Computational Prediction of Ginsenosides Targeting ADGRG3/GPR97 in Cancer and Immune Pathways: A Multi-Faceted In Silico Approach
Abstract
1. Introduction
2. Material and Methods
2.1. Gene Expression Analysis of ADGRG3 in Various Cancers
2.2. Virtual Screening
2.3. ADMET Analysis
2.4. Molecular Dynamics Simulation
2.5. Protein–Protein Interaction (PPI) and Pathway Enrichment Analyses
2.6. Expression Analysis of ADGRG3 in AML
3. Results
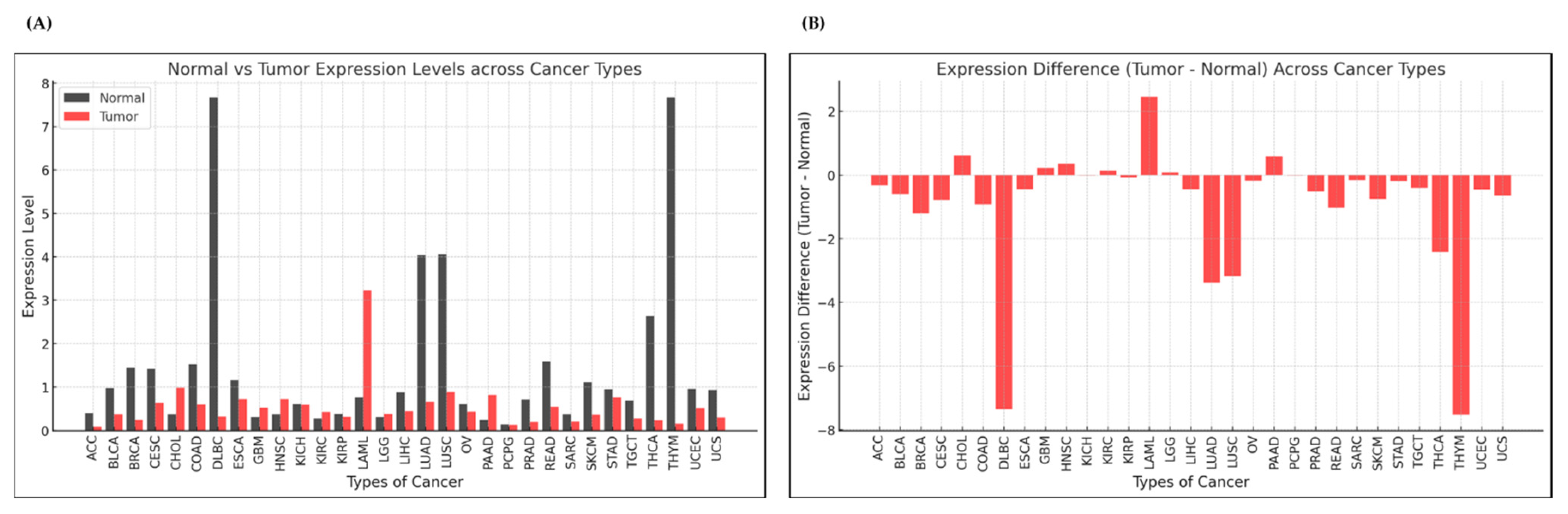
3.1. Expression Analysis of ADGRG3 Gene in Various Cancers
3.1.1. Pan-Cancer Analysis
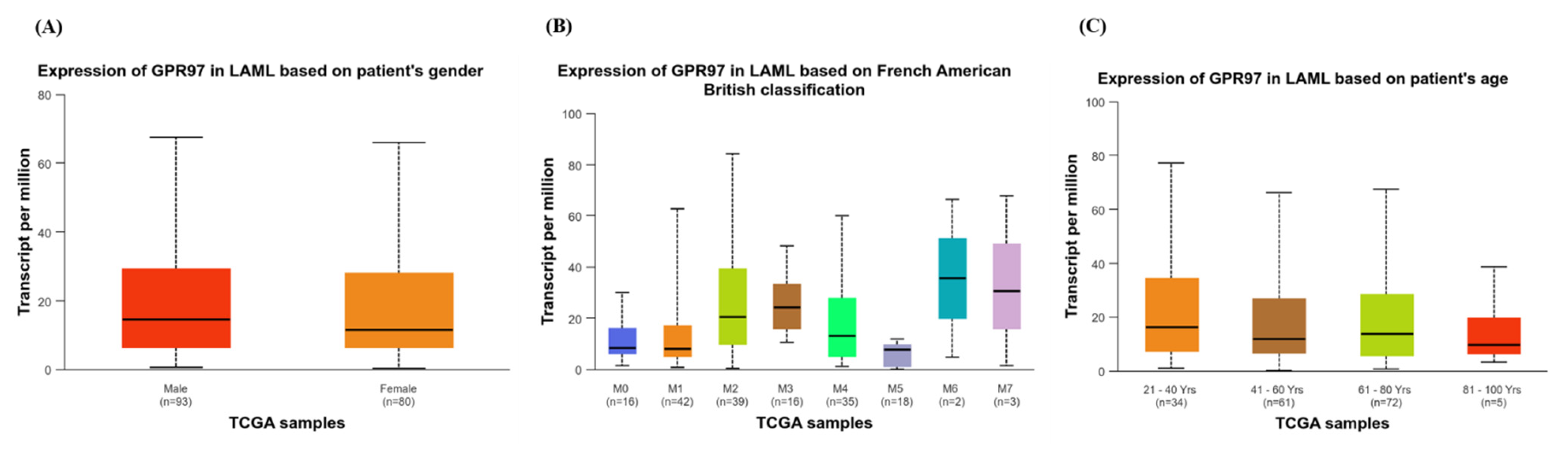
3.1.2. Expression of GPR97 in LAML Based on Clinical Attributes
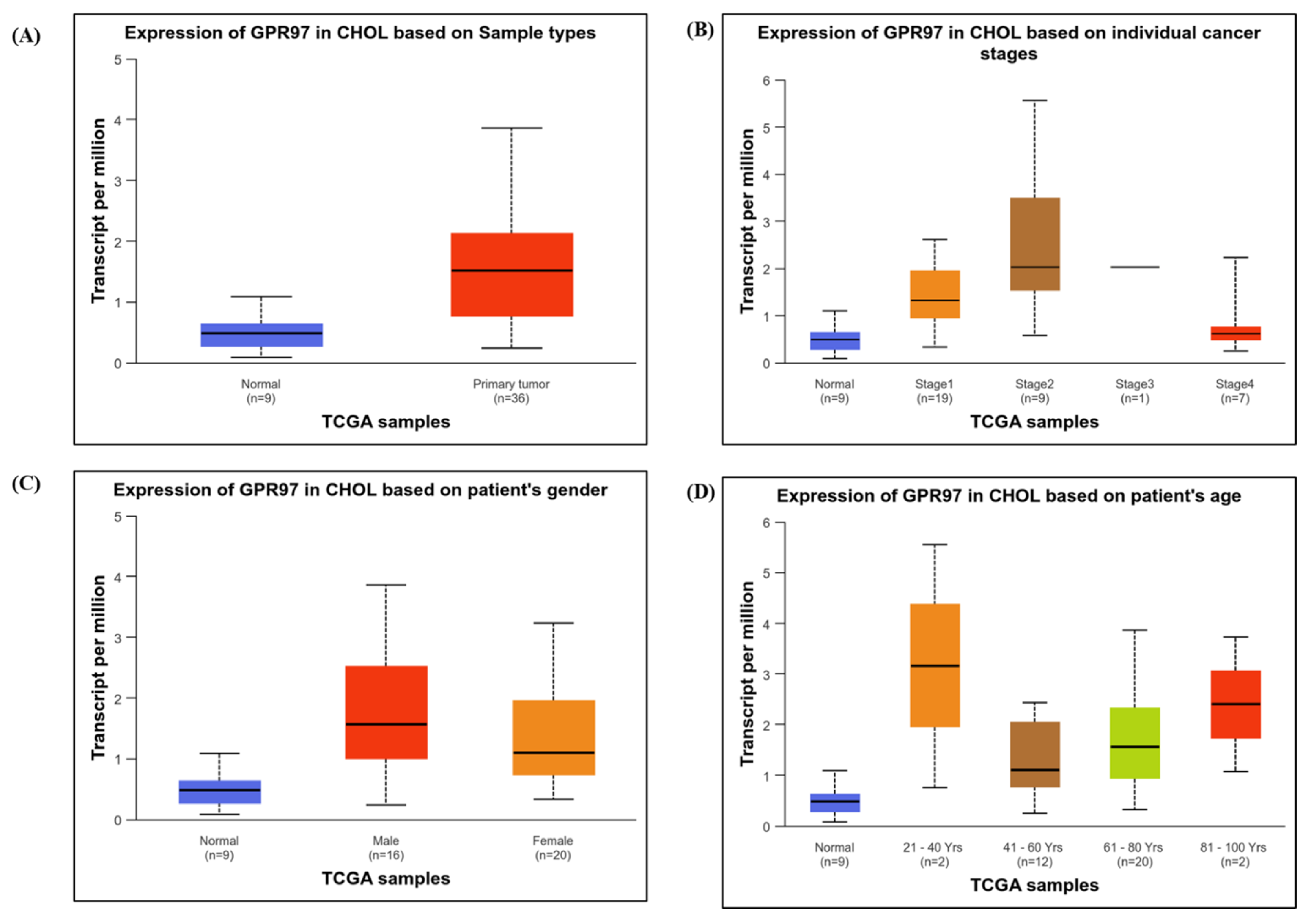
3.1.3. Expression of GPR97 in CHOL Based on Clinical Attributes
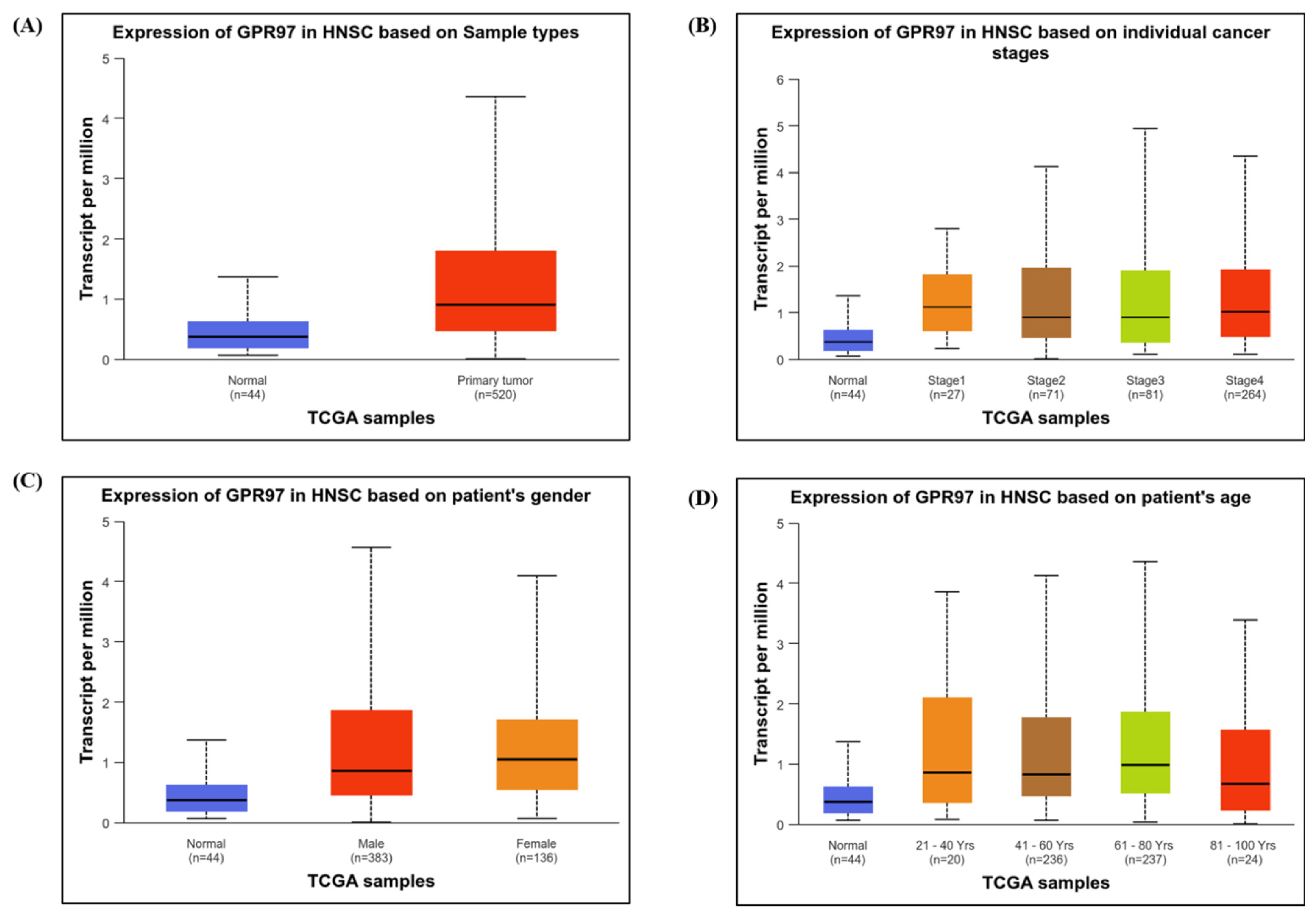
3.1.4. Expression of GPR97 in HNSC Based on Clinical Attributes
3.1.5. Expression of GPR97 in PAAD Based on Clinical Attributes
3.2. Survival and TME Analysis
3.3. Binding Affinity of Ginsenosides with ADGRG3
3.4. ADMET
3.5. Molecular Interaction Dynamics of Ginsenosides with ADGRG3
3.6. PPI and Pathway Analysis
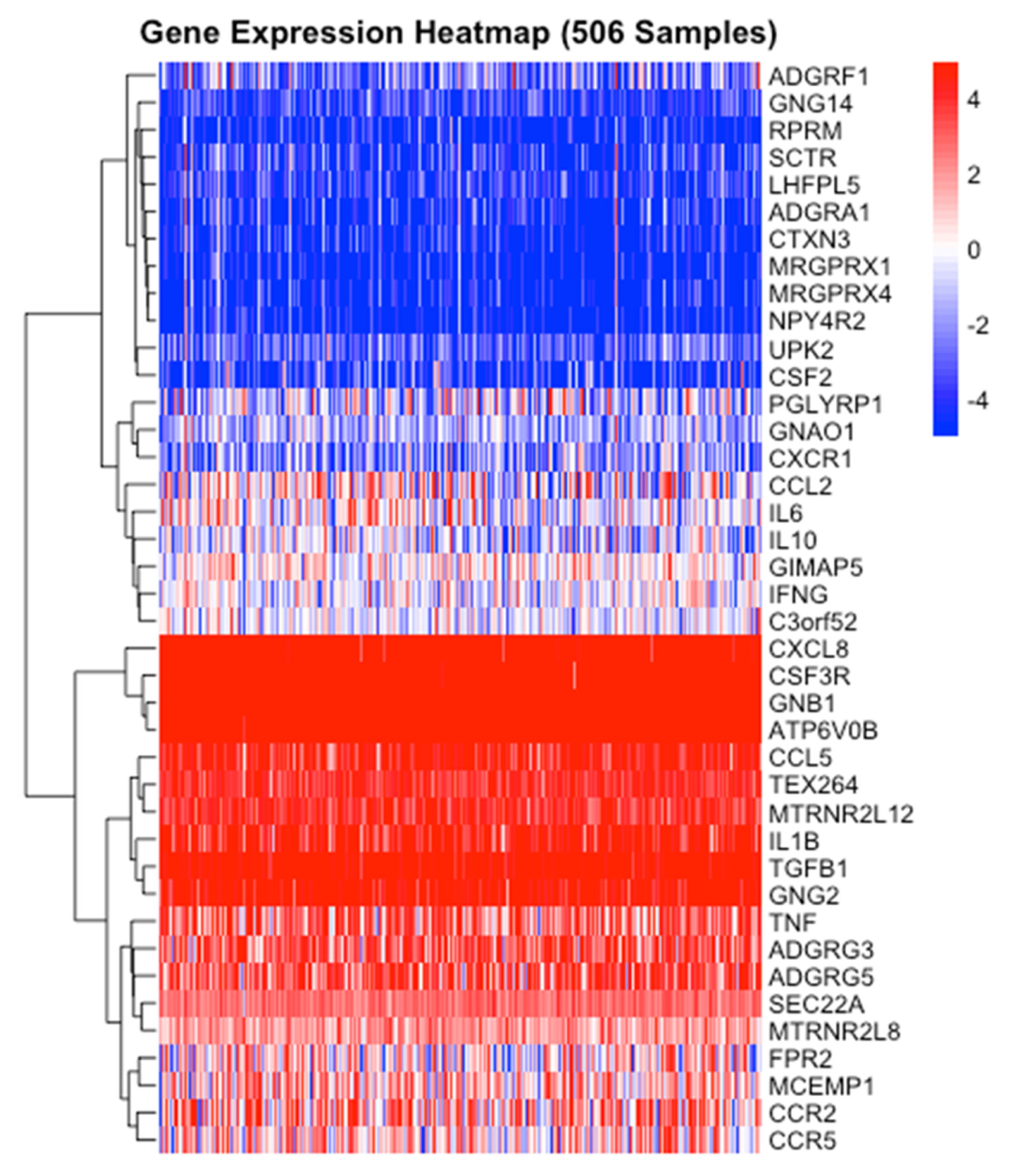
3.7. ADGRG3 Gene Expression and Other Interactions in Cancer and Inflammation
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wisler, J.W.; Rockman, H.A.; Lefkowitz, R.J. Biased G Protein–Coupled Receptor Signaling. Circulation 2018, 137, 2315–2317. [Google Scholar] [CrossRef] [PubMed]
- Finan, C.; Gaulton, A.; Kruger, F.A.; Lumbers, R.T.; Shah, T.; Engmann, J.; Galver, L.; Kelley, R.; Karlsson, A.; Santos, R.; et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017, 9, eaag1166. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yuan, J.; Hu, Z.; Zhang, Y.; Zhang, T.; Xu, M.; Long, M.; Fan, Y.; Tanyi, J.L.; Montone, K.T.; et al. Systematic illumination of druggable genes in cancer genomes. Cell Rep. 2022, 38, 110400. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Zhang, M.; Liu, Y.; Jang, H. AlphaFold, allosteric, and orthosteric drug discovery: Ways forward. Drug Discov. Today 2023, 28, 103551. [Google Scholar] [CrossRef]
- Radoux, C.J.; Vianello, F.; McGreig, J.; Desai, N.; Bradley, A.R. The druggable genome: Twenty years later. Front. Bioinform. 2022, 2, 958378. [Google Scholar] [CrossRef]
- Surh, Y.J. Reverse pharmacology applicable for botanical drug development—Inspiration from the legacy of traditional wisdom. J. Tradit. Complement. Med. 2011, 1, 5–7. [Google Scholar] [CrossRef]
- Santhanam, B.; Sluter, M.; Babu, M.M. Exploring GPCR signaling pathway networks as cancer therapeutic targets. Cell Genom. 2024, 4, 100560. [Google Scholar] [CrossRef]
- Bernatavicius, A.; Šícho, M.; Janssen, A.P.A.; Hassen, A.K.; Preuss, M.; van Westen, G.J.P. AlphaFold Meets De Novo Drug Design: Leveraging Structural Protein Information in Multitarget Molecular Generative Models. J. Chem. Inf. Model. 2024, 64, 8113–8122. [Google Scholar] [CrossRef]
