Predictive Models of Patient Severity in Intensive Care Units Based on Serum Cytokine Profiles: Advancing Rapid Analysis
Abstract
:1. Introduction
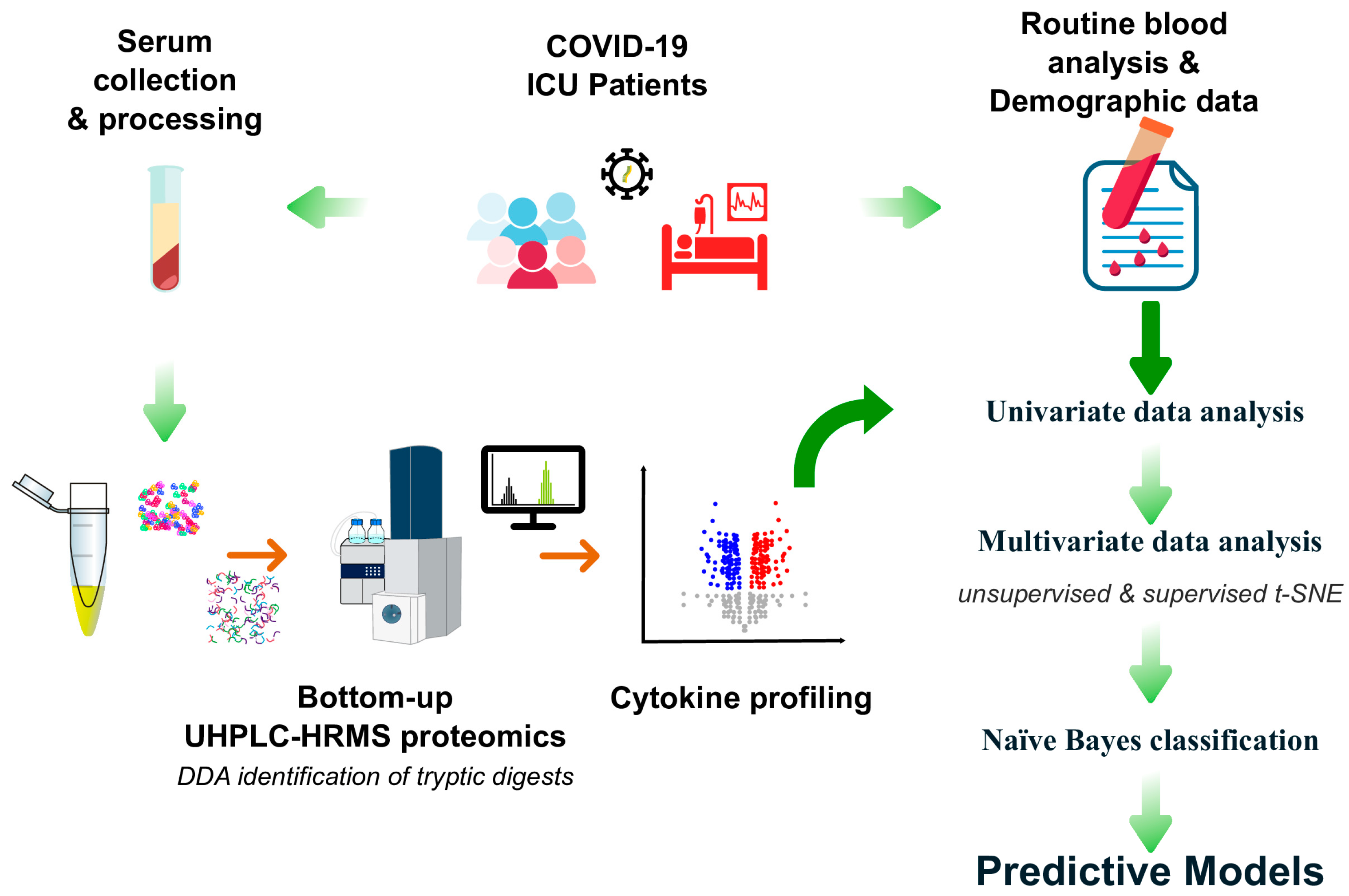
2. Material and Methods
2.1. Study Design
2.2. Demographics and Clinical Characteristics
2.3. Serum Cytokines
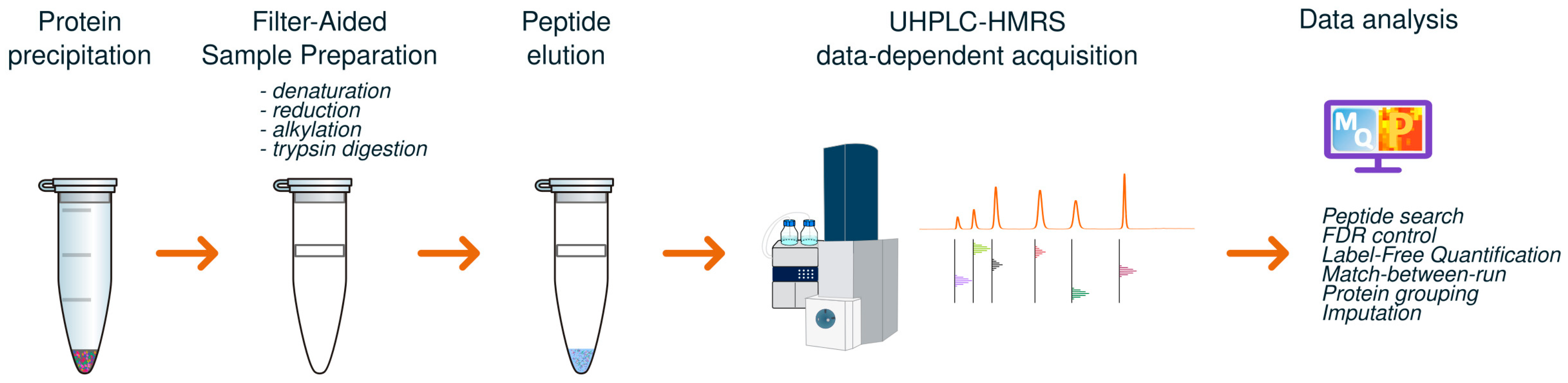
2.3.1. Filter-Assisted Sample Preparation (FASP) for Proteomics
2.3.2. UHPLC-HRMS Analysis
2.3.3. Data Processing
2.4. Routine Blood Analyses
2.5. Statistical Analysis
3. Results and Discussion
- Group A vs. B. This comparison reflects patients’ severity based on the need for IMV, highlighting the progression in severity from Group A (milder condition, without IMV) to Group B (patients requiring IMV).
- Group B vs. C. This comparison focuses on mortality in the ICU among patients requiring IMV. It distinguishes patients who survived (Group B) from those who died in the ICU in an average of 7 days after the current analysis (Group C), thereby isolating factors associated to mortality in the ICU.
- Group A + B vs. C. This analysis is similar to the previous one, except it combined Groups A and B (including both non-IMV and IMV patients), and compared them to Group C, providing a broader control group that included a higher diversity of patients.
