1. Introduction
Lung diseases pose a significant global health threat, with rising morbidity and mortality rates [
1]. According to the WHO, conditions like lung cancer and pneumonia are major contributors to the global disease burden [
2], particularly in industrialized regions where pollution, smoking, and unhealthy lifestyles exacerbate their prevalence [
3]. Additionally, an aging population has increased the incidence of lung diseases among the elderly [
4], placing a heavy burden on healthcare resources. Since many lung lesions are asymptomatic in the early stages [
5], timely intervention is often missed, highlighting the urgent need for improved early detection and accurate diagnostic methods in modern medical research and practice [
6].
The rapidly advancing technology of AI, especially the progressive refinement in deep learning methodologies, has really unveiled great potential within the medical domain [
7]. From predicting disease risks, to augmenting clinical diagnostics, AI is increasingly becoming an indispensable tool for coping with the traditional challenges inherent in medicine [
8]. In the field of medical image analysis, traditional machine learning methods have played an important role in lung lesion detection, particularly in early computer-aided diagnosis (CAD) systems. Common machine learning methods include Support Vector Machine (SVM), Random Forest (RF), k-Nearest Neighbors (KNN), and Extreme Gradient Boosting (XGBoost). These methods typically rely on manually extracted features, such as Gray-Level Co-occurrence Matrix (GLCM), morphological features, and histogram equalization features, followed by classification for lung lesion detection and categorization. However, the performance of these methods heavily depends on the quality of feature engineering. Manually designed features often fail to comprehensively capture the complex information in medical images, especially when dealing with large datasets or complex feature distributions, which may lead to poor generalization. Additionally, machine learning methods face significant challenges in handling domain shifts in data distributions, often resulting in a considerable performance drop when applied to target domain data [
9].
With the advancement of deep learning, convolutional neural network (CNN)-based methods have gradually replaced traditional handcrafted feature extraction approaches, allowing models to automatically learn multi-level feature representations and thereby enhancing the robustness and accuracy of lung lesion detection. In recent years, numerous state-of-the-art object detection methods have emerged, such as the YOLO series, Faster R-CNN, and SSD [
10]. These methods achieve fast and high-precision object detection by jointly optimizing feature extraction networks and detection heads, making them suitable for a wide range of complex scenarios [
11]. However, in the field of medical imaging, traditional object detection methods still face certain limitations, particularly when dealing with domain shifts in data distributions. These models’ heavy reliance on source domain data often leads to a significant drop in performance when applied to target domain data [
12]. Moreover, the morphology and size of lung lesions in medical images exhibit a high degree of variability, further increasing the difficulty of detection.
To mitigate these challenges, domain adaptation techniques have been incorporated into medical image analysis. This study contributes to the field by addressing these critical challenges and proposes the following advancements:
This study introduces the CNDAD—Net framework, which integrates domain adaptation and multi-scale feature fusion to address the challenges of cross-domain medical image analysis. This approach effectively mitigates performance degradation caused by cross-domain data distribution discrepancies, enhancing the model’s generalization and adaptability. The proposed framework is particularly suited for analyzing medical image data across different devices and organizations.
A novel multi-dimensional feature Cross-Fusion Block (CFB) is designed and rigorously validated, enabling the deep fusion of 2D, 3D, and domain-adapted features across multiple scales. Through bidirectional interactions among features of varying dimensions, the CFB module significantly improves the model’s capacity to detect diverse lesion regions, enhancing detection accuracy and robustness. This design reduces the likelihood of missed detections and false positives, offering scalable technical support for complex medical image analysis tasks.
The CNDAD—Net framework demonstrates superior performance on the LUNA16 dataset, substantially surpassing traditional target detection methods in precision, recall, and mAP. This achievement provides an innovative technological pathway for the intelligent analysis of medical images and the precise diagnosis of lung lesions, advancing the development and application of AI-driven diagnostic solutions in medicine.