- Liessmann, F.; von Bredow, L.; Meiler, J.; Liebscher, I. Targeting adhesion G protein-coupled receptors. Current status and future perspectives. Structure 2024, 32, 2188–2205. [Google Scholar] [CrossRef]
- Lin, H.H. Functional partnerships between GPI-anchored proteins and adhesion GPCRs. Bioessays 2023, 45, e2300115. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Schottlender, G.; Prieto, J.M.; Palumbo, M.C.; Castello, F.A.; Serral, F.; Sosa, E.J.; Turjanski, A.G.; Martì, M.A.; Fernández Do Porto, D. From drugs to targets: Reverse engineering the virtual screening process on a proteomic scale. Front. Drug Discov. 2022, 2, 969983. [Google Scholar] [CrossRef]
- Serrano-Marín, J.; Reyes-Resina, I.; Martínez-Pinilla, E.; Navarro, G.; Franco, R. Natural Compounds as Guides for the Discovery of Drugs Targeting G-Protein-Coupled Receptors. Molecules 2020, 25, 5060. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Wang, H.; Yu, L.; Xu, C.; Sun, H.; Lyu, Y.; Li, L.; Zhang, D.L. A correlation study of adhesion G protein-coupled receptors as potential therapeutic targets in Uterine Corpus Endometrial cancer. Int. Immunopharmacol. 2022, 108, 108743. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Chen, X.; Lu, S.; Kuang, Y.; Fei, J.; Wang, Z. Gpr97/Adgrg3 ameliorates experimental autoimmune encephalomyelitis by regulating cytokine expression. Acta Biochim. et Biophys. Sin. 2018, 50, 666–675. [Google Scholar] [CrossRef]
- Yang, J.; Wu, S.; Alachkar, H. Characterization of upregulated adhesion GPCRs in acute myeloid leukemia. Transl. Res. 2019, 212, 26–35. [Google Scholar] [CrossRef]
- Peng, Y.-M.; van de Garde, M.D.; Cheng, K.-F.; Baars, P.A.; Remmerswaal, E.B.; van Lier, R.A.; Mackay, C.R.; Lin, H.-H.; Hamann, J. Specific expression of GPR56 by human cytotoxic lymphocytes. J. Leukoc. Biol. 2011, 90, 735–740. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, X.; Wang, S.; Wang, J.; Du, B.; Wang, Z.; Liu, M.; Jiang, W.; Qian, M.; Ren, H. Gpr97 is dispensable for metabolic syndrome but is involved in macrophage inflammation in high-fat diet-induced obesity in mice. Sci. Rep. 2016, 6, 24649. [Google Scholar] [CrossRef]
- Tischner, D.; Grimm, M.; Kaur, H.; Staudenraus, D.; Carvalho, J.; Looso, M.; Günther, S.; Wanke, F.; Moos, S.; Siller, N. Single-cell profiling reveals GPCR heterogeneity and functional patterning during neuroinflammation. JCI Insight 2017, 2, e95063. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Zhang, H.-x.; Shen, C.; Lu, S.; Kuang, Y.; Wan, Y.; Wang, W.; Yan, H.; Dang, S. Gpr97 is essential for the follicular versus marginal zone B-lymphocyte fate decision. Cell Death Dis. 2013, 4, e853. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.; Noel, T.; Harris, M.; Ladds, G. Emerging roles of adhesion G protein-coupled receptors. Biochem. Soc. Trans. 2021, 49, 1695–1709. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Yu, Z.; Lu, H.; Jiang, P.; Qian, X.; Han, Y.; Qian, P. Adhesion GPCR ADGRE2 Maintains Proteostasis to Promote Progression in Acute Myeloid Leukemia. Cancer Res. 2024, 84, 2090–2108. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.F.; Li, X.J.; Zhang, H.Y. Natural products and drug discovery. Can thousands of years of ancient medical knowledge lead us to new and powerful drug combinations in the fight against cancer and dementia? EMBO Rep. 2009, 10, 194–200. [Google Scholar] [CrossRef]