3.1. Univariate Data Analysis
3.1.1. Cytokines
3.1.2. Routine Blood Analyses
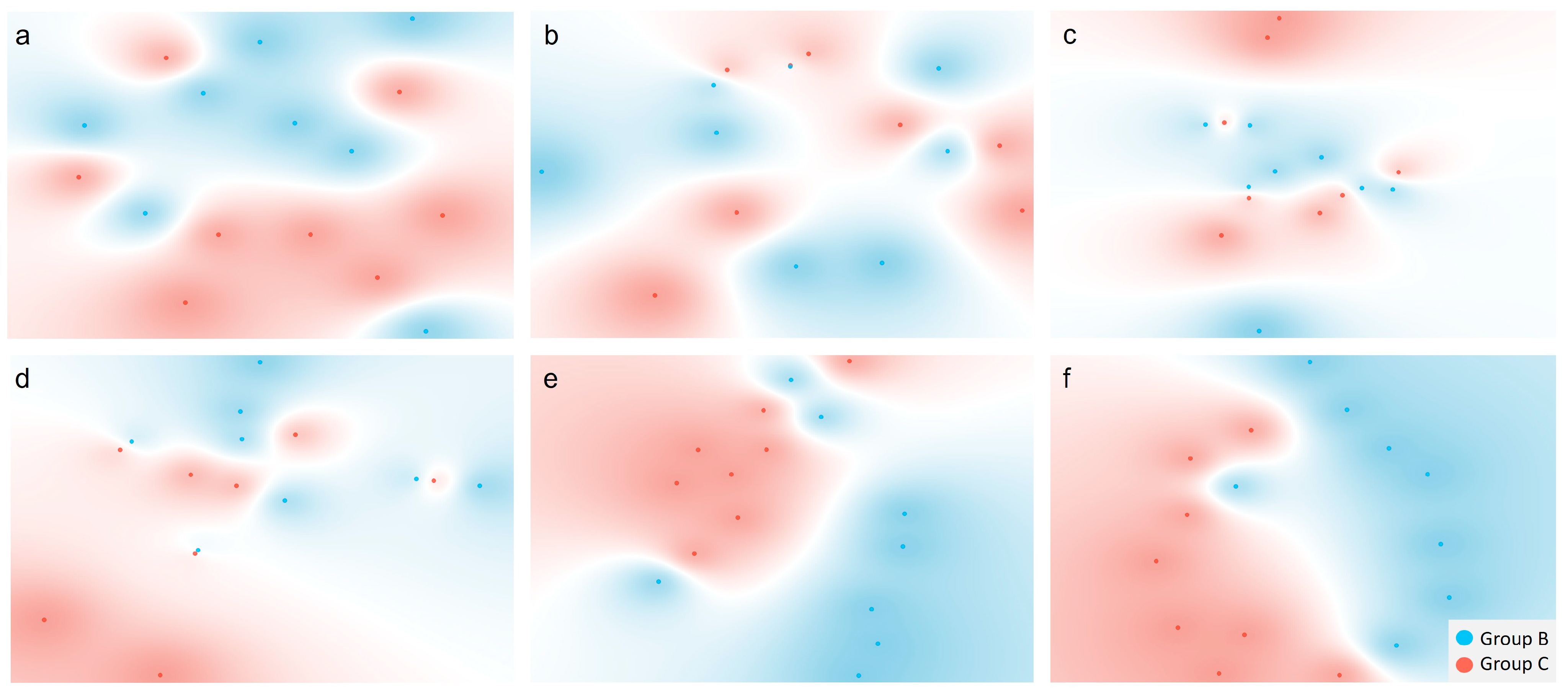
3.2. Multivariate Data Analysis
3.2.1. Comparisons Between Groups A and B
3.2.2. Comparisons Between Groups B and C
3.2.3. Comparisons Between the Combined Groups A and B and Group C
3.2.4. Comparisons Between Predictive Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pellathy, T.P.; Pinsky, M.R.; Hravnak, M. ICU Scoring Systems. Crit. Care Nurse 2021, 41, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Rapsang, A.G.; Shyam, D.C. Scoring systems in the intensive care unit: A compendium. Indian J. Crit. Care Med. 2014, 18, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Li, X.; Zhao, J.; Sun, S. Identification of hospitalized mortality of patients with COVID-19 by machine learning models based on blood inflammatory cytokines. Front. Public Health 2022, 10, 1001340. [Google Scholar] [CrossRef]
- Luo, Y.; Mao, L.; Yuan, X.; Xue, Y.; Lin, Q.; Tang, G.; Song, H.; Wang, F.; Sun, Z. Prediction Model Based on the Combination of Cytokines and Lymphocyte Subsets for Prognosis of SARS-CoV-2 Infection. J. Clin. Immunol. 2020, 40, 960. [Google Scholar] [CrossRef]
- Tulu, T.W.; Wan, T.K.; Chan, C.L.; Wu, C.H.; Woo, P.Y.M.; Tseng, C.Z.S.; Vodencarevic, A.; Menni, C.; Chan, K.H.K. Machine learning-based prediction of COVID-19 mortality using immunological and metabolic biomarkers. BMC Digit. Health 2023, 1, 6. [Google Scholar]
- Nguyen, H.T.T.; Le-Quy, V.; Ho, S.V.; Thomsen, J.H.D.; Stoico, M.P.; Tong, H.V.; Nguyen, N.L.; Krarup, H.B.; Nguyen, S.H.; Tran, V.Q.; et al. Outcome prediction model and prognostic biomarkers for COVID-19 patients in Vietnam. ERJ Open Res. 2023, 9, 00481–02022. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wen, L.; Lei, W.; Ning, Z. Added value of systemic inflammation markers in predicting pulmonary infection in stroke patients A retrospective study by machine learning analysis. Medicine 2021, 100, e28439. [Google Scholar] [CrossRef]
- Alanazi, A.; Aldakhil, L.; Aldhoayan, M.; Aldosari, B. Machine Learning for Early Prediction of Sepsis in Intensive Care Unit (ICU) Patients. Medicina 2023, 59, 1276. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Lai, K.; Chen, Z.; Wan, J.; Xu, Y. Machine learning for the prediction of acute kidney injury in critical care patients with acute cerebrovascular disease. Ren. Fail. 2022, 44, 43–53. [Google Scholar] [CrossRef]
- Andrews, H.S.; Herman, J.D.; Gandhi, R.T. Treatments for COVID-19. Annu. Rev. Med. 2024, 75, 145–157. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar]
- Wiśniewski, J.R.; Mann, M. A Proteomics Approach to the Protein Normalization Problem: Selection of Unvarying Proteins for MS-Based Proteomics and Western Blotting. J. Proteome Res. 2016, 15, 2321–2326. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Anowar, F.; Sadaoui, S.; Selim, B. Conceptual and empirical comparison of dimensionality reduction algorithms (PCA, KPCA, LDA, MDS, SVD, LLE, ISOMAP, LE, ICA, t-SNE). Comput. Sci. Rev. 2021, 40, 100378. [Google Scholar] [CrossRef]
- Al-Aidaroos, K.M.; Bakar, A.A.; Othman, Z. Medical data classification with Naive Bayes approach. Inf. Technol. J. 2012, 11, 1166–1174. [Google Scholar] [CrossRef]
- Domingos, P.; Pazzani, M. On the Optimality of the Simple Bayesian Classifier under Zero-One Loss. Mach. Learn. 1997, 29, 103–130. [Google Scholar] [CrossRef]
- Pohjolainen, V.; Ryynänen, O.P.; Räsänen, P.; Roine, R.P.; Koponen, S.; Karlsson, H. Bayesian prediction of treatment outcome in anorexia nervosa: A preliminary study. Nord. J. Psychiatry 2015, 69, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Guido, R.; Ferrisi, S.; Lofaro, D.; Conforti, D. An Overview on the Advancements of Support Vector Machine Models in Healthcare Applications: A Review. Information 2024, 15, 235. [Google Scholar] [CrossRef]
- Reddy, G.S.; Chittineni, S. Entropy based C4.5-SHO algorithm with information gain optimization in data mining. PeerJ Comput. Sci. 2021, 7, 1–22. [Google Scholar] [CrossRef]
- Chen, Y.; Bai, M.; Zhang, Y.; Liu, J.; Yu, D. Proactively selection of input variables based on information gain factors for deep learning models in short-term solar irradiance forecasting. Energy 2023, 284, 129261. [Google Scholar] [CrossRef]
- Odhiambo Omuya, E.; Onyango Okeyo, G.; Waema Kimwele, M. Feature Selection for Classification using Principal Component Analysis and Information Gain. Expert. Syst. Appl. 2021, 174, 114765. [Google Scholar] [CrossRef]
- Prasetiyowati, M.I.; Maulidevi, N.U.; Surendro, K. Determining threshold value on information gain feature selection to increase speed and prediction accuracy of random forest. J. Big Data 2021, 8, 84. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Soltani-Zangbar, M.S.; Parhizkar, F.; Ghaedi, E.; Tarbiat, A.; Motavalli, R.; Alizadegan, A.; Aghebati-Maleki, L.; Rostamzadeh, D.; Yousefzadeh, Y.; Jadideslam, G.; et al. A comprehensive evaluation of the immune system response and type-I Interferon signaling pathway in hospitalized COVID-19 patients. Cell Commun. Signal 2022, 20, 106. [Google Scholar] [CrossRef]
