Abd El Kader, I.; Xu, G.; Shuai, Z.; Saminu, S.; Javaid, I.; Salim Ahmad, I. Differential deep convolutional neural network model for brain tumor classification. Brain Sci. 2021, 11, 352. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, R.A.; Karar, M.E.; Elshaer, Z.; Coburger, J.; Wirtz, C.R.; Burgert, O.; Mathis-Ullrich, F. Explainability of deep neural networks for MRI analysis of brain tumors. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- Micallef, N.; Seychell, D.; Bajada, C.J. Exploring the u-net++ model for automatic brain tumor segmentation. IEEE Access 2021, 9, 125523–125539. [Google Scholar] [CrossRef]
- Athisayamani, S.; Antonyswamy, R.S.; Sarveshwaran, V.; Almeshari, M.; Alzamil, Y.; Ravi, V. Feature extraction using a residual deep convolutional neural network (ResNet-152) and optimized feature dimension reduction for MRI brain tumor classification. Diagnostics 2023, 13, 668. [Google Scholar] [CrossRef]
- Choi, Y.S.; Bae, S.; Chang, J.H.; Kang, S.-G.; Kim, S.H.; Kim, J.; Rim, T.H.; Choi, S.H.; Jain, R.; Lee, S.-K. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro-Oncology 2021, 23, 304–313. [Google Scholar] [CrossRef]
- Hu, M.; Zhong, Y.; Xie, S.; Lv, H.; Lv, Z. Fuzzy system based medical image processing for brain disease prediction. Front. Neurosci. 2021, 15, 714318. [Google Scholar] [CrossRef] [PubMed]
- Conte, G.M.; Weston, A.D.; Vogelsang, D.C.; Philbrick, K.A.; Cai, J.C.; Barbera, M.; Sanvito, F.; Lachance, D.H.; Jenkins, R.B.; Tobin, W.O. Generative adversarial networks to synthesize missing T1 and FLAIR MRI sequences for use in a multisequence brain tumor segmentation model. Radiology 2021, 299, 313–323. [Google Scholar] [CrossRef]
- Khan, A.H.; Abbas, S.; Khan, M.A.; Farooq, U.; Khan, W.A.; Siddiqui, S.Y.; Ahmad, A. Intelligent model for brain tumor identification using deep learning. Appl. Comput. Intell. Soft Comput. 2022, 2022, 8104054. [Google Scholar] [CrossRef]
- Archana, K.; Komarasamy, G. A novel deep learning-based brain tumor detection using the Bagging ensemble with K-nearest neighbor. J. Intell. Syst. 2023, 32, 20220206. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Wang, Y.; Xia, Y. Inter-slice context residual learning for 3D medical image segmentation. IEEE Trans. Med. Imaging 2020, 40, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Gunasekara, S.R.; Kaldera, H.; Dissanayake, M.B. A systematic approach for MRI brain tumor localization and segmentation using deep learning and active contouring. J. Healthc. Eng. 2021, 2021, 6695108. [Google Scholar] [CrossRef] [PubMed]
- Anantharajan, S.; Gunasekaran, S.; Subramanian, T.; Venkatesh, R. MRI brain tumor detection using deep learning and machine learning approaches. Meas. Sens. 2024, 31, 101026. [Google Scholar] [CrossRef]
- Chen, G.; Ru, J.; Zhou, Y.; Rekik, I.; Pan, Z.; Liu, X.; Lin, Y.; Lu, B.; Shi, J. MTANS: Multi-scale mean teacher combined adversarial network with shape-aware embedding for semi-supervised brain lesion segmentation. NeuroImage 2021, 244, 118568. [Google Scholar] [CrossRef]