- Nair, N.U.; Greninger, P.; Zhang, X.; Friedman, A.A.; Amzallag, A.; Cortez, E.; Sahu, A.D.; Lee, J.S.; Dastur, A.; Egan, R.K.; et al. A landscape of response to drug combinations in non-small cell lung cancer. Nat. Commun. 2023, 14, 3830. [Google Scholar] [CrossRef]
- Bishayee, A.; Ahmed, S.; Brankov, N.; Perloff, M. Triterpenoids as potential agents for the chemoprevention and therapy of breast cancer. Front. Biosci. 2011, 16, 980–996. [Google Scholar] [CrossRef]
- Aly, S.H.; Elbadry, A.M.M.; Doghish, A.S.; El-Nashar, H.A.S. Unveiling the pharmacological potential of plant triterpenoids in breast cancer management: An updated review. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2024, 397, 5571–5596. [Google Scholar] [CrossRef]
- Câmara, J.S.; Perestrelo, R.; Ferreira, R.; Berenguer, C.V.; Pereira, J.A.M.; Castilho, P.C. Plant-Derived Terpenoids: A Plethora of Bioactive Compounds with Several Health Functions and Industrial Applications—A Comprehensive Overview. Molecules 2024, 29, 3861. [Google Scholar] [CrossRef]
- Li, Y.-X.; Kim, S.-K. Triterpenoids as Anticancer Drugs from Marine Sponges. In Handbook of Anticancer Drugs from Marine Origin; Kim, S.-K., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 15–27. [Google Scholar]
- Shin, B.K.; Kwon, S.W.; Park, J.H. Chemical diversity of ginseng saponins from Panax ginseng. J. Ginseng Res. 2015, 39, 287–298. [Google Scholar] [CrossRef]
- Wang, P.; Song, T.; Shi, R.; He, M.; Wang, R.; Lv, J.; Jiang, M. Triterpenoids From Alisma Species: Phytochemistry, Structure Modification, and Bioactivities. Front. Chem. 2020, 8, 363. [Google Scholar] [CrossRef]
- Goddard, Z.R.; Searcey, M.; Osbourn, A. Advances in triterpene drug discovery. Trends Pharmacol. Sci. 2024, 45, 964–968. [Google Scholar] [CrossRef]
- Strüh, C.M.; Jäger, S.; Kersten, A.; Schempp, C.M.; Scheffler, A.; Martin, S.F. Triterpenoids Amplify Anti-Tumoral Effects of Mistletoe Extracts on Murine B16.F10 Melanoma In Vivo. PLoS ONE 2013, 8, e62168. [Google Scholar] [CrossRef]
- Esmeeta, A.; Adhikary, S.; Dharshnaa, V.; Swarnamughi, P.; Ummul Maqsummiya, Z.; Banerjee, A.; Pathak, S.; Duttaroy, A.K. Plant-derived bioactive compounds in colon cancer treatment: An updated review. Biomed. Pharmacother. 2022, 153, 113384. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Lu, J.-J.; Huang, M.-Q.; Bao, J.-L.; Chen, X.-P.; Wang, Y.-T. Terpenoids: Natural products for cancer therapy. Expert Opin. Investig. Drugs 2012, 21, 1801–1818. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.C.; Mathiyalagan, R.; Nahar, J.; Ramadhania, Z.M.; Kong, B.M.; Lee, D.-W.; Choi, S.K.; Lee, C.S.; Boopathi, V.; Yang, D.U.; et al. Transcriptome expression profile of compound-K-enriched red ginseng extract (DDK-401) in Korean volunteers and its apoptotic properties. Front. Pharmacol. 2022, 13, 999192. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.P. Ginsenosides chemistry, biosynthesis, analysis, and potential health effects. Adv. Food Nutr. Res. 2009, 55, 1–99. [Google Scholar] [CrossRef]