- Costela-Ruiz, V.J.; Illescas-Montes, R.; Puerta-Puerta, J.M.; Ruiz, C.; Melguizo-Rodríguez, L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020, 54, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, W.; Yin, J.; Ouyang, Y.; Pang, L.; Feng, Y.; Qiao, L.; Guo, X.; Shi, H.; Jin, R.; et al. The inhibition of IL-2/IL-2R gives rise to CD8+ T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia. Cell Death Dis. 2020, 11, 429. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.L.; Borba, H.H.L.; Bonetti, A.d.F.; Leonart, L.P.; Pontarolo, R. Cytokines and Interferons: Types and Functions. In Autoantibodies and Cytokines; Khan, W.A., Ed.; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar]
- Naeini, L.G.; Abbasi, L.; Karimi, F.; Kokabian, P.; Abdi Abyaneh, F.; Naderi, D. The Important Role of Interleukin-2 in COVID-19. J. Immunol. Res. 2023, 2023, 7097329. [Google Scholar]
- Madureira, G.; Soares, R. The misunderstood link between SARS-CoV-2 and angiogenesis. A narrative review. Pulmonology 2023, 29, 323–331. [Google Scholar] [CrossRef]
- Sun, Z.Y.; Xia, H.G.; Zhu, D.Q.; Deng, L.M.; Zhu, P.Z.; Wang, D. Bin. Clinical significance of mechanical ventilation on ischemic-reperfusion injury caused by lung chest trauma and VEGF expression levels in peripheral blood. Exp. Ther. Med. 2017, 14, 2531–2535. [Google Scholar] [CrossRef]
- Jøntvedt Jørgensen, M.; Holter, J.C.; Christensen, E.E.; Schjalm, C.; Tonby, K.; Pischke, S.E.; Jenum, S.; Skeie, L.G.; Nur, S.; Lind, A.; et al. Increased interleukin-6 and macrophage chemoattractant protein-1 are associated with respiratory failure in COVID-19. Sci. Rep. 2020, 10, 21697. [Google Scholar] [CrossRef]
- Mahrous, A.A.; Hassanien, A.A.; Atta, M.S. Predictive value of C-reactive protein in critically ill patients who develop acute lung injury. Egypt. J. Chest Dis. Tuberc. 2015, 64, 225–236. [Google Scholar] [CrossRef]
- Bhattacharya, B.; Prashant, A.; Vishwanath, P.; Suma, M.N.; Nataraj, B. Prediction of outcome and prognosis of patients on mechanical ventilation using body mass index, SOFA score, C-Reactive protein, and serum albumin. Indian J. Crit. Care Med. 2011, 15, 87. [Google Scholar]
- Russo, A.; Tellone, E.; Barreca, D.; Ficarra, S.; Laganà, G. Implication of COVID-19 on Erythrocytes Functionality: Red Blood Cell Biochemical Implications and Morpho-Functional Aspects. Int. J. Mol. Sci. 2022, 23, 2171. [Google Scholar] [CrossRef]
- Lechuga, G.C.; Morel, C.M.; De-Simone, S.G. Hematological alterations associated with long COVID-19. Front. Physiol. 2023, 14, 1203472. [Google Scholar] [CrossRef]
- Ceccato, A.; Panagiotarakou, M.; Ranzani, O.T.; Martin-Fernandez, M.; Almansa-Mora, R.; Gabarrus, A.; Bueno, L.; Cilloniz, C.; Liapikou, A.; Ferrer, M.; et al. Lymphocytopenia as a Predictor of Mortality in Patients with ICU-Acquired Pneumonia. J. Clin. Med. 2019, 8, 843. [Google Scholar] [CrossRef] [PubMed]
- Toori, K.U.; Qureshi, M.A.; Chaudhry, A. Lymphopenia: A useful predictor of COVID-19 disease severity and mortality. Pak. J. Med. Sci. 2021, 37, 1988. [Google Scholar] [CrossRef]
- Deng, Q.; Hu, B.; Zhang, Y.; Wang, H.; Zhou, X.; Hu, W.; Cheng, Y.; Yan, J.; Ping, H.; Zhou, Q. Suspected myocardial injury in patients with COVID-19: Evidence from front-line clinical observation in Wuhan, China. Int. J. Cardiol. 2020, 311, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Chen, T.; Mui, D.; Ferrari, V.; Jagasia, D.; Scherrer-Crosbie, M.; Chen, Y.; Han, Y. Cardiovascular manifestations and treatment considerations in COVID-19. Heart 2020, 106, 1132–1141. [Google Scholar] [CrossRef]
- McKenna, E.; Wubben, R.; Isaza-Correa, J.M.; Melo, A.M.; Mhaonaigh, A.U.; Conlon, N.; O’Donnell, J.S.; Ní Cheallaigh, C.; Hurley, T.; Stevenson, N.J.; et al. Neutrophils in COVID-19: Not Innocent Bystanders. Front. Immunol. 2022, 13, 864387. [Google Scholar] [CrossRef]
- Heidari-Beni, F.; Vahedian-Azimi, A.; Shojaei, S.; Rahimi-Bashar, F.; Shahriary, A.; Johnston, T.P.; Sahebkar, A. The Level of Procalcitonin in Severe COVID-19 Patients: A Systematic Review and Meta-Analysis. Adv. Exp. Med. Biol. 2021, 1321, 277–286. [Google Scholar]
- Henry, B.M.; Aggarwal, G.; Wong, J.; Benoit, S.; Vikse, J.; Plebani, M.; Lippi, G. Lactate dehydrogenase levels predict coronavirus disease 2019 (COVID-19) severity and mortality: A pooled analysis. Am. J. Emerg. Med. 2020, 38, 1722. [Google Scholar] [CrossRef]
- Lopes-Pacheco, M.; Silva, P.L.; Cruz, F.F.; Battaglini, D.; Robba, C.; Pelosi, P.; Morales, M.M.; Caruso Neves, C.; Rocco, P.R.M. Pathogenesis of Multiple Organ Injury in COVID-19 and Potential Therapeutic Strategies. Front. Physiol. 2021, 12, 593223. [Google Scholar] [CrossRef]
- Feld, J.; Tremblay, D.; Thibaud, S.; Kessler, A.; Naymagon, L. Ferritin levels in patients with COVID-19: A poor predictor of mortality and hemophagocytic lymphohistiocytosis. Int. J. Lab. Hematol. 2020, 42, 773–779. [Google Scholar] [CrossRef]
- Kaushal, K.; Kaur, H.; Sarma, P.; Bhattacharyya, A.; Sharma, D.J.; Prajapat, M.; Pathak, M.; Kothari, A.; Kumar, S.; Rana, S.; et al. Serum ferritin as a predictive biomarker in COVID-19. A systematic review, meta-analysis and meta-regression analysis. J. Crit. Care 2022, 67, 172. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, X.; Fan, Q.; Liu, H.; Liu, X.; Liu, Z.; Zhang, Z. D-dimer levels on admission to predict in-hospital mortality in patients with COVID-19. J. Thromb. Haemost. 2020, 18, 1324–1329. [Google Scholar] [CrossRef] [PubMed]