- Gumaei, A.; Hassan, M.M.; Hassan, M.R.; Alelaiwi, A.; Fortino, G. A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Jia, Z.; Chen, D. Brain tumor identification and classification of MRI images using deep learning techniques. IEEE Access 2020. [Google Scholar] [CrossRef]
- Ullah, M.S.; Khan, M.A.; Almujally, N.A.; Alhaisoni, M.; Akram, T.; Shabaz, M. BrainNet: A fusion assisted novel optimal framework of residual blocks and stacked autoencoders for multimodal brain tumor classification. Sci. Rep. 2024, 14, 5895. [Google Scholar] [CrossRef]
- Tripathy, S.; Singh, R.; Ray, M. Automation of brain tumor identification using efficientnet on magnetic resonance images. Procedia Comput. Sci. 2023, 218, 1551–1560. [Google Scholar] [CrossRef]
- Srinivasan, S.; Francis, D.; Mathivanan, S.K.; Rajadurai, H.; Shivahare, B.D.; Shah, M.A. A hybrid deep CNN model for brain tumor image multi-classification. BMC Med. Imaging 2024, 24, 21. [Google Scholar] [CrossRef]
- Tahir, B.; Iqbal, S.; Usman Ghani Khan, M.; Saba, T.; Mehmood, Z.; Anjum, A.; Mahmood, T. Feature enhancement framework for brain tumor segmentation and classification. Microsc. Res. Tech. 2019, 82, 803–811. [Google Scholar] [CrossRef]
- Wadhwa, A.; Bhardwaj, A.; Verma, V.S. A review on brain tumor segmentation of MRI images. Magn. Reson. Imaging 2019, 61, 247–259. [Google Scholar] [CrossRef]
- Farchi, N.R.K.T.A. Automatic classification of preprocessed mri brain tumors images using deep convolutional neural network. IJTPE J. 2023, 15, 68–73. [Google Scholar]
- Chaudhary, J.; Rani, R.; Kamboj, A. Deep learning-based approach for segmentation of glioma sub-regions in MRI. Int. J. Intell. Comput. Cybern. 2020, 13, 389–406. [Google Scholar] [CrossRef]
- Ramamoorthy, M.; Qamar, S.; Manikandan, R.; Jhanjhi, N.Z.; Masud, M.; AlZain, M.A. Earlier detection of brain tumor by pre-processing based on histogram equalization with neural network. Healthcare 2022, 10, 1218. [Google Scholar] [CrossRef]
- Kiranmayee, B.; Rajinikanth, T.; Nagini, S. Enhancement of SVM based MRI brain image classification using pre-processing techniques. Indian J. Sci. Technol. 2016, 9, 1–7. [Google Scholar] [CrossRef]
- Lavanya, N.; Nagasundaram, S. Improving Brain Tumor MRI Images with Pre-Processing Techniques for Noise Removal. In Proceedings of the 2023 International Conference on Sustainable Communication Networks and Application (ICSCNA), Theni, India, 15–17 November 2023; pp. 1530–1538. [Google Scholar]
- Harish, S.; Ahammed, G.A. Integrated modelling approach for enhancing brain MRI with flexible pre-processing capability. Int. J. Electr. Comput. Eng. 2019, 9, 2416. [Google Scholar]
- Poornachandra, S.; Naveena, C. Pre-processing of mr images for efficient quantitative image analysis using deep learning techniques. In Proceedings of the 2017 International Conference on Recent Advances in Electronics and Communication Technology (ICRAECT), Bangalore, India, 16–17 March 2017; pp. 191–195. [Google Scholar]