- Mathiyalagan, R.; Murugesan, M.; Ramadhania, Z.M.; Nahar, J.; Manivasagan, P.; Boopathi, V.; Jang, E.-S.; Yang, D.C.; Conde, J.; Thambi, T. Triterpenoid saponin-based supramolecular host-guest injectable hydrogels inhibit the growth of melanoma via ROS-mediated apoptosis. Mater. Sci. Eng. R Rep. 2024, 160, 100824. [Google Scholar] [CrossRef]
- Murugesan, M.; Mathiyalagan, R.; Boopathi, V.; Kong, B.M.; Choi, S.-K.; Lee, C.-S.; Yang, D.C.; Kang, S.C.; Thambi, T. Production of Minor Ginsenoside CK from Major Ginsenosides by Biotransformation and Its Advances in Targeted Delivery to Tumor Tissues Using Nanoformulations. Nanomaterials 2022, 12, 3427. [Google Scholar] [CrossRef]
- Im, D.S.; Nah, S.Y. Yin and Yang of ginseng pharmacology: Ginsenosides vs gintonin. Acta Pharmacol. Sin. 2013, 34, 1367–1373. [Google Scholar] [CrossRef]
- Leung, K.W.; Wong, A.S. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Morshed, M.N.; Akter, R.; Karim, M.R.; Iqbal, S.; Kang, S.C.; Yang, D.C. Bioconversion, Pharmacokinetics, and Therapeutic Mechanisms of Ginsenoside Compound K and Its Analogues for Treating Metabolic Diseases. Curr. Issues Mol. Biol. 2024, 46, 2320–2342. [Google Scholar] [CrossRef] [PubMed]
- Ouma, R.B.O.; Ngari, S.M.; Kibet, J.K. A review of the current trends in computational approaches in drug design and metabolism. Discov. Public Health 2024, 21, 108. [Google Scholar] [CrossRef]
- Yue, P.Y.K.; Mak, N.K.; Cheng, Y.K.; Leung, K.W.; Ng, T.B.; Fan, D.T.P.; Yeung, H.W.; Wong, R.N.S. Pharmacogenomics and the Yin/Yang actions of ginseng: Anti-tumor, angiomodulating and steroid-like activities of ginsenosides. Chin. Med. 2007, 2, 6. [Google Scholar] [CrossRef]
- Anuraga, G.; Tang, W.C.; Phan, N.N.; Ta, H.D.K.; Liu, Y.H.; Wu, Y.F.; Lee, K.H.; Wang, C.Y. Comprehensive Analysis of Prognostic and Genetic Signatures for General Transcription Factor III (GTF3) in Clinical Colorectal Cancer Patients Using Bioinformatics Approaches. Curr. Issues Mol. Biol. 2021, 43, 2–20. [Google Scholar] [CrossRef]
- Ewell, S.M.; Burton, H.; Mochona, B. In Silico Screening of 1,3,4-Thiadiazole Derivatives as Inhibitors of Vascular Endothelial Growth Factor Receptor-2 (VEGFR-2). Curr. Issues Mol. Biol. 2024, 46, 11220–11235. [Google Scholar] [CrossRef]
- Kritsi, E.; Christodoulou, P.; Tsiaka, T.; Georgiadis, P.; Zervou, M. A Computational Approach for the Discovery of Novel DNA Methyltransferase Inhibitors. Curr. Issues Mol. Biol. 2024, 46, 3394–3407. [Google Scholar] [CrossRef]
- Opo, F.; Alkarim, S.; Alrefaei, G.I.; Molla, M.H.R.; Alsubhi, N.H.; Alzahrani, F.; Ahammad, F. Pharmacophore-Model-Based Virtual-Screening Approaches Identified Novel Natural Molecular Candidates for Treating Human Neuroblastoma. Curr. Issues Mol. Biol. 2022, 44, 4838–4858. [Google Scholar] [CrossRef] [PubMed]