- Kangro, K.; Wolberg, A.S.; Flick, M.J. Fibrinogen, Fibrin, and Fibrin Degradation Products in COVID-19. Curr. Drug Targets 2022, 23, 1602. [Google Scholar]
- Patel, D.; Kher, V.; Desai, B.; Lei, X.; Cen, S.; Nanda, N.; Gholamrezanezhad, A.; Duddalwar, V.; Varghese, B.; Oberai, A.A. Machine learning based predictors for COVID-19 disease severity. Sci. Rep. 2021, 11, 4673. [Google Scholar] [CrossRef] [PubMed]
- Osawa, E.A.; Maciel, A.T. An algorithm to predict the need for invasive mechanical ventilation in hospitalized COVID-19 patients: The experience in Sao Paulo. Acute Crit. Care 2022, 37, 580–591. [Google Scholar] [CrossRef]
- Nicholson, C.J.; Wooster, L.; Sigurslid, H.H.; Li, R.H.; Jiang, W.; Tian, W.; Lino Cardenas, C.L.; Malhotra, R. Estimating risk of mechanical ventilation and in-hospital mortality among adult COVID-19 patients admitted to Mass General Brigham: The VICE and DICE scores. eClinicalMedicine 2021, 33, 100765. [Google Scholar] [CrossRef]
- Chen, G.; Zhao, X.; Chen, X.; Liu, C. Early decrease in blood lymphocyte count is associated with poor prognosis in COVID-19 patients: A retrospective cohort study. BMC Pulm. Med. 2023, 23, 453. [Google Scholar] [CrossRef]
- Yeşilyurt, A.Ö.; Bayrakçi, S.; Ayhan, N.A.; Bulut, Y.; Firat, A.; Saygılı, N.B.; Seydaoğlu, G.; Özyilmaz, E. Prognostic importance of monitoring the lymphocyte count in critically ill COVID-19 patients during the ICU stay. J. Crit. Care 2024, 81, 154673. [Google Scholar] [CrossRef]
- Wang, S.; Sheng, Y.; Tu, J.; Zhang, L. Association between peripheral lymphocyte count and the mortality risk of COVID-19 inpatients. BMC Pulm. Med. 2021, 21, 55. [Google Scholar] [CrossRef]
- Michels, E.H.A.; Appelman, B.; de Brabander, J.; van Amstel, R.B.E.; van Linge, C.C.A.; Chouchane, O.; Reijnders, T.D.Y.; Schuurman, A.R.; Sulzer, T.A.L.; Klarenbeek, A.M.; et al. Host Response Changes and Their Association with Mortality in COVID-19 Patients with Lymphopenia. Am. J. Respir. Crit. Care Med. 2024, 209, 402–416. [Google Scholar] [CrossRef]
- Linssen, J.; Ermens, A.; Berrevoets, M.; Seghezzi, M.; Previtali, G.; Brugge, S.; Russcher, H.; Verbon, A.; Gillis, J.; Riedl, J.; et al. A novel haemocytometric COVID-19 prognostic score developed and validated in an observational multicentre European hospital-based study. eLife 2020, 9, e63195. [Google Scholar] [CrossRef]
- Anderberg, S.B.; Luther, T.; Berglund, M.; Larsson, R.; Rubertsson, S.; Lipcsey, M.; Larsson, A.; Frithiof, R.; Hultström, M. Increased levels of plasma cytokines and correlations to organ failure and 30-day mortality in critically ill COVID-19 patients. Cytokine 2021, 138, 155389. [Google Scholar] [CrossRef]
- dos Santos Medeiros, S.M.D.F.R.; Sousa Lino, B.M.N.; Perez, V.P.; Sousa, E.S.S.; Campana, E.H.; Miyajima, F.; Carvalho-Silva, W.H.V.; Dejani, N.N.; de Sousa Fernandes, M.S.; Yagin, F.H.; et al. Predictive biomarkers of mortality in patients with severe COVID-19 hospitalized in intensive care unit. Front. Immunol. 2024, 15, 1416715. [Google Scholar] [CrossRef]
- Sharma, J.; Rajput, R.; Bhatia, M.; Arora, P.; Sood, V. Clinical Predictors of COVID-19 Severity and Mortality: A Perspective. Front. Cell Infect. Microbiol. 2021, 11, 674277. [Google Scholar] [CrossRef]
- Lin, W.C.; Lin, C.F.; Chen, C.L.; Chen, C.W.; Lin, Y.S. Prediction of outcome in patients with acute respiratory distress syndrome by bronchoalveolar lavage inflammatory mediators. Exp. Biol. Med. 2010, 235, 57–65. [Google Scholar] [CrossRef]
- Hennus, M.P.; Van Vught, A.J.; Brabander, M.; Brus, F.; Jansen, N.J.; Bont, L.J. Mechanical Ventilation Drives Inflammation in Severe Viral Bronchiolitis. PLoS ONE 2013, 8, e83035. [Google Scholar] [CrossRef]
- Jang, H.J.; Leem, A.Y.; Chung, K.S.; Ahn, J.Y.; Jung, J.Y.; Kang, Y.A.; Park, M.S.; Kim, Y.S.; Lee, S.H. Soluble IL-2R Levels Predict in-Hospital Mortality in COVID-19 Patients with Respiratory Failure. J. Clin. Med. 2021, 10, 4242. [Google Scholar] [CrossRef]
- Karlsson, S.; Pettilä, V.; Tenhunen, J.; Lund, V.; Hovilehto, S.; Ruokonen, E. Vascular endothelial growth factor in severe sepsis and septic shock. Anesth. Analg. 2008, 106, 1820–1826. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Huang, F.; Yang, Y.; Wang, F.; Yuan, J.; Zhang, Z.; Qin, Y.; Li, X.; Zhao, D.; et al. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl. Sci. Rev. 2020, 7, 1003–1011. [Google Scholar] [CrossRef]
- Smail, S.W.; Babaei, E.; Amin, K.; Abdulahad, W.H. Serum IL-23, IL-10, and TNF-α predict in-hospital mortality in COVID-19 patients. Front. Immunol. 2023, 14, 1145840. [Google Scholar] [CrossRef]
- Mwangi, V.I.; Netto, R.L.A.; de Morais, C.E.P.; Silva, A.S.; Silva, B.M.; Lima, A.B.; Neves, J.C.F.; Borba, M.G.S.; Val, F.F.A.; de Almeida, A.C.G.; et al. Temporal patterns of cytokine and injury biomarkers in hospitalized COVID-19 patients treated with methylprednisolone. Front. Immunol. 2023, 16, 1229611. [Google Scholar] [CrossRef]
- Silva, M.J.A.; Ribeiro, L.R.; Gouveia, M.I.M.; Marcelino, B.d.R.; dos Santos, C.S.; Lima, K.V.B.; Lima, L.N.G.C. Hyperinflammatory Response in COVID-19: A Systematic Review. Viruses 2023, 15, 553. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Montalvá, A.; Álvarez-Sierra, D.; Martínez-Gallo, M.; Perurena-Prieto, J.; Arrese-Muñoz, I.; Ruiz-Rodríguez, J.C.; Espinosa-Pereiro, J.; Bosch-Nicolau, P.; Martínez-Gómez, X.; Antón, A.; et al. Exposing and Overcoming Limitations of Clinical Laboratory Tests in COVID-19 by Adding Immunological Parameters; A Retrospective Cohort Analysis and Pilot Study. Front. Immunol. 2022, 13, 902837. [Google Scholar] [CrossRef] [PubMed]