- Archa, S.; Kumar, C.S. Segmentation of brain tumor in MRI images using CNN with edge detection. In Proceedings of the 2018 International Conference on Emerging Trends and Innovations in Engineering and Technological Research (ICETIETR), Ernakulam, India, 11–13 July 2018; pp. 1–4. [Google Scholar]
- Ali, T.M.; Nawaz, A.; Ur Rehman, A.; Ahmad, R.Z.; Javed, A.R.; Gadekallu, T.R.; Chen, C.-L.; Wu, C.-M. A sequential machine learning-cum-attention mechanism for effective segmentation of brain tumor. Front. Oncol. 2022, 12, 873268. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.; Liu, S.; Di Ieva, A. Spherical coordinates transformation pre-processing in Deep Convolution Neural Networks for brain tumor segmentation in MRI. Med. Biol. Eng. Comput. 2022, 60, 121–134. [Google Scholar] [CrossRef]
- Biratu, E.S.; Schwenker, F.; Ayano, Y.M.; Debelee, T.G. A survey of brain tumor segmentation and classification algorithms. J. Imaging 2021, 7, 179. [Google Scholar] [CrossRef]
- Kapoor, L.; Thakur, S. A survey on brain tumor detection using image processing techniques. In Proceedings of the 2017 7th International Conference on Cloud Computing, Data Science & Engineering-Confluence, Noida, India, 12–13 January 2017; pp. 582–585. [Google Scholar]
- Aggarwal, M.; Tiwari, A.K.; Sarathi, M.P.; Bijalwan, A. An early detection and segmentation of Brain Tumor using Deep Neural Network. BMC Med. Inform. Decis. Mak. 2023, 23, 78. [Google Scholar] [CrossRef]




| Web of Science | IEEE | Scopus | |
|---|---|---|---|
| Keyword: | “brain tumor” | “brain tumor” | “brain tumor” |
| Criterion applied: | “publication topic” | “publishing topic” | “keyword” |
| Value of criterion: | Image processing with a medical image | Analysis of medical image | Image processing in the medical field |
| Records returned: | 113 | 101 | 58 |
| Record excluded: | 14 | 18 | 17 |
| Records extracted: | 99 | 83 | 41 |
| Task | Stepping | Excluded Papers | ||
|---|---|---|---|---|
| Web of Science | IEEE | Scopus | ||
| Filter | Counting articles, reviews, and conference papers | 6 | 4 | 2 |
| Removal duplicated | Exclusion based on duplicates in the same database | 5 | 4 | 10 |
| Exclusion based on duplicates between the three databases | 15 | 13 | 0 | |
| Evaluation | Analyzing the abstract papers | 28 | 22 | 10 |
| Citations and the year of publication of papers | 12 | 29 | 10 | |
| No. | Authors | Approach | Type |
|---|---|---|---|
| 1 | Díaz-Pernas et al., 2021 [17] | CNN, sliding mechanism | Practical |
| 2 | Ghafourian et al., 2023 [18] | Social spider optimization (SSO) algorithm | Practical |
| 3 | Menze et al., 2021 [19] | U-Net, intensity normalization, spatial harmonization | Conceptual |
| 4 | Kurdi et al., 2023 [20] | Fuzzy c-means clustering | Practical |
| 5 | Vankdothu & Hameed, 2022 [5] | K-means clustering, gray level co-occurrence matrix (GLCM) | Practical |
| 6 | Daimary et al., 2020 [21] | CNN, U-Net | Practical |
| 7 | Reddy et al., 2018 [22] | K-means clustering | Practical |