The remainder of this paper is structured as follows:
Section 2 reviews related work on object detection, transfer learning, and domain adaptation.
Section 3 presents the proposed CNDAD—Net framework in detail. Experimental setup and results are elaborated upon in
Section 4, followed by concluding remarks at the end.
2. Related Works
2.1. Target Detection Study
Target detection, a cornerstone task in computer vision, involves identifying the bounding boxes of targets and determining their respective classes. While traditional methods rely on handcrafted features and operators, deep learning approaches employ deep neural networks to autonomously learn target features from data, exhibiting robust nonlinear fitting capabilities. Classical CNN-based target detection methods are generally categorized into two-stage and one-stage approaches. Notable examples of two-stage methods include Faster R-CNN [
13] and R-FCN [
14], while prominent one-stage methods comprise YOLO [
15], SSD [
16], and RetinaNet [
17]. Two-stage methods are typically characterized by higher accuracy, whereas one-stage methods prioritize speed. To address specific challenges in target detection, several advancements have been proposed. Lin et al. introduced Feature Pyramid Networks (FPNs), which enhance the detection of small targets by integrating features across different depths in a top-down hierarchical structure [
18]. For target deformation information extraction, Dai et al. developed Deformable Convolutional Networks (DCNs), incorporating deformable convolution and deformable Region of Interest (RoI) pooling mechanisms, enabling the adaptive spatial sampling of targets [
19]. Additionally, Zhang S. et al. dynamically define positive and negative samples based on IoU between ground-truth and predicted bounding boxes, addressing the issue of instance allocation in detection tasks [
20]. The collection of labeled datasets and the training of target detection models from scratch remain time-intensive and laborious. To alleviate these challenges, various methodologies have been explored, including semi-supervised learning, few-shot learning, and transfer learning. These approaches significantly reduce the reliance on large labeled datasets while enabling effective manipulation of rendering factors such as weather conditions and time of capture. Notably, comparisons using synthetic data on urban road scenarios have demonstrated that synthetic datasets, when optimized, can outperform real-world datasets, thereby enhancing the overall performance of target detection systems.
It can be found from the above studies that significant advancements have been made in object detection technology, particularly with the introduction of deep learning methods, which have greatly improved detection accuracy and efficiency. Two-stage methods, such as Faster R-CNN and R-FCN, excel in detection precision, while one-stage methods, such as YOLO, SSD, and RetinaNet, offer advantages in computational speed. Additionally, various improvements, including Feature Pyramid Networks (FPNs), Deformable Convolutional Networks (DCNs), and adaptive positive–negative sample assignment strategies, have enhanced the adaptability and robustness of object detection across different scenarios. However, in the medical imaging domain, the high cost of data acquisition and annotation poses significant challenges for model training and optimization. To address these issues, researchers have explored approaches such as semi-supervised learning, self-supervised learning, few-shot learning, and transfer learning to mitigate data scarcity while improving model generalization and detection performance. Future research directions will focus on efficiently leveraging limited medical data and integrating multimodal information to further enhance detection accuracy and stability.
2.2. Target Detection in the Adaptive Perspective of the Transfer Learning Domain
Transfer learning is about using labeled data from one area (the source domain) to help analyze data in another area (the target domain), aiming to transfer knowledge and improve performance in the new context. By leveraging pre-existing datasets, transfer learning reduces the labor-intensive process of collecting and annotating target domain data, thereby streamlining the application of deep learning algorithms. Among various transfer learning techniques, domain adaptation stands out by assuming a shared feature and category space between source and target domains, with differences confined to feature distribution.