| Groups | p-Value A vs. B | p-Value B vs. C | p-Value A + B vs. C | |||||
|---|---|---|---|---|---|---|---|---|
| Variables | A Discharged, no IMV (n = 8) | B Discharged, IMV (n = 8) | A + B All Discharged (n = 16) | C Deceased, IMV (n = 8) | ||||
| Median age (IQR); years | 52.00 (43.25–62.00) | 55.00 (51.00–66.75) | 53.50 (50.25–62.25) | 65.00 (60.50–67.75) | 0.328 | 0.234 | 0.061 | |
| Median BMI, kg/m2 | 27.68 (26.91–33.95) (n = 6) | 31.18 (25.44–35.66) | 29.08 (26.93–34.95) (n = 14) | 29.39 (26.12–33.41) (n = 7) | 0.587 | 0.610 | 0.799 | |
| Comorbidities; n (%) | 6 (75.0) | 7 (87.5) | 13 (81.3) | 5 (62.5) | - | 0.569 | 0.362 | |
| Arterial hypertension; n (%) | 2 (25.0) | 6 (75.0) | 8 (50.0) | 3 (37.5) | 0.132 | 0.315 | 0.679 | |
| Obesity; n (%) | 2 (25.0) | 4 (50.0) | 6 (37.5) | 2 (25.0) | 0.608 | 0.608 | 0.667 | |
| Diabetes; n (%) | 2 (25.0) | 3 (37.5) | 5 (31.3) | 3 (37.5) | - | - | - | |
| Dyslipidemia; n (%) | 2 (25.0) | 2 (25.0) | 4 (25.0) | 2 (25.0) | - | - | - | |
| Chronic respiratory disease; n (%) | 2 (25.0) | 2 (25.0) | 4 (25.0) | 3 (37.5) | - | - | 0.647 | |
| IMV; n (%) | 0 (100.0) | 8 (100.0) | 8 (50.0) | 8 (100.0) | <0.001 | - | 0.022 | |
| HFO; n (%) | 8 (100.0) | 2 (25.0) | 10 (62.5) | 2 (25.0) | 0.007 | - | 0.193 | |
| Origin | - | - | 0.631 * | |||||
| Portugal | 6 (75.0) | 6 (75.0) | 12 (75.0) | 7 (87.5) | ||||
| Other European countries | 0 (0.0) | 1 (12.5) | 1 (6.25) | 0 (0.0) | ||||
| Africa | 1 (12.5) | 0 (0.0) | 1 (6.25) | 1 (12.5) | ||||
| Asia | 1 (12.5) | 1 (12.5) | 2 (12.5) | 0 (0.0) | ||||
| Median time between ICU admission and sample collection; days | 5.00 (1.50–6.00) | 5.50 (3.00–9.75) | 5.00 (3.00–7.00) | 4.50 (3.25–7.75) | 0.382 | 0.798 | 0.834 | |
| Median time between sample collection and death (IQR); days | - | - | - | 5.00 (4.25–11.50) | - | - | - | |
| Median ICU length of stay (IQR); days | 7.00 (5.00–8.75) | 13.50 (10.50–22.00) | 10.00 (6.50–14.50) | 12.50 (8.00–15.25) | 0.002 | 0.328 | 0.490 | |
| Groups | Outcome (log2 (LFQ Intensity)) | p-Value A vs. B | p-Value B vs. C | p-Value A + B vs. C | ||||
|---|---|---|---|---|---|---|---|---|
| Variables | A Discharged, no IMV (n = 8) | B Discharged, IMV (n = 8) | A + B All Discharged (n = 16) | C Deceased, IMV (n = 8) | ||||
| CCL3 | 12.82 (12.02–13.33) | 14.32 (12.39–14.56) | 13.25 (12.22–14.33) | 13.68 (12.38–14.08) | 0.130 | 0.328 | 0.834 | |
| CCL4 | 16.21 (15.27–17.75) | 17.14 (15.75–17.57) | 16.58 (15.70–17.57) | 17.71 (15.67–18.39) | 0.574 | 0.505 | 0.264 | |
| CCL5 | 14.59 (13.00–15.12) | 13.23 (11.98–14.36) | 13.68 (12.85–15.05) | 13.50 (12.33–14.03) | 0.195 | 0.878 | 0.490 | |
| CCL11 | 18.39 (17.49–18.83) | 17.69 (16.54–18.37) | 17.84 (17.28–18.65) | 17.40 (16.70–18.42) | 0.195 | 0.878 | 0.350 | |
| CXCL8 | 14.09 (12.95–16.13) | 14.11 (13.37–15.31) | 14.11 (13.34–15.71) | 13.82 (12.81–15.48) | 1.000 | 0.645 | 0.569 | |
| CXCL9 | 17.39 (16.87–17.51) | 16.37 (15.16–17.43) | 17.06 (15.50–17.51) | 16.10 (15.58–16.37) | 0.279 | 0.721 | 0.093 | |
| CXCL10 | 18.04 (17.85–18.30) | 17.56 (17.29–18.22) | 17.96 (17.52–18.29) | 17.55 (17.37–17.87) | 0.105 | 0.959 | 0.136 | |
| IFN-α | 13.29 (12.15–13.79) | 14.53 (13.01–17.22) | 13.62 (12.55–15.85) | 15.82 (13.07–18.76) | 0.234 | 0.442 | 0.172 | |
| IFN-β | 14.87 (14.40–15.01) | 13.88 (13.19–14.96) | 14.67 (13.48–15.01) | 13.48 (12.38–13.77) | 0.279 | 0.328 | 0.081 | |
| IFN-γ | 15.44 (14.67–15.97) | 14.94 (13.86–15.37) | 15.14 (14.42–15.59) | 14.26 (14.02–14.81) | 0.161 | 0.161 | 0.019 | |
| IL-1α | 16.41 (15.40–16.82) | 15.89 (13.60–16.10) | 15.96 (15.25–16.73) | 15.85 (14.29–16.28) | 0.279 | 0.721 | 0.653 | |
| IL-1β | 12.67 (12.52–13.43) | 13.02 (12.47–13.89) | 12.95 (12.52–13.68) | 12.75 (11.31–13.70) | 0.442 | 0.505 | 0.569 | |
| IL-1RN | 12.08 (11.42–13.00) | 13.24 (12.67–13.47) | 12.77 (12.00–13.35) | 13.66 (12.13–13.85) | 0.050 | 0.442 | 0.172 | |
| IL-2R | 12.83 (11.81–13.08) | 12.73 (11.19–13.74) | 12.83 (11.79–13.29) | 11.08 (10.31–12.30) | 0.878 | 0.105 | 0.016 | |
| IL-6 | 13.21 (10.42–14.25) | 12.98 (10.69–14.03) | 13.12 (10.50–14.23) | 13.34 (12.11–13.49) | 0.878 | 0.645 | 0.881 | |
| IL-7 | 15.44 (15.00–16.14) | 14.41 (12.79–16.38) | 15.39 (13.77–16.24) | 15.13 (12.81–15.48) | 0.382 | 0.878 | 0.610 | |
| IL-10 | 14.70 (13.01–14.91) | 14.92 (14.13–15.58) | 14.79 (13.96–15.05) | 14.36 (13.36–16.96) | 0.328 | 0.798 | 0.881 | |
| IL-13 | 13.06 (11.20–16.62) | 12.29 (11.90–12.97) | 12.61 (11.38–15.09) | 14.39 (12.78–15.03) | 0.505 | 0.105 | 0.238 | |
| IL-15 | 17.65 (17.26–17.84) | 17.35 (16.81–17.83) | 17.54 (16.97–17.84) | 17.15 (16.50–17.71) | 0.328 | 0.505 | 0.238 | |
| CSF2 | 14.44 (13.38–14.89) | 13.94 (13.10–14.72) | 14.20 (13.38–14.80) | 14.17 (13.28–14.60) | 0.574 | 0.798 | 0.928 | |
| EGF | 13.01 (12.63–15.71) | 16.04 (13.38–16.93) | 14.60 (12.78–16.78) | 14.48 (12.56–15.85) | 0.234 | 0.382 | 0.834 | |
| HGF | 14.16 (13.52–15.06) | 13.47 (11.82–14.07) | 13.86 (13.26–14.40) | 14.16 (13.69–14.73) | 0.105 | 0.105 | 0.383 | |
| TNF-α | 13.42 (12.74–13.92) | 13.15 (12.87–13.89) | 13.27 (12.87–13.91) | 13.26 (12.42–13.67) | 0.721 | 0.645 | 0.528 | |