| 8 | Ramalakshmi et al., 2024 [23] | Gray standard normalization, regional binary patterns | Practical |
| 9 | Pereira et al., 2016 [24] | CNN, intensity normalization, bias field correction | Practical |
| 10 | Balamurugan & Gnanamanoharan, 2023 [25] | Hybrid FCM-GMM (fuzzy c-means and Gaussian mixture model) | Practical |
| 11 | Liu et al., 2021 [26] | U-Net, context-guided attentive CRFs | Practical |
| 12 | Lin et al., 2023 [27] | Modality-correlated cross-attention (MCCA), trans and CNN feature calibration (TCFC), transformer | Practical |
| 13 | Al-Masni & Kim, 2021 [28] | U-Net, inversion recovery (IR) | Practical |
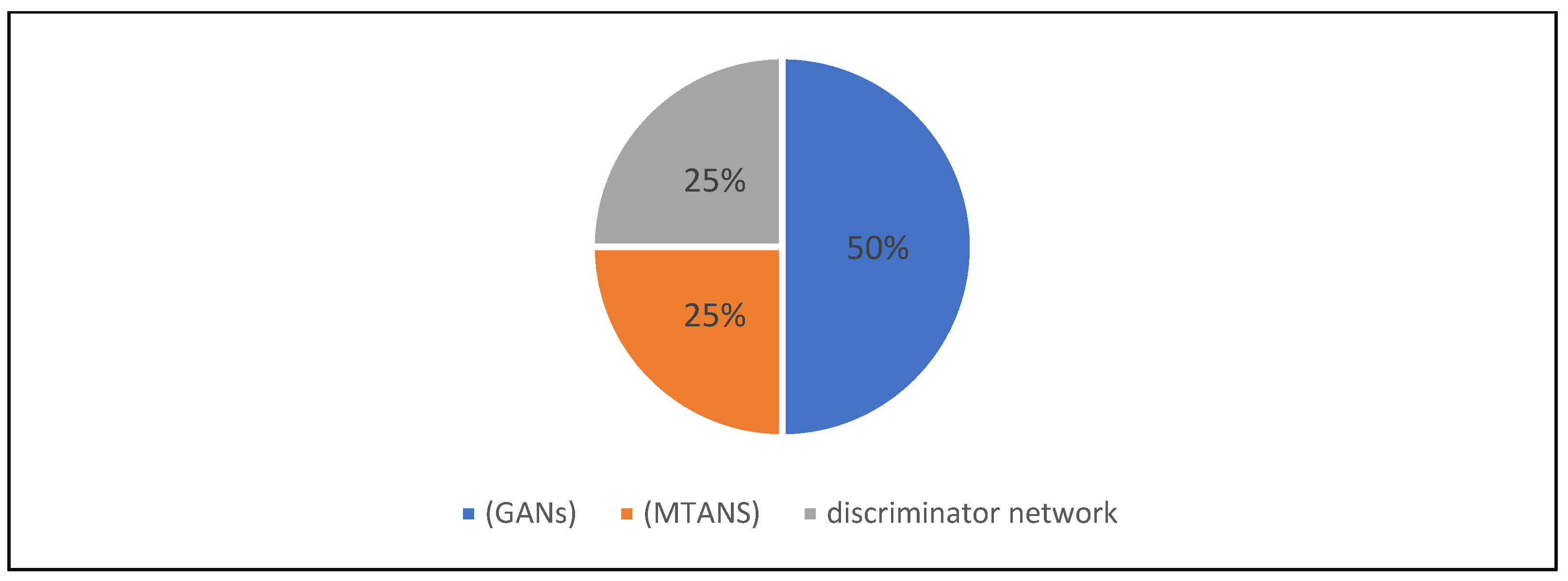
| 14 | Zhang et al., 2021 [29] | GANS for knowledge transfer, cross-modality feature transition (CMFT) process, modalities, and a cross-modality feature fusion (CMFF) | Practical |
| 15 | Xu et al., 2023 [30] | Primary feature conservation (PFC) strategy and compact split-attention (CSA) | Practical |
| 16 | Peng et al., 2022 [31] | 3d U-Net | Practical |
| 17 | Ladkat et al., 2022 [32] | 3D attention U-Net, a mathematical model for pixel enhancement | Practical |
| 18 | Preetha et al., 2021 [33] | CNN | Practical |
| 19 | Xu et al., 2022 [34] | CNN, transformer | Practical |
| 20 | Telrandhe et al., 2016 [35] | K-means clustering, median filtering, and skull masking | Practical |
| 21 | Abd El Kader et al., 2021 [36] | CNN | Practical |
| 22 | Zeineldin et al., 2022 [37] | Neuroxai framework | Conceptual |
| 23 | Micallef et al., 2021 [38] | U-Net | Practical |
| 24 | Athisayamani et al., 2023 [39] | Canny Mayfly algorithm (ACMA), spatial gray level dependence matrix (SGLDM) | Practical |
| 25 | Choi et al., 2021 [40] | CNN | Practical |
| 26 | Hu et al., 2021 [41] | HPU-Net, fuzzy c-means clustering | Practical |
| 27 | Conte et al., 2021 [42] | U-Net | Practical |
| 28 | Khan et al., 2022 [43] | CNN | Practical |
| 29 | Archana & Komarasamy, 2023 [44] | U-Net | Practical |
| 30 | Zhang et al., 2020 [45] | Context residual module, inter-slice context information | Practical |
| 31 | Gunasekara et al., 2021 [46] | CNN, Chan–Vese algorithm | Conceptual |
| 32 | Anantharajan et al., 2024 [47] | Fuzzy c-means clustering, adaptive contrast enhancement algorithm (ACEA), and a median filter | Practical |
| 33 | Chen et al., 2021 [48] | Teacher model with adversarial learning, signed distance maps (SDM) | Practical |
| 34 | Gumaei et al., 2019 [49] | The paper does not focus on image segmentation but utilizes a principal component analysis-normalized GIST (PCA-NGIST) feature extraction method without segmentation. | Practical |
| 35 | Jia & Chen, 2020 [50] | FAHS-SVM, skull stripping, morphological operations, and wavelet transformation | Practical |
| 36 | MS Ullah et al. (2024) [51] | ResNet-50 and Stacked Autoencoders | Practical |
| 37 | Tripathy et al., 2023 [52] | Cropping and converting to grayscale, the binary thresholding method | Practical |
| 38 | Srinivasan S et al., 2024 [53] | Deep Convolutional Neural Network (CNN) | Practical |