- Sadybekov, A.V.; Katritch, V. Computational approaches streamlining drug discovery. Nature 2023, 616, 673–685. [Google Scholar] [CrossRef]
- Morshed, M.N.; Karim, M.R.; Akter, R.; Iqbal, S.; Mathiyalagan, R.; Ahn, J.C.; Yang, D.C.; Song, J.H.; Kang, S.C.; Yang, D.U. Potential of Gut Microbial Metabolites in Treating Osteoporosis and Obesity: A Network Pharmacology and Bioinformatics Approach. Med. Sci. Monit. 2024, 30, e942899. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. Współczesna Onkol. 2015, 19, A68–A77. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Ping, Y.-Q.; Mao, C.; Xiao, P.; Zhao, R.-J.; Jiang, Y.; Yang, Z.; An, W.-T.; Shen, D.-D.; Yang, F.; Zhang, H.; et al. Structures of the glucocorticoid-bound adhesion receptor GPR97–Go complex. Nature 2021, 589, 620–626. [Google Scholar] [CrossRef]
- Piao, X.; Zhang, H.; Kang, J.P.; Yang, D.U.; Li, Y.; Pang, S.; Jin, Y.; Yang, D.C.; Wang, Y. Advances in Saponin Diversity of Panax ginseng. Molecules 2020, 25, 3452. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol. Biol. 2017, 1607, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2018, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. DoGSiteScorer: A web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Fu, L.; Shi, S.; Yi, J.; Wang, N.; He, Y.; Wu, Z.; Peng, J.; Deng, Y.; Wang, W.; Wu, C.; et al. ADMETlab 3.0: An updated comprehensive online ADMET prediction platform enhanced with broader coverage, improved performance, API functionality and decision support. Nucleic Acids Res. 2024, 52, W422–W431. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef] [PubMed]
- Timalsina, P.; Charles, K.; Mondal, A.M. STRING PPI Score to Characterize Protein Subnetwork Biomarkers for Human Diseases and Pathways. In Proceedings of the 2014 IEEE International Conference on Bioinformatics and Bioengineering, Boca Raton, FL, USA, 10–12 November 2014; pp. 251–256. [Google Scholar]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Rashid, N.; Paul, A.; Islam, S.; Sajib, S.; Nasirujjaman, K.; Hoque, K.; Reza, M. Studies on antioxidant potential, phytochemical properties and toxicity of four popular medicinal plants of Bangladesh. J. Bio-Sci. 2017, 25, 27–37. [Google Scholar] [CrossRef]
- Barrett, T.; Suzek, T.O.; Troup, D.B.; Wilhite, S.E.; Ngau, W.C.; Ledoux, P.; Rudnev, D.; Lash, A.E.; Fujibuchi, W.; Edgar, R. NCBI GEO: Mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005, 33, D562–D566. [Google Scholar] [CrossRef]
- Lasry, A.; Nadorp, B.; Fornerod, M.; Nicolet, D.; Wu, H.; Walker, C.J.; Sun, Z.; Witkowski, M.T.; Tikhonova, A.N.; Guillamot-Ruano, M.; et al. An inflammatory state remodels the immune microenvironment and improves risk stratification in acute myeloid leukemia. Nat. Cancer 2023, 4, 27–42. [Google Scholar] [CrossRef]