| VEGF | 14.27 (11.81–14.41) | 14.03 (13.67–14.44) | 14.14 (13.67–14.41) | 13.49 (13.38–13.94) | 0.721 | 0.083 | 0.038 | |
| PARK7 | 14.81 (14.61–15.60) | 14.55 (13.24–15.52) | 14.81 (13.81–15.54) | 14.35 (14.00–14.77) | 0.442 | 0.878 | 0.291 | |
| Normality Range | Outcome | p-Value A vs. B | p-Value B vs. C | p-Value A + B vs. C | |||||
|---|---|---|---|---|---|---|---|---|---|
| Variables | A Discharged, no IMV (n = 8) | B Discharged, IMV (n = 8) | A + B All Discharged (n = 16) | C Deceased, IMV (n = 8) | |||||
| Neutrophil count, ×109/L | 2.0–8.5 | 7.71 (5.99–10.98) | 7.86 (6.00–18.08) | 7.71 (6.00–11.40) | 9.81 (5.77–16.01) | 0.721 | 1.000 | 0.697 | |
| Eosinophil count, ×109/L | 0.0–0.6 | 0.09 (0.02–0.17); n = 7 | 0.11 (0.02–0.15) | 0.09 (0.02–0.15); n = 15 | 0.05 (001–0.17); n = 6 | 1.000 | 0.573 | 0.470 | |
| Lymphocyte count, ×109/L | 0.9–3.5 | 1.26 (1.13–1.41) | 1.32 (0.74–1.68) | 1.27 (0.94–1.53) | 0.61 (0.54–0.75) | 0.798 | 0.015 | 0.003 | |
| RBC count, ×1012/L | 4.4–5.9 | 4.39 (4.15–4.85) | 3.88 (3.79–4.35) | 4.19 (3.87–4.49) | 3.71 (2.85–3.94) | 0.028 | 0.105 | 0.005 | |
| Ferritin, ng/mL | 30–340 | 1221.40 (503.00–6552.75); n = 5 | 1591.10 (1111.80–3719.30); n = 5 | 1432.45 (849.48–5525.00); n = 10 | 1776.60 (1395.40-.); n = 3 | 0.690 | 0.571 | 0.573 | |
| Platelet count, ×109/L | 150–450 | 309.50 (220.75–380.75) | 255.50 (166.25–357.75) | 290.00 (209.75–360.50) | 288.00 (253.00–303.50) | 0.505 | 0.798 | 0.787 | |
| Fibrinogen, g/L | 2.00–4.00 | 6.10 (4.80-.); n = 3 | 6.20 (4.50–7.33); n = 6 | 6.10 (5.10–7.85); n = 9 | 4.60 (3.90-.); n = 3 | 0.905 | 0.714 | 0.482 | |
| D-dimers, µg/L | <230 | 1805.00 (273.00–4043.75); n = 6 | 2104.50 (623.25–3123.50) | 2104.50 (456.75–3089.50); n = 14 | 987.00 (397.00–3019.00); n = 7 | 0.755 | 0.613 | 0.636 | |
| Procalcitonin, ng/mL | <0.06 | 0.09 (0.05–0.16); n = 6 | 0.30 (0.12–1.54); n = 6 | 0.14 (0.05–0.39); n = 12 | 0.29 (0.08-.); n = 3 | 0.132 | 0.905 | 0.536 | |
| CRP, mg/L | <5 | 46.50 (16.25–116.38) | 206.65 (97.03–239.73) | 98.35 (37.65–220.73) | 154.05 (91.63–274.40) | 0.028 | 1.000 | 0.238 | |
| LDH, U/L | 125–220 | 518.00 (322.00–615.00); n = 7 | 388.00 (354.00–478.00); n = 7 | 392.50 (346.00–581.25); n = 14 | 422.50 (357.25–526.25); n = 6 | 0.535 | 0.628 | 1.000 | |
| Creatinine, mg/dL | 0.72–1.25 | 0.73 (0.66–0.84) | 0.75 (0.63–1.00) | 0.74 (0.65–0.85) | 0.96 (0.65–1.42) | 1.000 | 0.382 | 0.214 | |
| hs-cTn I, pg/mL | <34.2 | 2.15 (1.90–3.35); n = 6 | 9.95 (3.30–257.98); n = 6 | 3.40 (1.95–12.73); n = 12 | 43.00 (7.55–285.30); n = 5 | 0.026 | 0.429 | 0.037 | |
| Dataset | Feature Selection Mode | Feature Subsets | AUC | Accuracy | Precision | Recall | Specificity |
|---|---|---|---|---|---|---|---|
| Cytokines | Full dataset | All 25 cytokines | 0.588 | 0.700 | 0.708 | 0.700 | 0.700 |
| Information gain | IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF, IL-15, IL-13, IL-1α, IFN-γ | 0.777 | 0.725 | 0.730 | 0.725 | 0.725 | |
| IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF, IL-15, IL-13, IL-1α | 0.790 | 0.700 | 0.708 | 0.700 | 0.700 | ||
| IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF, IL-15, IL-13 | 0.795 | 0.725 | 0.756 | 0.725 | 0.725 | ||
| IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF, IL-15 | 0.804 | 0.700 | 0.738 | 0.700 | 0.700 | ||
| IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF | 0.810 | 0.750 | 0.798 | 0.750 | 0.750 | ||
| IL-1RN, CXCL10, CCL3, CXCL9, IL-7 | 0.746 | 0.700 | 0.738 | 0.700 | 0.700 | ||
| IL-1RN, CXCL10, CCL3, CXCL9 | 0.730 | 0.625 | 0.633 | 0.625 | 0.625 | ||
| IL-1RN, CXCL10, CCL3 | 0.744 | 0.650 | 0.652 | 0.650 | 0.650 | ||
| IL-1RN, CXCL10 | 0.665 | 0.575 | 0.577 | 0.575 | 0.575 | ||
| IL-1RN | 0.605 | 0.550 | 0.551 | 0.550 | 0.550 | ||
| Univariate Analysis | IL-1RN | 0.605 | 0.550 | 0.551 | 0.550 | 0.550 | |
| Routine blood analyses | Full dataset | All 9 laboratory biomarkers | 0.396 | 0.350 | 0.350 | 0.350 | 0.350 |
| Information gain | LDH, RBCs, CRP, lymphocytes, neutrophils, creatinine, platelets, D-dimers | 0.401 | 0.450 | 0.449 | 0.450 | 0.450 | |
| LDH, RBCs, CRP, lymphocytes, neutrophils, creatinine, platelets | 0.414 | 0.400 | 0.399 | 0.400 | 0.400 | ||
| LDH, RBCs, CRP, lymphocytes, neutrophils, creatinine | 0.511 | 0.475 | 0.475 | 0.475 | 0.475 | ||
| LDH, RBCs, CRP, lymphocytes, neutrophils | 0.561 | 0.475 | 0.475 | 0.475 | 0.475 | ||
| LDH, RBCs, CRP, lymphocytes | 0.628 | 0.500 | 0.500 | 0.500 | 0.500 | ||
| LDH, RBCs, CRP | 0.647 | 0.525 | 0.525 | 0.525 | 0.525 | ||
| LDH, RBCs | 0.565 | 0.575 | 0.580 | 0.575 | 0.575 | ||
| LDH | 0.419 | 0.525 | 0.528 | 0.525 | 0.525 | ||
| Univariate Analysis | CRP | 0.624 | 0.675 | 0.699 | 0.675 | 0.675 | |
| RBC | 0.708 | 0.650 | 0.665 | 0.650 | 0.650 | ||
| CRP, RBCs | 0.744 | 0.575 | 0.577 | 0.575 | 0.575 | ||
| Cytokines and Routine blood analyses / Hybrid data | Full dataset | All 34 features | 0.535 | 0.550 | 0.551 | 0.550 | 0.550 |
| Information gain | LDH, RBCs, CRP, IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF, IL-15 | 0.879 | 0.800 | 0.800 | 0.800 | 0.800 | |
| LDH, RBCs, CRP, IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF | 0.891 | 0.850 | 0.850 | 0.850 | 0.850 | ||
| LDH, RBCs, CRP, IL-1RN, CXCL10, CCL3, CXCL9, IL-7 | 0.851 | 0.750 | 0.750 | 0.750 | 0.750 | ||