| 39 | Tahir et al., 2019 [54] | Otsu method wavelet denoising and histogram equalization | Practical |
| 40 | Wadhwa et al., 2019 [55] | The paper reviews multiple approaches, focusing on a hybrid model combining fully convolutional neural networks (FCNN) with conditional random fields (CRF) | Conceptual |
| 41 | Farchi, 2023 [56] | The paper focuses on classification, not segmentation, but employs image preprocessing techniques such as resizing, grayscale conversion, image smoothing, and enhancement prior to classification | Practical |
| 42 | Chaudhary et al., 2020 [57] | Binary segmentation | Practical |
| 43 | Ramamoorthy et al., 2022 [58] | Otsu method | Practical |
| 44 | Kiranmayee et al., 2016 [59] | Fuzzy c-means clustering, thresholding method, Watershed segmentation | Practical |
| 45 | Lavanya & Nagasundaram, 2023 [60] | Filters | Practical |
| 46 | Harish & Ahammed, 2019 [61] | The segmentation aspect is not the focus of the paper. Instead, it discusses image enhancement techniques to improve the quality of brain MRI images | Practical |
| 47 | Poornachandra & Naveena, 2017 [62] | N4ITK algorithm | Conceptual |
| 48 | Archa & Kumar, 2018 [63] | CNN, wavelet transform, median filtering | Practical |
| 49 | Ali et al., 2022 [64] | U-Net, Markov random field (MRF) | Practical |
| 50 | Russo et al., 2022 [65] | CNN, transforming Cartesian coordinates into spherical coordinates | Practical |
| 51 | Biratu et al., 2021 [66] | CNN | Conceptual |
| 52 | Kapoor & Thakur, 2017 [67] | K-means clustering, Fuzzy C-Means clustering, Genetic Algorithms, thresholding method, Watershed segmentation | Conceptual |
| 53 | Aggarwal et al., 2023 [68] | Resnet | Practical |
| No. | Authors | TT Mechanism | Evaluation Metrics | Segmentation Approach |
|---|---|---|---|---|
| 1 | Díaz-Pernas et al., 2021 [17] | Split 80-20, cross-validation method | DICE score, predicted tumor type accuracy score, and sensitivity | CNN, sliding mechanism |
| 2 | Ghafourian et al., 2023 [18] | Split 70-30, cross-validation method | Accuracy, sensitivity, specificity, F1 score | Social spider optimization (SSO) algorithm |
| 3 | Menze et al., 2021 [19] | Augmentation | Hausdorff distance, volumetric mismatch | U-Net, intensity normalization, spatial harmonization |
| 4 | Kurdi et al., 2023 [20] | MATLAB TOOL, | Pixel accuracy, accuracy, sensitivity, specificity, error rate | Fuzzy c-means clustering |
| 5 | Vankdothu & Hameed, 2022 [5] | Split 70-30 | DICE score, accuracy, Sensitivity, specificity | K-means clustering, Gray level co-occurrence matrix (GLCM) |
| 6 | Daimary et al., 2020 [21] | Split 60-40 | DICE score, accuracy | CNN, U-Net |
| 7 | Reddy et al., 2018 [22] | DICE score, accuracy, precision, recall, true positive (TP), true negative (TN), false positive (FP), false negative (FN) | K-means clustering | |
| 8 | Ramalakshmi et al., 2024 [23] | False classification ratio, accuracy, Sensitivity, specificity | Gray standard normalization, regional binary patterns | |