- Morshed, M.N.; Awais, M.; Akter, R.; Park, J.; Ling, L.; Kong, B.M.; Yang, D.C.; Yang, D.U.; Kang, S.C.; Jung, S.-K. Exploring the therapeutic potential of Terminalia ferdinandiana (Kakadu Plum) in targeting obesity-induced Type 2 diabetes and chronic inflammation: An in silico and experimental study. S. Afr. J. Bot. 2024, 171, 32–44. [Google Scholar] [CrossRef]
- Stark, J.L.; Powers, R. Application of NMR and MD in structure-based drug discovery. Top. Curr. Chem. 2012, 326, 1–34. [Google Scholar] [CrossRef]
- Balupuri, A.; Gadhe, C.G.; Balasubramanian, P.K.; Kothandan, G.; Cho, S.J. In silico study on indole derivatives as anti HIV-1 agents: A combined docking, molecular dynamics and 3D-QSAR study. Arch. Pharm. Res. 2014, 37, 1001–1015. [Google Scholar] [CrossRef]
- Zhao, H.; Caflisch, A. Molecular dynamics in drug design. Eur. J. Med. Chem. 2015, 91, 4–14. [Google Scholar] [CrossRef]
- Duan, Z.; Deng, J.; Dong, Y.; Zhu, C.; Li, W.; Fan, D. Anticancer effects of ginsenoside Rk3 on non-small cell lung cancer cells: In vitro and in vivo. Food Funct. 2017, 8, 3723–3736. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Ye, Y.; Xiao, L.; Duan, X.; Zhang, Y.; Zhang, H. Anticancer effects of ginsenoside Rg3 (Review). Int. J. Mol. Med. 2017, 39, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Guan, Y. Ginsenosides in cancer: A focus on the regulation of cell metabolism. Biomed. Pharmacother. 2022, 156, 113756. [Google Scholar] [CrossRef] [PubMed]
- Dana, S.M.M.A.; Meghdadi, M.; Kakhki, S.K.; Khademi, R. Anti-leukemia effects of ginsenoside monomer: A narrative review of pharmacodynamics study. Curr. Ther. Res. 2024, 100, 100739. [Google Scholar] [CrossRef]
- Park, S.E.; Park, C.; Kim, S.H.; Hossain, M.A.; Kim, M.Y.; Chung, H.Y.; Son, W.S.; Kim, G.-Y.; Choi, Y.H.; Kim, N.D. Korean red ginseng extract induces apoptosis and decreases telomerase activity in human leukemia cells. J. Ethnopharmacol. 2009, 121, 304–312. [Google Scholar] [CrossRef]
- Ma, S.; Huang, Q.; Hu, Q.; Gao, R.; Lan, J.; Yu, X.; Zhao, Y.; Shen, F.; Mi, A.; Wang, B. Ginsenoside Rk3 Inhibits the Extramedullary Infiltration of Acute Monocytic Leukemia Cell via miR-3677-5p/CXCL12 Axis. Evid. Based Complement. Altern. Med. 2022, 2022, 3065464. [Google Scholar] [CrossRef]
- Liu, H.; Yang, J.; Du, F.; Gao, X.; Ma, X.; Huang, Y.; Xu, F.; Niu, W.; Wang, F.; Mao, Y.; et al. Absorption and disposition of ginsenosides after oral administration of Panax notoginseng extract to rats. Drug Metab. Dispos. 2009, 37, 2290–2298. [Google Scholar] [CrossRef]
- Kim, H.J.; Oh, T.K.; Kim, Y.H.; Lee, J.; Moon, J.M.; Park, Y.S.; Sung, C.M. Pharmacokinetics of Ginsenoside Rb1, Rg3, Rk1, Rg5, F2, and Compound K from Red Ginseng Extract in Healthy Korean Volunteers. Evid. Based Complement. Altern. Med. 2022, 2022, 8427519. [Google Scholar] [CrossRef]
- Xie, H.-T.; Wang, G.-J.; Zhao, X.-C.; Sun, J.-G. Study on uptake and metabolism of ginsenoside Rg3. Chin. J. Clin. Pharmacol. Ther. 2020, 9, 257–260. [Google Scholar]