| LDH, RBCs, CRP, IL-1RN, CXCL10, CCL3, CXCL9 | 0.827 | 0.700 | 0.700 | 0.700 | 0.700 | ||
| LDH, RBCs, CRP, IL-1RN, CXCL10, CCL3 | 0.800 | 0.725 | 0.726 | 0.725 | 0.725 | ||
| LDH, RBCs, CRP, IL-1RN, CXCL10 | 0.764 | 0.650 | 0.652 | 0.650 | 0.650 | ||
| LDH, RBCs, CRP, IL-1RN | 0.652 | 0.550 | 0.551 | 0.550 | 0.550 | ||
| LDH, RBCs, CRP | 0.647 | 0.525 | 0.525 | 0.525 | 0.525 | ||
| LDH, RBCs | 0.565 | 0.575 | 0.580 | 0.575 | 0.575 | ||
| LDH | 0.419 | 0.525 | 0.528 | 0.525 | 0.525 | ||
| Univariate Analysis | IL-1RN, RBCs, CRP | 0.730 | 0.700 | 0.700 | 0.700 | 0.700 |
| Friedman Test | Overall p-Values from the Friedman Test | p-Values Adjusted by the Bonferroni Correction for Multiple Tests | |||
|---|---|---|---|---|---|
| Features | Cytokines vs. Routine Blood Analyses | Cytokines vs. Hybrid Data | Routine Blood Analyses vs. Hybrid Data | ||
| Full datasets | 0.368 | 0.472 | 0.999 | 0.999 | |
| Subsets achieved by the information gain algorithm | 0.001 | 0.007 | 0.999 | 0.003 | |
| Dataset | Feature Selection Mode | Feature Subsets | AUC | Accuracy | Precision | Recall | Specificity |
|---|---|---|---|---|---|---|---|
| Cytokines | Full dataset | All 25 cytokines | 0.445 | 0.450 | 0.450 | 0.450 | 0.450 |
| Information gain | HGF, IL-10, IL-2R, IL-13, IL-7, CXCL10, PARK7, CXCL9, IFN-γ, VEGF | 0.731 | 0.650 | 0.650 | 0.650 | 0.650 | |
| HGF, IL-10, IL-2R, IL-13, IL-7, CXCL10, PARK7, CXCL9, IFN-γ | 0.744 | 0.700 | 0.708 | 0.700 | 0.700 | ||
| HGF, IL-10, IL-2R, IL-13, IL-7, CXCL10, PARK7, CXCL9 | 0.740 | 0.600 | 0.601 | 0.600 | 0.600 | ||
| HGF, IL-10, IL-2R, IL-13, IL-7, CXCL10, PARK7 | 0.746 | 0.725 | 0.726 | 0.725 | 0.725 | ||
| HGF, IL-10, IL-2R, IL-13, IL-7, CXCL10 | 0.731 | 0.650 | 0.652 | 0.650 | 0.650 | ||
| HGF, IL-10, IL-2R, IL-13, IL-7 | 0.719 | 0.650 | 0.652 | 0.650 | 0.650 | ||
| HGF, IL-10, IL-2R, IL-13 | 0.728 | 0.700 | 0.708 | 0.700 | 0.700 | ||
| HGF, IL-10, IL-2R | 0.724 | 0.725 | 0.726 | 0.725 | 0.725 | ||
| HGF, IL-10 | 0.772 | 0.775 | 0.813 | 0.775 | 0.775 | ||
| HGF | 0.713 | 0.750 | 0.750 | 0.750 | 0.750 | ||
| Univariate Analysis | VEGF | 0.541 | 0.575 | 0.575 | 0.575 | 0.575 | |
| Routine blood analyses | Full dataset | All 9 laboratory biomarkers | 0.489 | 0.475 | 0.472 | 0.475 | 0.475 |
| Information gain | Lymphocytes, platelets, RBCs, D-dimers, eosinophils, LDH, creatinine, CRP | 0.606 | 0.575 | 0.585 | 0.575 | 0.575 | |
| Lymphocytes, platelets, RBCs, D-dimers, eosinophils, LDH, creatinine | 0.634 | 0.650 | 0.656 | 0.650 | 0.650 | ||
| Lymphocytes, platelets, RBCs, D-dimers, eosinophils, LDH | 0.613 | 0.600 | 0.610 | 0.600 | 0.600 | ||
| Lymphocytes, platelets, RBCs, D-dimers, eosinophils | 0.634 | 0.600 | 0.610 | 0.600 | 0.600 | ||
| Lymphocytes, platelets, RBCs, D-dimers | 0.591 | 0.525 | 0.525 | 0.525 | 0.525 | ||
| Lymphocytes, platelets, RBCs | 0.690 | 0.625 | 0.633 | 0.625 | 0.625 | ||
| Lymphocytes, platelets | 0.684 | 0.650 | 0.679 | 0.650 | 0.650 | ||
| Lymphocytes | 0.708 | 0.725 | 0.730 | 0.725 | 0.725 | ||
| Cytokines and Routine blood analyses/Hybrid data | Full dataset | All 34 variables | 0.448 | 0.450 | 0.450 | 0.450 | 0.450 |
| Information gain | Lymphocytes, IL-2R, HGF, platelets, IL-10, eosinophils, IL-7, CXCL10, PARK7, CXCL9 | 0.768 | 0.650 | 0.656 | 0.650 | 0.650 | |
| Lymphocytes, IL-2R, HGF, platelets, IL-10, eosinophils, IL-7, CXCL10, PARK7 | 0.765 | 0.675 | 0.675 | 0.675 | 0.675 | ||
| Lymphocytes, IL-2R, HGF, platelets, IL-10, eosinophils, IL-7, CXCL10 | 0.765 | 0.675 | 0.679 | 0.675 | 0.675 | ||
| Lymphocytes, IL-2R, HGF, platelets, IL-10, eosinophils, IL-7 | 0.758 | 0.700 | 0.708 | 0.700 | 0.700 | ||
| Lymphocytes, IL-2R, HGF, platelets, IL-10, eosinophils | 0.774 | 0.625 | 0.628 | 0.625 | 0.625 | ||
| Lymphocytes, IL-2R, HGF, platelets, IL-10 | 0.770 | 0.675 | 0.679 | 0.675 | 0.675 | ||
| Lymphocytes, IL-2R, HGF, platelets | 0.746 | 0.625 | 0.628 | 0.625 | 0.625 | ||
| Lymphocytes, IL-2R, HGF | 0.774 | 0.725 | 0.730 | 0.725 | 0.725 | ||
| Lymphocytes, IL-2R | 0.725 | 0.675 | 0.687 | 0.675 | 0.675 | ||
| Lymphocytes | 0.708 | 0.725 | 0.730 | 0.725 | 0.725 | ||
| Univariate Analysis | VEGF; Lymphocytes | 0.636 | 0.625 | 0.633 | 0.625 | 0.625 |
| Friedman Test | Overall p-Values from the Friedman Test | p-Values Adjusted by the Bonferroni Correction for Multiple Tests | |||
|---|---|---|---|---|---|
| Features | Cytokines vs. Routine Blood Analyses | Cytokines vs. Hybrid Data | Routine Blood Analyses vs. Hybrid Data | ||
| Full datasets | 0.368 | 0.472 | 0.999 | 0.999 | |
| Subsets achieved by the information gain algorithm | <0.001 | 0.055 | 0.297 | <0.001 | |
| Dataset | Feature Selection Mode | Feature Subsets | AUC | Accuracy | Precision | Recall | Specificity |
|---|---|---|---|---|---|---|---|
| Cytokines | Full dataset | All 25 cytokines | 0.717 | 0.660 | 0.777 | 0.660 | 0.757 |
| Information gain | CXCL9, IFN-γ, VEGF, IL-2R, IL-1RN, CCL5, IL-1β, PARK7, IL-15, IL-10 | 0.727 | 0.640 | 0.768 | 0.640 | 0.743 | |
| CXCL9, IFN-γ, VEGF, IL-2R, IL-1RN, CCL5, IL-1β, PARK7, IL-15 | 0.713 | 0.600 | 0.714 | 0.600 | 0.700 | ||
| CXCL9, IFN-γ, VEGF, IL-2R, IL-1RN, CCL5, IL-1β, PARK7 | 0.717 | 0.660 | 0.777 | 0.660 | 0.757 | ||
| CXCL9, IFN-γ, VEGF, IL-2R, IL-1RN, CCL5, IL-1β | 0.740 | 0.660 | 0.777 | 0.660 | 0.757 | ||
| CXCL9, IFN-γ, VEGF, IL-2R, IL-1RN, CCL5 | 0.777 | 0.620 | 0.759 | 0.620 | 0.730 | ||