| 9 | Pereira et al., 2016 [24] | Cross-validation method | DICE score, Sensitivity, Positive Predictive Value (PPV) | CNN, intensity normalization, bias field correction |
| 10 | Balamurugan & Gnanamanoharan, 2023 [25] | Split 70-30 | Accuracy, sensitivity, specificity, precision, recall, F-score, DICE Similarity Index (DSI) | Hybrid FCM-GMM (fuzzy c-means and Gaussian mixture model) |
| 11 | Liu et al., 2021 [26] | Cross-validation method | Sensitivity, specificity, Hausdorff 95 distance | U-Net, context-guided attentive CRFs |
| 12 | Lin et al., 2023 [27] | Split 80-10-10 | DICE score, sensitivity, Hausdorff 95 distance | Modality-correlated cross-attention (MCCA), Trans and CNN feature calibration (TCFC), transformer |
| 13 | Al-Masni & Kim, 2021 [28] | Split 80-20 | DICE score, accuracy, sensitivity, specificity, Jaccard index, Matthews correlation coefficient (MCC), and area under the curve (AUC) | U-Net, inversion recovery (IR) |
| 14 | Zhang et al., 2021 [29] | Split 80-20 | DICE, score, sensitivity, specificity, Hausdorff 95 distance | Gans for knowledge transfer, cross-modality feature Transition (CMFT) process, modalities, and a cross-modality feature fusion (CMFF) |
| 15 | Xu et al., 2023 [30] | Split 70-10-20, augmentation, Adam optimization | DICE score, accuracy, precision, recall, mean intersection over union (MIOU) | Primary feature conservation (PFC) strategy and compact split-attention (CSA) |
| 16 | Peng et al., 2022 [31] | Split 80-20, augmentation | DICE score, ICC | 3D U-Net |
| 17 | Ladkat et al., 2022 [32] | Split 77-23 | Sensitivity, specificity, Hausdorff95 distance | 3D attention U-Net, a mathematical model for pixel enhancement |
| 18 | Preetha et al., 2021 [33] | Split 8-0-20, cross-validation method | Dice score, C-index, SSIM | CNN |
| 19 | Xu et al., 2022 [34] | Adam optimization | Hausdorff 95 distance, mean Intersection over union (MIOU) | CNN, transformer |
| 20 | Telrandhe et al., 2016 [35] | SVM | Accuracy | K-means clustering, median filtering, and skull masking |
| 21 | Abd El Kader et al., 2021 [36] | Split 8-0-20, cross-validation method | DICE score, accuracy, sensitivity, specificity, F1 score, precision | CNN |
| 22 | Zeineldin et al., 2022 [37] | Augmentation, Z-score | DICE score | Neuroxai framework |
| 23 | Micallef et al., 2021 [38] | Split 80-20, cross-validation method, augmentation | DICE score, sensitivity, specificity, Hausdorff95 distance | U-Net |
| 24 | Athisayamani et al., 2023 [39] | Split 70-15-15, augmentation | Accuracy, sensitivity, specificity, and recall | Canny Mayfly algorithm (ACMA), spatial gray level dependence matrix (SGLDM) |
| 25 | Choi et al., 2021 [40] | Accuracy, precision, and recall | CNN | |
| 26 | Hu et al., 2021 [41] | Cross-validation method | DICE score, Jaccard coefficient | HPU-Net, fuzzy c-means clustering |
| 27 | Conte et al., 2021 [42] | Split 64-20-16, cross-validation method | DICE score | U-Net |
| 28 | Khan et al., 2022 [43] | Split 87-13, cross-validation method | Accuracy, error rate | CNN |