- Qu, L.; Liu, Y.; Deng, J.; Ma, X.; Fan, D. Ginsenoside Rk3 is a novel PI3K/AKT-targeting therapeutics agent that regulates autophagy and apoptosis in hepatocellular carcinoma. J. Pharm. Anal. 2023, 13, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, G.; Gao, Y.; Gao, L.; Kang, Y.; Zhao, Y.; Zhao, L.; Li, S. Total minor ginsenosides exert anti-fatigue effects via antioxidant, anti-inflammatory, regulating gut microbiota and serum metabolism. Life Sci. 2024, 359, 123231. [Google Scholar] [CrossRef]
- Harada, N.; Arahori, Y.; Okuyama, M.; Luis, P.B.; Joseph, A.I.; Kitakaze, T.; Goshima, N.; Schneider, C.; Inui, H.; Yamaji, R. Curcumin activates G protein-coupled receptor 97 (GPR97) in a manner different from glucocorticoid. Biochem. Biophys. Res. Commun. 2022, 595, 41–46. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Fu, J.; Cai, Y.; Cheng, H.; Cui, X.; Sun, M.; Liu, M.; Zhang, X. Ginsenoside compound K induces ferroptosis via the FOXO pathway in liver cancer cells. BMC Complement. Med. Ther. 2024, 24, 174. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, Y.; Zhu, Y.; Li, X. Anti-cancer effects of ginsenoside compound k on pediatric acute myeloid leukemia cells. Cancer Cell Int. 2013, 13, 24. [Google Scholar] [CrossRef]
- Guan, X.; Yuan, Y.; Wang, G.; Zheng, R.; Zhang, J.; Dong, B.; Ran, N.; Hsu, A.C.; Wang, C.; Wang, F. Ginsenoside Rg3 ameliorates acute exacerbation of COPD by suppressing neutrophil migration. Int. Immunopharmacol. 2020, 83, 106449. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Q.; Zheng, Q.H.; Chen, H.; Chen, L.; Xu, J.B.; Chen, M.Y.; Lu, D.; Wang, Z.H.; Tong, H.F.; Lin, S. Ginsenoside Rg3 inhibition of vasculogenic mimicry in pancreatic cancer through downregulation of VE-cadherin/EphA2/MMP9/MMP2 expression. Int. J. Oncol. 2014, 45, 1065–1072. [Google Scholar] [CrossRef]
- Hou, Y.; Meng, X.; Sun, K.; Zhao, M.; Liu, X.; Yang, T.; Zhang, Z.; Su, R. Anti-cancer effects of ginsenoside CK on acute myeloid leukemia in vitro and in vivo. Heliyon 2022, 8, e12106. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yuan, Z.; Sun, Y.; Bu, Y.; Li, W.; Fei, Z. Ginsenoside Rg3 enhances the anti-proliferative activity of erlotinib in pancreatic cancer cell lines by downregulation of EGFR/PI3K/Akt signaling pathway. Biomed. Pharmacother. 2017, 96, 619–625. [Google Scholar] [CrossRef]
- Zeng, D.; Wang, J.; Kong, P.; Chang, C.; Li, J.; Li, J. Ginsenoside Rg3 inhibits HIF-1α and VEGF expression in patient with acute leukemia via inhibiting the activation of PI3K/Akt and ERK1/2 pathways. Int. J. Clin. Exp. Pathol. 2014, 7, 2172–2178. [Google Scholar]
- Cui, Z.Y.; Jo, E.; Jang, H.J.; Hwang, I.H.; Lee, K.B.; Yoo, H.S.; Park, S.J.; Jung, M.K.; Lee, Y.W.; Jang, I.S. Modified Ginseng Extract Induces Apoptosis in HepG2 Cancer Cells by Blocking the CXCL8-Mediated Akt/Nuclear Factor-κB Signaling Pathway. Am. J. Chin. Med. 2018, 46, 1645–1662. [Google Scholar] [CrossRef] [PubMed]