Current domain-adaptive target detection methods can be broadly classified into adversarial feature learning, image translation, domain randomization, and pseudo-label self-training approaches [
21]. For instance, Li et al. [
22] introduced an adversarial close matching (ATM) method, addressing equilibrium selection in adversarial domain adaptation and aligning source and target domain distributions. Similarly, Kang et al. [
23] proposed a contrast-adaptive network (CAN) for unsupervised domain adaptation, explicitly measuring intra-class and inter-class domain discrepancies and employing an alternating optimization strategy to enhance model generalizability. When source domain data are inaccessible, Yang et al. [
24] proposed the generalized source-free domain adaptation (G-SFDA) method, which integrates clustering strategies to achieve passive domain adaptation. Building on this, Li et al. [
25] relaxed previous assumptions about shared convolutional spaces in domain adaptation, while Zhuo et al. [
26] developed the domain-conditioned adaptive network (DCAN), which employs attention mechanisms to activate specific convolutional channels and uncover critical cross-domain knowledge. Furthermore, Ma et al. [
27] introduced the active universal domain adaptation network (AUAN), which progressively aligns source and target domains while utilizing a clustering non-transferable gradient embedding strategy to address private class samples in the target domain. Saito et al. [
28] proposed a novel approach using a one-to-many classifier trained on labeled source data, learning thresholds from source samples to adapt an open-set classifier for the target domain by minimizing class entropy.
The above studies underscore the advancements achieved in target detection through first-order and second-order methods, particularly in multi-target detection. However, challenges persist due to disparities in data types and class imbalances. Domain-adaptive methods address these issues by mapping data from disparate domains into a shared feature space, aligning domain-specific data distributions to facilitate consistent target detection tasks across different data types. Building on these insights, this paper focuses on designing a target detection algorithm for lung lesions by integrating domain adaptation techniques within a transfer learning framework. The proposed approach aims to improve detection accuracy, ensuring enhanced performance in the analysis of lung lesion images.
3. Methodology
3.1. CNN and Resnet for Feature Extraction
CNNs are deep learning architectures specifically designed to process grid-structured data, such as images. Their core strength lies in the efficient extraction of localized features from input data while significantly reducing the number of parameters, making them highly effective for handling high-dimensional datasets. The extraction of local features is accomplished through the application of convolutional kernels (filters), which scan the input image systematically. This process operates as follows:
where
x is the image, each corner marker represents its corresponding pixel position, while
y represents its associated output. Building on this foundation, further feature extraction is achieved through the application of the ReLU activation function. Typically, a single layer of a CNN is insufficient; instead, multiple layers are interconnected via pooling layers and fully connected layers to enhance the model’s overall performance and its capacity to leverage extracted features effectively [
29].
Beyond traditional CNNs, to address the varying applicability of features across volumes and networks, we accelerate feature integration at the multi-feature fusion layer by employing the ResNet architecture. ResNet, an enhanced deep CNN framework, overcomes the challenges of gradient vanishing and network degradation in deeper architectures through the introduction of “residual connections”. These connections simplify the training of deep networks while delivering superior performance. Unlike traditional deep networks that directly approximate the objective function
H(
x), ResNet introduces a residual function, defined as follows:
We make the network learn the residuals , and pass the input directly to the output via jump connections, where is the residual function, which is fitted by the convolutional layer. This design enables the network layers to concentrate on learning subtle differences between inputs and outputs, thereby enhancing the efficiency and performance of deep networks. The convolutional layers in CNNs extract fundamental features—such as edges and textures—from lung CT images via localized receptive fields, laying the groundwork for detailed lesion area identification. Through successive layers of convolutional stacking, the network progressively captures higher-level features, including the morphology and density characteristics of lesion regions.
ResNet further refines the deep feature extraction process by leveraging residual connections to mitigate the issue of gradient vanishing, thereby ensuring effective training of deep networks. In domain-adaptive scenarios, ResNet delivers stable and accurate feature representations, facilitating the reduction in distributional discrepancies between source and target domains [
30]. Moreover, the multi-scale feature extraction layer integrates convolutional operations at varying scales, enhancing the network’s ability to detect diverse lesions and bolstering robustness and detection accuracy in complex environments. To strengthen the model’s generalization capabilities and ensure robustness across different domains, we employ domain-adaptive feature transfer through CT images with varying levels and gray scales. This approach preserves model performance by harmonizing feature representations across heterogeneous domains.