| 29 | Archana & Komarasamy, 2023 [44] | Split 80-20 | DICE score, accuracy, F1 score, precision, recall | U-Net |
| 30 | Zhang et al., 2020 [45] | Split 75-0-25, cross-validation, augmentation | Hausdorff95 distance | Context residual module, inter-slice context information |
| 31 | Gunasekara et al., 2021 [46] | Split 80-0-20, cross-validation | DICE score, peak signal-to-noise ratio | CNN, Chan–Vese algorithm |
| 32 | Anantharajan et al., 2024 [47] | SVM | Accuracy, sensitivity, specificity, peak signal-to-noise ratio, Jaccard coefficient (JC) | Fuzzy c-means clustering, Adaptive Contrast Enhancement Algorithm (ACEA), and a median filter |
| 33 | Chen et al., 2021 [48] | Split 80-0-20, cross-validation method | False positive rate (FPR), true positive rate, positive predictive value (PPV) | Teacher model with adversarial learning, signed distance maps (SDM) |
| 34 | Jia & Chen, 2020 [50] | SVM | Accuracy, sensitivity, specificity | FAHS-SVM, skull stripping, morphological operations, and wavelet transformation |
| 35 | Tripathy et al., 2023 [52] | Split 67-13-20 | DICE score, accuracy, sensitivity, specificity, F1 score, precision | Cropping and converting to grayscale, binary thresholding |
| 36 | Srinivasan S et al., 2024 [53] | 5-fold cross-validation Dataset split into 60 training, 20 validation, 20 testing | Accuracy, Sensitivity, Specificity, Precision, ROC-AUC | classification task only (binary, multi-type, and tumor grade) |
| 37 | Tahir et al., 2019 [54] | Split 90-0-10, cross-validation method, SVM | Accuracy, sensitivity, specificity | Wavelet denoising and histogram equalization |
| 38 | Ramamoorthy et al., 2022 [58] | Accuracy, specificity, precision | Otsu method | |
| 39 | Kiranmayee et al., 2016 [59] | SVM | DICE score, accuracy | Fuzzy c-means clustering, thresholding method, Watershed segmentation |
| 40 | Archa & Kumar, 2018 [63] | Augmentation | DICE score, accuracy | CNN, wavelet transform, median filtering |
| 41 | Ali et al., 2022 [64] | Accuracy, sensitivity, precision | UNET, Markov Random Field (MRF) | |
| 42 | Russo et al., 2022 [65] | Split 80-20, cross-validation method, augmentation | DICE score, sensitivity, specificity, Hausdorff 95 distance | CNN, transforming Cartesian coordinates to spherical coordinates, Canny |
| 43 | Aggarwal et al., 2023 [68] | Split 34-46 | Sensitivity, specificity, Jaccard coefficient, peak signal-to-noise ratio, mean square error (MSE) | Resnet |
| No. | Authors | Accuracy | Sensitivity | Specifity | Recall OR (TPR) | F1 Score | Precision OR (ppv) | Dice Score | Hausdorff 95 Distance | Other Classification Metrics | Other Segmentation Metrics |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Díaz-Pernas et al., 2021 [17] | X | X | X | |||||||
| 2 | Ghafourian et al., 2023 [18] | X | X | X | X | ||||||
| 3 | Menze et al., 2021 [19] | X | |||||||||
| 4 | Kurdi et al., 2023 [20] | X | X | X | X | X | |||||
| 5 | Vankdothu & Hameed, 2022 [5] | X | X | X | X | ||||||