- Leem, K.H.; Kim, S.A.; Park, H.J. Antimania-Like Effect of Panax ginseng Regulating the Glutamatergic Neurotransmission in REM-Sleep Deprivation Rats. BioMed Res. Int. 2020, 2020, 3636874. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, S.; van Leeuwen, D.; Roshanzamir, F.; Pandit, S.; Shi, L.; Sasanian, N.; Nielsen, J.; Esbjörner, E.K.; Mijakovic, I. Ginsenoside Rg3 Reduces the Toxicity of Graphene Oxide Used for pH-Responsive Delivery of Doxorubicin to Liver and Breast Cancer Cells. Pharmaceutics 2023, 15, 391. [Google Scholar] [CrossRef] [PubMed]


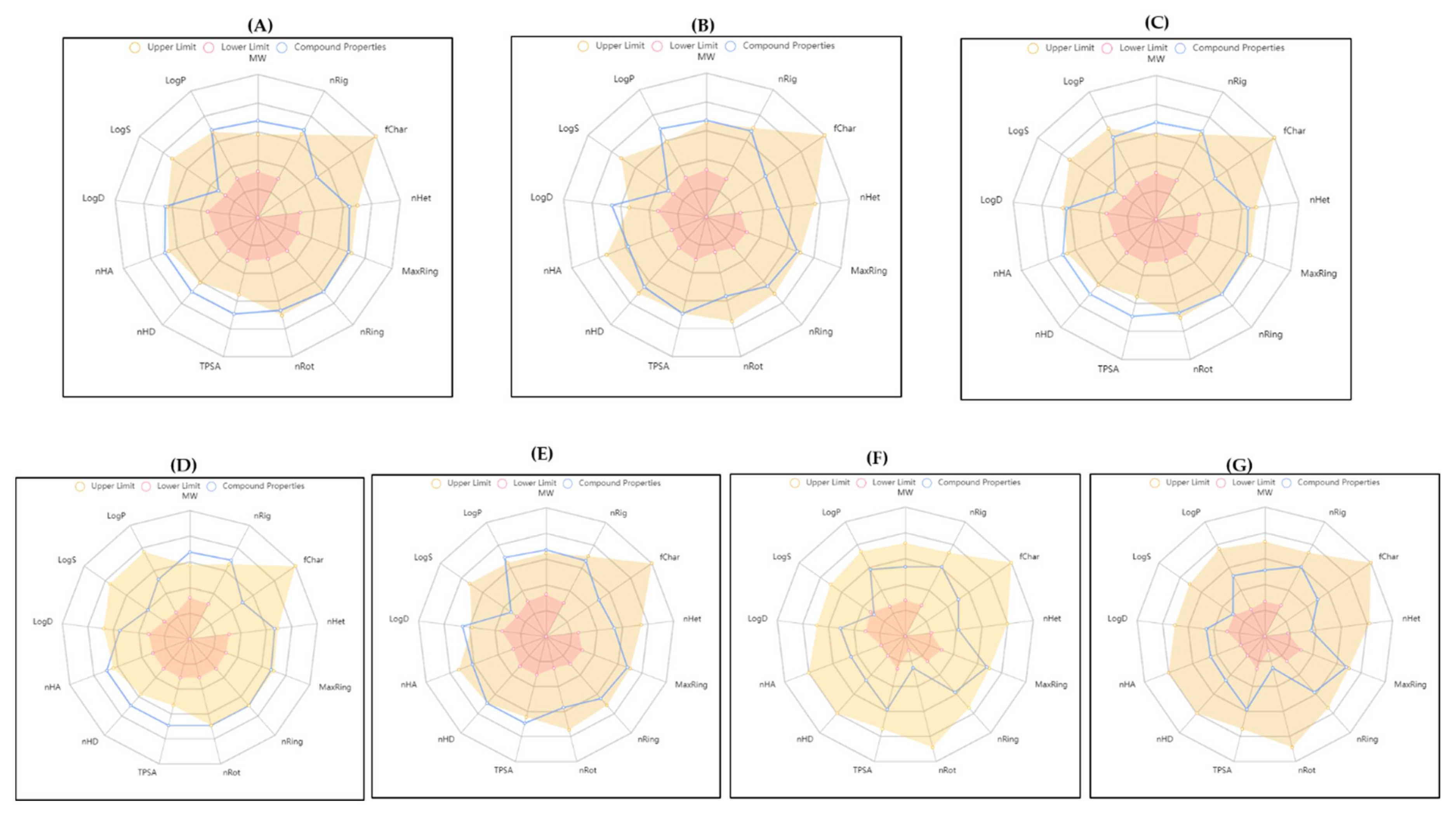

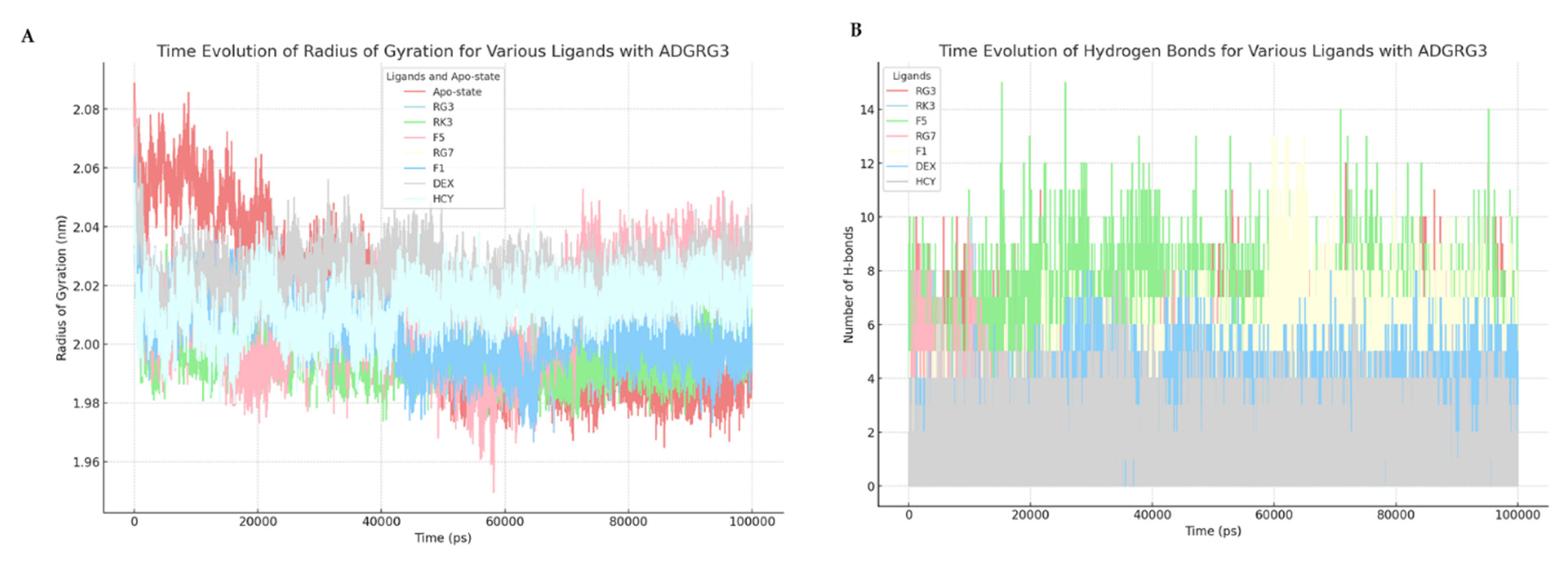
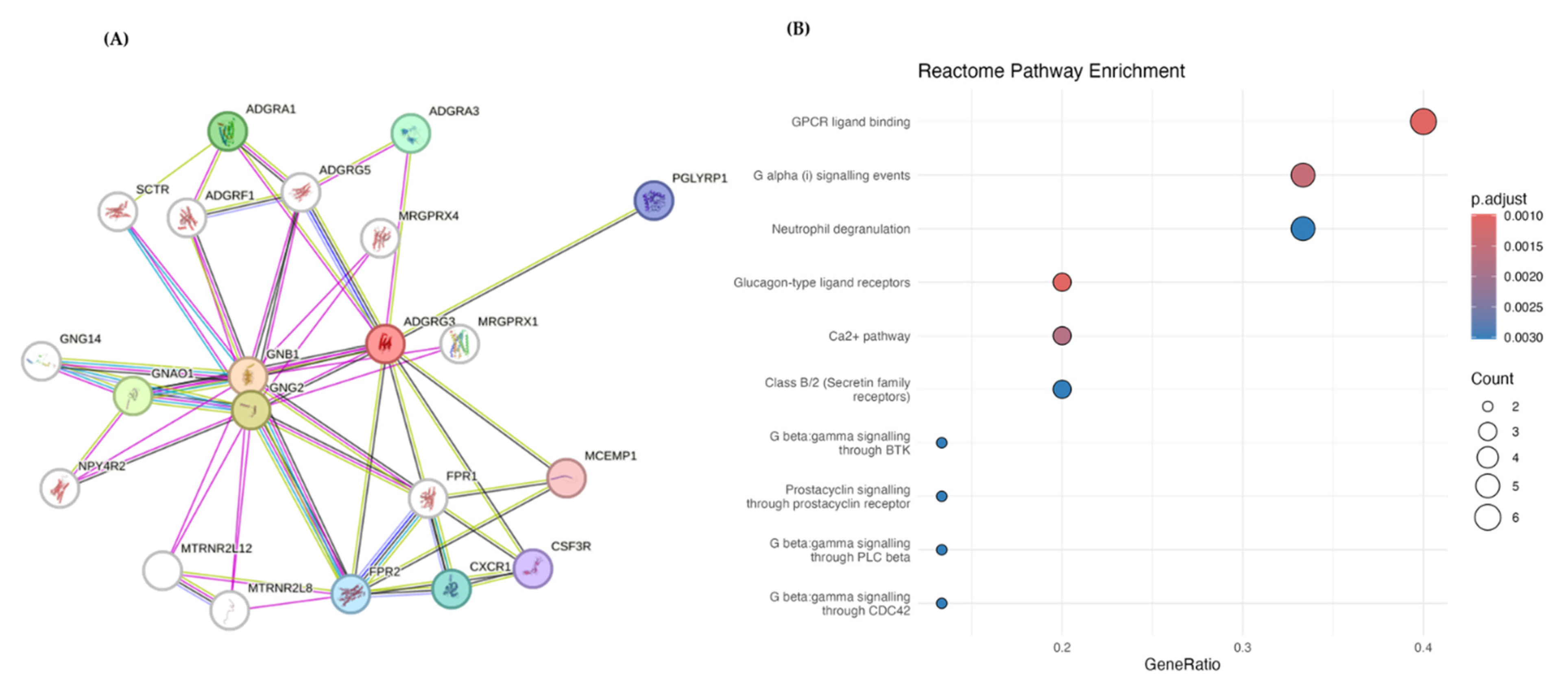

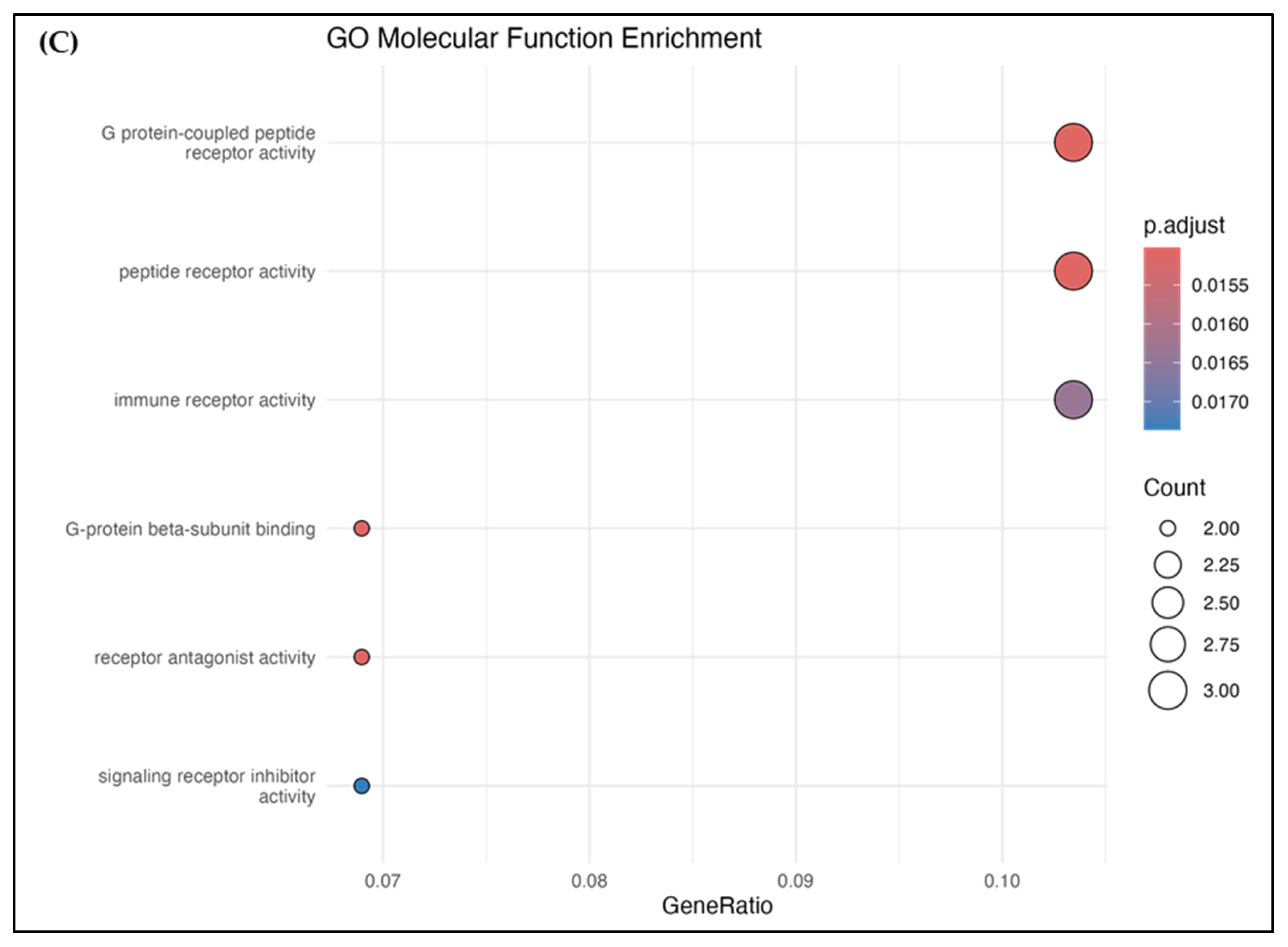
| Protein | Compound | Binding Energy (kcal/mol) | Hydrogen Bond Interactions | Other Interactions | No. of Hydrogen Bonds |
|---|---|---|---|---|---|
| ADGRG3 | Ginsenoside Rg3 | −10.7 | TYR 406, ARG 409, TYR 432, ALA 493, PHE 495 | HIS 436, TRP 490 | 5 |
| Ginsenoside Rk3 | −10.6 | TYR 406 | PHE 345, TRP 421, TRP 490, ALA 493, ASN 510 | 1 | |
| Ginsenoside F5 | −10.5 | SER 272, CYS 276, ALA 493, ASN 510 | LEU 349, TYR 406, ILE 494, PHE 525 | 4 | |
| Ginsenoside Rg7 | −10.4 | TYR 406, ARG 409, ALA 493, ASN 510 | LEU 349, ILE 494 | 4 | |
| Ginsenoside F1 | −10.3 | GLY 357, TYR 438, THR 442 | VAL 385, LEU 389, PHE 361 | 3 | |
| Control | Dexamethasone | −10.3 | PHE 506, ASN 510 | ALA 493 | 2 |
| HCY (hydrocortisone) | −10.4 | ASN 510 | LEU 319, PHE 345, TRP 490, ALA 493, PHE 506 | 1 |
| Complex | ΔVdwaals (kcal/mol) | ΔEEL (kcal/mol) | ΔEPB (kcal/mol) | ΔENPOLAR (kcal/mol) | ΔEDISPER (kcal/mol) | ΔGGas (kcal/mol) | ΔGSolv (kcal/mol) | ΔGTotal (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| ADGRG3–Ginsenoside Rg3 | −52.8947 | −14.1053 | 35 | −9 | 0.9 | −67 | 26.9 | −40.1 |
| ADGRG3–Ginsenoside Rk3 | −43.3333 | −8.66667 | 20 | −5 | 0.5 | −52 | 15.5 | −36.5 |
| ADGRG3–Ginsenoside F5 | −60.0481 | −17.4519 | 52.7 | −12 | 1.2 | −77.5 | 41.9 | −35.6 |
| ADGRG3–Ginsenoside Rg7 | −46.2609 | −9.73913 | 25 | −6 | 0.6 | −56 | 19.6 | −36.4 |
| ADGRG3–Ginsenoside F1 | −58.0645 | −13.9355 | 40 | −8 | 1 | −72 | 33 | −39 |
| ADGRG3–Dexamethasone | −45.6522 | −10.3478 | 21.5 | −6 | 0.6 | −56 | 16.1 | −39.9 |
| ADGRG3–HCY (hydrocortisone) | −45.8333 | −9.16667 | 20 | −5 | 0.5 | −55 | 15.5 | −39.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J. Computational Prediction of Ginsenosides Targeting ADGRG3/GPR97 in Cancer and Immune Pathways: A Multi-Faceted In Silico Approach. Appl. Sci. 2025, 15, 4332. https://doi.org/10.3390/app15084332
Lu J. Computational Prediction of Ginsenosides Targeting ADGRG3/GPR97 in Cancer and Immune Pathways: A Multi-Faceted In Silico Approach. Applied Sciences. 2025; 15(8):4332. https://doi.org/10.3390/app15084332
Chicago/Turabian StyleLu, Jing. 2025. "Computational Prediction of Ginsenosides Targeting ADGRG3/GPR97 in Cancer and Immune Pathways: A Multi-Faceted In Silico Approach" Applied Sciences 15, no. 8: 4332. https://doi.org/10.3390/app15084332
APA StyleLu, J. (2025). Computational Prediction of Ginsenosides Targeting ADGRG3/GPR97 in Cancer and Immune Pathways: A Multi-Faceted In Silico Approach. Applied Sciences, 15(8), 4332. https://doi.org/10.3390/app15084332