3.2. Transfer Learning and Domain Adaptation
Transfer learning leverages the inherent similarities between datasets, tasks, or models to transfer knowledge and methodologies acquired in a large data-rich domain to a data-scarce domain. This approach facilitates the resolution of new problems more efficiently and effectively, often accelerating learning processes and improving overall performance. For transfer learning, it is assumed that there is a source domain
and a target domain
, and the goal of transfer learning is to utilize the source domain knowledge
to improve the model performance on the target domain
. The target domain model is initialized with pre-trained model parameters
and the parameters are updated with target domain data:
where
is the loss in target domain and
is the model. Domain adaptation is a focused branch of transfer learning that tackles the problem of different feature distributions between the source and target domains. By adjusting the model’s feature representations, it enables smooth knowledge transfer between these domains [
31]. A key part of domain adaptation is reducing the gap between their feature distributions. In this study, we use Maximum Mean Discrepancy (MMD), which is calculated as follows:
where
is the feature mapping function, and
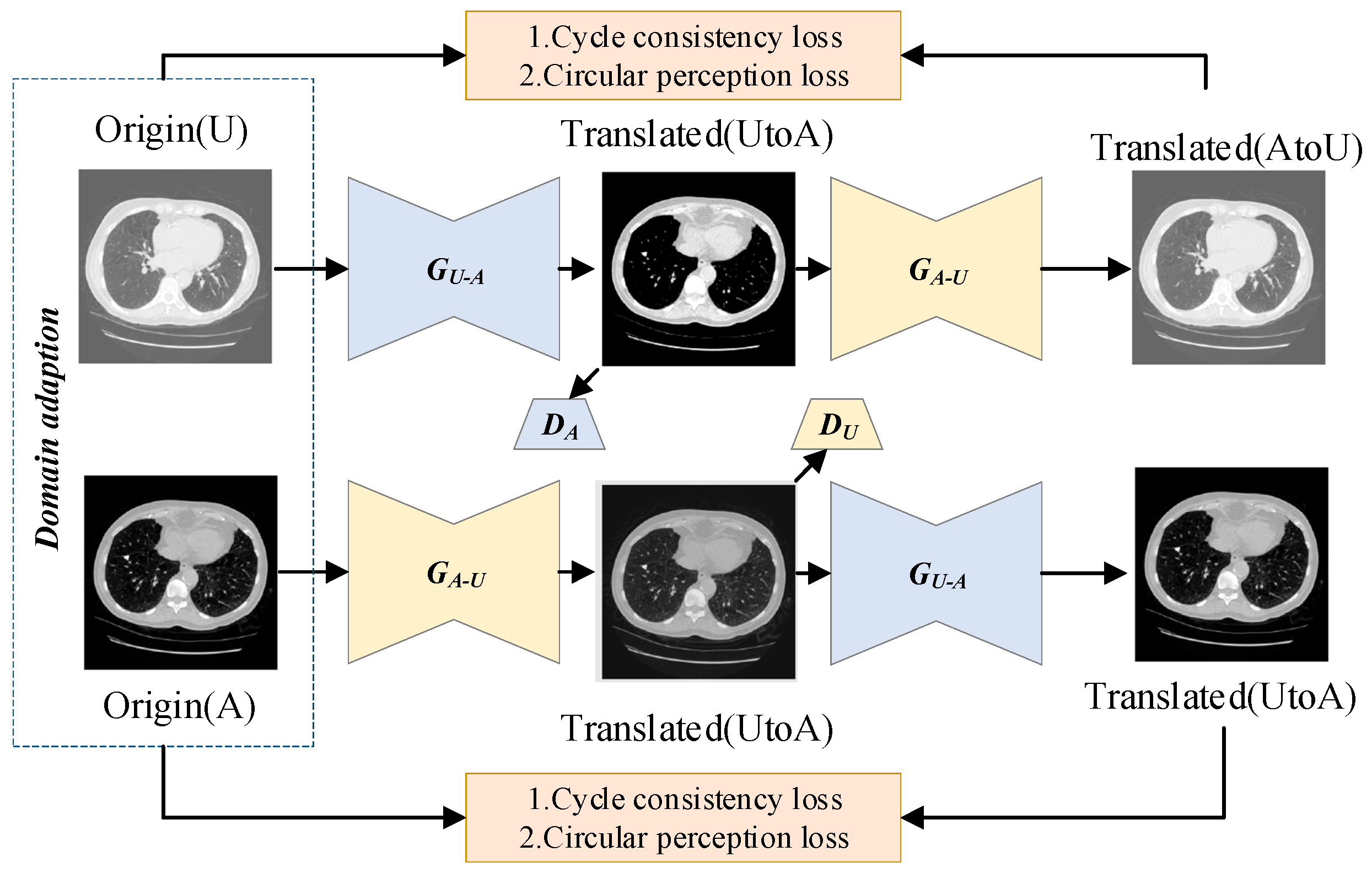
represents sample numbers in the source and target domains. After calculating the coarse-grained similarity between high-resolution and low-resolution images using the MMD metric, images with high similarity can be matched based on their similarity scores and then passed into a translation network. This module generates high-resolution images by harnessing the powerful capabilities of the image-to-image translation network. Drawing inspiration from the remarkable success of CycleGAN in image translation tasks, the domain transformation module adopts a network structure similar to CycleGAN. It consists of two key components: a coarse-grained similarity computation module and an image translation network for domain transformation.
The coarse-grained similarity computation serves to identify input images with high similarity, which facilitates network convergence and enhances the performance of the translation network. The overall workflow is depicted in
Figure 1.
The adversarial loss function ensures that the generator produces samples that align with the target domain’s distribution. However, relying only on adversarial loss cannot fully guarantee a one-to-one correspondence between the input image and the desired output image. To overcome this limitation and provide additional regularization for the translation network’s training, the cyclic consistency loss function is introduced:
The L1 norm constraints
and
, as well as
and
, are computed. The above cyclic consistency loss function compares images in pixel space, which is not sufficient to recover all texture information. Therefore, perceptual consistency loss function is introduced:
where
is the standard
norm and
is the feature extractor. Given the variations in data formats, errors, and clarity differences arising from different hospitals, equipment, and timeframes, it is challenging to achieve uniformity across datasets. Inspired by transfer learning, this paper introduces a transfer learning framework designed to enhance data performance across diverse sources. This framework effectively improves the quality of heterogeneous datasets, ensuring better performance and generating clearer outputs by refining and enhancing older, lower-quality data in contrast to traditional methods.