| 6 | Daimary et al., 2020 [21] | X | X | ||||||||
| 7 | Reddy et al., 2018 [22] | X | X | X | X | X | |||||
| 8 | Ramalakshmi et al., 2024 [23] | X | X | X | X | ||||||
| 9 | Pereira et al., 2016 [24] | X | X | X | |||||||
| 10 | Balamurugan & Gnanamanoharan, 2023 [25] | X | X | X | X | X | X | X | |||
| 11 | Liu et al., 2021 [26] | X | X | X | |||||||
| 12 | Lin et al., 2023 [27] | X | X | X | |||||||
| 13 | Al-Masni & Kim, 2021 [28] | X | X | X | X | X | |||||
| 14 | Zhang et al., 2021 [29] | X | X | X | X | ||||||
| 15 | Xu et al., 2023 [30] | X | X | X | X | X | |||||
| 16 | Peng et al., 2022 [31] | X | X | ||||||||
| 17 | Ladkat et al., 2022 [32] | X | X | X | |||||||
| 18 | Preetha et al., 2021 [33] | X | X | X | |||||||
| 19 | Xu et al., 2022 [34] | X | X | ||||||||
| 20 | Telrandhe et al., 2016 [35] | X | |||||||||
| 21 | Abd El Kader et al., 2021 [36] | X | X | X | X | X | |||||
| 22 | Zeineldin et al., 2022 [37] | X | |||||||||
| 23 | Micallef et al., 2021 [38] | X | X | X | X | ||||||
| 24 | Athisayamani et al., 2023 [39] | X | X | X | X | ||||||
| 25 | Choi et al., 2021 [40] | X | X | X | |||||||
| 26 | Hu et al., 2021 [41] | X | X | ||||||||
| 27 | Conte et al., 2021 [42] | X | |||||||||
| 28 | Khan et al., 2022 [43] | X | X | ||||||||
| 29 | Archana & Komarasamy, 2023 [44] | X | X | X | X | X | |||||
| 30 | Zhang et al., 2020 [45] | X | |||||||||
| 31 | Gunasekara et al., 2021 [46] | X | X | ||||||||
| 32 | Anantharajan et al., 2024 [47] | X | X | X | X | ||||||
| 33 | Chen et al., 2021 [48] | X | X | X | |||||||
| 34 | Jia & Chen, 2020 [50] | X | X | X | |||||||
| 35 | Tripathy et al., 2023 [52] | X | X | X | X | X | X | ||||
| 36 | Srinivasan S et al., 2024 [53] | X | X | X | X | X | |||||
| 37 | Tahir et al., 2019 [54] | X | X | X | |||||||
| 38 | Ramamoorthy et al., 2022 [58] | X | X | X | |||||||
| 39 | Kiranmayee et al., 2016 [59] | X | X | ||||||||
| 40 | Archa & Kumar, 2018 [63] | X | X | ||||||||
| 41 | Ali et al., 2022 [64] | X | X | X | |||||||
| 42 | Russo et al., 2022 [65] | X | X | X | X | ||||||
| 43 | Aggarwal et al., 2023 [68] | X | X | X | X |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Ashoor, A.; Lilik, F.; Nagy, S. A Systematic Analysis of Neural Networks, Fuzzy Logic and Genetic Algorithms in Tumor Classification. Appl. Sci. 2025, 15, 5186. https://doi.org/10.3390/app15095186
Al-Ashoor A, Lilik F, Nagy S. A Systematic Analysis of Neural Networks, Fuzzy Logic and Genetic Algorithms in Tumor Classification. Applied Sciences. 2025; 15(9):5186. https://doi.org/10.3390/app15095186
Chicago/Turabian StyleAl-Ashoor, Ahmed, Ferenc Lilik, and Szilvia Nagy. 2025. "A Systematic Analysis of Neural Networks, Fuzzy Logic and Genetic Algorithms in Tumor Classification" Applied Sciences 15, no. 9: 5186. https://doi.org/10.3390/app15095186
APA StyleAl-Ashoor, A., Lilik, F., & Nagy, S. (2025). A Systematic Analysis of Neural Networks, Fuzzy Logic and Genetic Algorithms in Tumor Classification. Applied Sciences, 15(9), 5186. https://doi.org/10.3390/app15095186