| CXCL9, IFN-γ, VEGF, IL-2R, IL-1RN | 0.768 | 0.660 | 0.777 | 0.660 | 0.757 | ||
| CXCL9, IFN-γ, VEGF, IL-2R | 0.664 | 0.620 | 0.680 | 0.620 | 0.680 | ||
| CXCL9, IFN-γ, VEGF | 0.629 | 0.620 | 0.700 | 0.620 | 0.697 | ||
| CXCL9, IFN-γ | 0.697 | 0.600 | 0.650 | 0.600 | 0.650 | ||
| CXCL9 | 0.667 | 0.600 | 0.594 | 0.600 | 0.550 | ||
| Univariate Analysis | IFN-β | 0.642 | 0.600 | 0.594 | 0.600 | 0.550 | |
| IFN-γ | 0.614 | 0.580 | 0.584 | 0.580 | 0.553 | ||
| IL-2R | 0.663 | 0.680 | 0.674 | 0.680 | 0.680 | ||
| VEGF | 0.532 | 0.500 | 0.504 | 0.500 | 0.467 | ||
| IFN-β; IFN-γ; IL-2R; VEGF | 0.675 | 0.640 | 0.665 | 0.640 | 0.660 | ||
| Routine blood analyses | Full dataset | All 9 laboratory biomarkers | 0.728 | 0.620 | 0.664 | 0.620 | 0.663 |
| Information gain | Lymphocytes, RBCs, eosinophils, creatinine, LDH, platelets, CRP, neutrophils | 0.740 | 0.620 | 0.664 | 0.620 | 0.663 | |
| Lymphocytes, RBCs, eosinophils, creatinine, LDH, platelets, CRP | 0.745 | 0.700 | 0720 | 0.700 | 0.717 | ||
| Lymphocytes, RBCs, eosinophils, creatinine, LDH, platelets | 0.787 | 0.760 | 0.766 | 0.760 | 0.757 | ||
| Lymphocytes, RBCs, eosinophils, creatinine, LDH | 0.837 | 0.800 | 0.814 | 0.800 | 0.817 | ||
| Lymphocytes, RBCs, eosinophils, creatinine | 0.842 | 0.760 | 0.766 | 0.760 | 0.757 | ||
| Lymphocytes, RBCs, eosinophils | 0.830 | 0.740 | 0.774 | 0.740 | 0.777 | ||
| Lymphocytes, RBCs | 0.887 | 0.820 | 0.840 | 0.820 | 0.847 | ||
| Lymphocytes | 0.743 | 0.660 | 0.663 | 0.660 | 0.640 | ||
| Univariate Analysis | RBCs | 0.813 | 0.840 | 0.845 | 0.840 | 0.843 | |
| Cytokines and Routine blood analyses / Hybrid data | Full dataset | All 34 variables | 0.820 | 0.750 | 0.857 | 0.750 | 0.875 |
| Information gain | Lymphocytes, CXCL9, RBCs, IFN-γ, VEGF, IL-2R, IL-1RN, CCL5, IL-1β, PARK7 | 0.825 | 0.680 | 0.786 | 0.680 | 0.770 | |
| Lymphocytes, CXCL9, RBCs, IFN-γ, VEGF, IL-2R, IL-1RN, CCL5, IL-1β | 0.853 | 0.700 | 0.795 | 0.700 | 0.783 | ||
| Lymphocytes, CXCL9, RBCs, IFN-γ, VEGF, IL-2R, IL-1RN, CCL5 | 0.882 | 0.640 | 0.768 | 0.640 | 0.743 | ||
| Lymphocytes, CXCL9, RBCs, IFN-γ, VEGF, IL-2R, IL-1RN | 0.885 | 0.660 | 0.777 | 0.660 | 0.757 | ||
| Lymphocytes, CXCL9, RBCs, IFN-γ, VEGF, IL-2R | 0.818 | 0.660 | 0.777 | 0.660 | 0.757 | ||
| Lymphocytes, CXCL9, RBCs, IFN-γ, VEGF | 0.859 | 0.600 | 0.750 | 0.600 | 0.717 | ||
| Lymphocytes, CXCL9, RBCs, IFN-γ | 0.871 | 0.700 | 0.795 | 0.700 | 0.783 | ||
| Lymphocytes, CXCL9, RBCs | 0.875 | 0.780 | 0.833 | 0.780 | 0.837 | ||
| Lymphocytes, CXCL9 | 0.787 | 0.780 | 0.814 | 0.780 | 0.820 | ||
| Univariate Analysis | IFN-β; IFN-γ; IL-2R; VEGF; lymphocytes; RBCs | 0.823 | 0.680 | 0.673 | 0.680 | 0.770 |
| Overall p-Values from the Friedman Test | p-Values Adjusted by the Bonferroni Correction for Multiple Tests | ||||
|---|---|---|---|---|---|
| Features | Cytokines vs. Routine Blood Analyses | Cytokines vs. Hybrid Data | Routines Blood Analyses vs. Hybrid Data | ||
| Full datasets | 0.368 | 0.999 | 0.472 | 0.999 | |
| Subsets achieved by the information gain algorithm | <0.001 | 0.029 | <0.001 | 0.716 | |
| Groups | Target | Best Serum Cytokines | Best Routine Blood Analyses | Best Hybrid Data |
|---|---|---|---|---|
| A vs. B | Need for IMV | IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF (AUC = 0.810; R = Sp = 0.750) | LDH, RBCs (AUC = 0.565; R = Sp = 0.575) | LDH, RBCs, CRP, IL-1RN, CXCL10, CCL3, CXCL9, IL-7, HGF (AUC = 0.891; R = Sp = 0.850) |
| B vs. C | Mortality in the ICU, among patients with IMV | IL-10, HGF (AUC = 0.772; R = Sp = 0.775) | Lymphocytes (AUC = 0.708; R = Sp = 0.725) | Lymphocytes, IL-2R, HGF (AUC = 0.774; R = Sp = 0.725) |
| A + B vs. C | Mortality in the ICU, among patients with/ without IMV | CXCL9, IFN-γ, VEGF, IL-2R, IL-1RN (AUC = 0.768; R = 0.660; Sp = 0.757) | Lymphocytes, RBCs (AUC = 0.887; R = 0.820; Sp = 0.847) | Lymphocytes, CXCL9, RBCs (AUC = 0.875; R = 0.780; Sp = 0.837) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rekowski, C.P.V.; Fonseca, T.A.H.; Araújo, R.; Martins, A.; Pinto, I.; Oliveira, M.C.; Justino, G.C.; Bento, L.; Calado, C.R.C. Predictive Models of Patient Severity in Intensive Care Units Based on Serum Cytokine Profiles: Advancing Rapid Analysis. Appl. Sci. 2025, 15, 4823. https://doi.org/10.3390/app15094823
Rekowski CPV, Fonseca TAH, Araújo R, Martins A, Pinto I, Oliveira MC, Justino GC, Bento L, Calado CRC. Predictive Models of Patient Severity in Intensive Care Units Based on Serum Cytokine Profiles: Advancing Rapid Analysis. Applied Sciences. 2025; 15(9):4823. https://doi.org/10.3390/app15094823
Chicago/Turabian StyleRekowski, Cristiana P. Von, Tiago A. H. Fonseca, Rúben Araújo, Ana Martins, Iola Pinto, M. Conceição Oliveira, Gonçalo C. Justino, Luís Bento, and Cecília R. C. Calado. 2025. "Predictive Models of Patient Severity in Intensive Care Units Based on Serum Cytokine Profiles: Advancing Rapid Analysis" Applied Sciences 15, no. 9: 4823. https://doi.org/10.3390/app15094823
APA StyleRekowski, C. P. V., Fonseca, T. A. H., Araújo, R., Martins, A., Pinto, I., Oliveira, M. C., Justino, G. C., Bento, L., & Calado, C. R. C. (2025). Predictive Models of Patient Severity in Intensive Care Units Based on Serum Cytokine Profiles: Advancing Rapid Analysis. Applied Sciences, 15(9), 4823. https://doi.org/10.3390/app15094823