3.3. The Establishment of CNDAD—Net
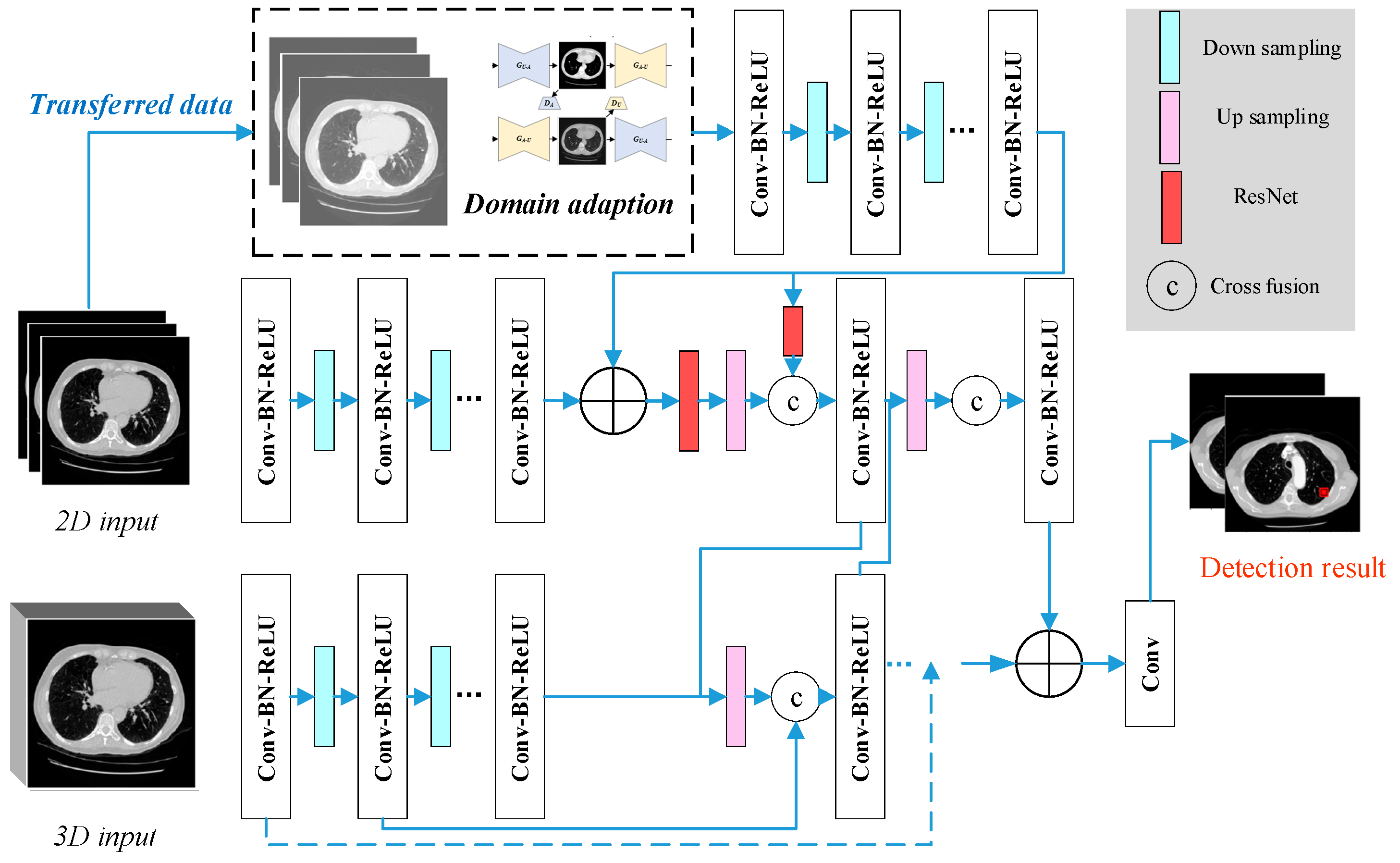
Upon finalizing the design of the various model modules, we proceeded with the construction of the model, taking into account the overall characteristics of lung CT data and the specific dataset properties. To enhance feature representation, we integrated features across different scales, fusing 2D and 3D data features. Building on this foundation, we further incorporated data from the transfer domain to construct the CNDAD—Net framework. The overall structure of CNDAD—Net is illustrated in
Figure 2.
CNDAD—Net, as shown in
Figure 2, employs a classical encoding–decoding structure for the initial feature extraction from input data. This architecture captures local information through the encoder while recovering global features via the decoder, effectively preserving detailed information and capturing contextual semantics, thus establishing a robust foundation for subsequent processing. To address the feature distribution discrepancies between source and target domains, the domain-adapted transfer method detailed in
Section 3.2 is employed. This approach leverages a domain discriminator within a GAN to generate domain-adapted data and extract their unique features. During adversarial training, the domain discriminator classifies features from the source and target domains, while the main network iteratively refines its feature representations based on the discriminator’s feedback. This process gradually minimizes the distributional gap between domains, thereby enhancing the model’s adaptability to target domain data and supporting lesion detection in environments with complex data distributions.
Following feature extraction, the model incorporates a multidimensional feature cross-fusion strategy to fully exploit the inherent correlations among 2D features, 3D features, and domain-adapted features. Specifically, the Cross-Fusion Block (CFB) is introduced to achieve multi-scale fusion of 2D and 3D feature maps. Each CFB module bridges the output features of corresponding layers in the 2D and 3D subnetwork decoders, facilitating bidirectional feature integration. This significantly enhances the network’s ability to perceive and localize lesion regions. Furthermore, the design of the CFB module allows the model to effectively capture multi-scale lesion information at various levels, rendering the fused features more discriminative and robust. By integrating these innovations, CNDAD—Net achieves the precise target detection of lung lesion sites through the cross-fusion of multidimensional features, setting a new benchmark for performance in this domain.
4. Experiment Results and Analysis
4.1. Dataset and Experiment Setup
LUNA16 (LUng Nodule Analysis 2016) is a benchmark dataset specifically designed for lung nodule detection tasks, derived from the LIDC-IDRI (The Lung Image Database Consortium Image Collection) dataset [
32,
33]. It is widely used in the field of pulmonary CT image analysis to evaluate the performance of various lung nodule detection algorithms. Each nodule in the dataset has been annotated by multiple radiologists, following a consistency scoring system (ranging from 1 to 5) that reflects the level of suspicion assigned by the radiologists. Only nodules with diameters between 3 mm and 30 mm are included in the dataset [
34].
The dataset contains a significant proportion of non-nodular regions, or background areas, posing substantial challenges for detection algorithms. This necessitates meticulous attention to data annotations. In this study, we aim to enhance model performance by leveraging domain adaptation techniques within a transfer learning framework to expand the detection range and improve robustness.
For evaluation metrics, pulmonary nodule detection commonly employs precision, recall, F1-score, and mean average precision (mAP), which are calculated as follows:
The AP is determined by plotting the precision–recall (P-R) curve. The area under the P-R curve represents the AP. The mAP is calculated by averaging the AP values across all categories. Since this study focuses exclusively on the single-category detection of lung nodules,
n = 1
n = 1, simplifying the calculation to a single AP value. Following the construction of the model, further analysis was conducted. For method comparison, we selected the mainstream YOLOv7 model and compared its performance with the improved YOLO-KMeans model proposed by Yin et al. [
32], the DETR model introduced by Dai et al. [
35], as well as the foundational models Faster R-CNN [
13] and SSD [
16]. Among them, Yin et al.’s method can be regarded as a benchmark method which employs the most classical YOLO. In addition to comparative analysis, ablation experiments were performed to evaluate the contributions of different components within the proposed framework. Specifically, we examined the performance of the CND—Net without domain adaptation, as well as the C2D—Net and C3D—Net models, which utilize single 2D and 3D data features, respectively, without employing the proposed multi-scale and multidimensional data fusion techniques. These experiments provided critical insights into the impact of domain adaptation and feature fusion on the overall model performance.
And image data requires a large computational overhead, so the experimental environment built is as
Table 1:
For model training, we adopted a deep learning-based detection network that leverages a 3D convolutional neural network (3D-CNN) backbone, pre-trained on medical image datasets to enhance feature extraction. Given the domain shift challenge in medical imaging, we incorporated transfer learning with domain adaptation to improve detection performance across different CT scan distributions. Specifically, an adversarial domain adaptation strategy was implemented to align feature distributions between source and target domains. The model was trained using the Adam optimizer, with an initial learning rate of 1e−4, batch size of 16, and training of 50 epochs. To mitigate class imbalance between nodules and non-nodule regions, we employed focal loss and hard negative mining techniques.
4.2. Experiment Result and Result Analysis
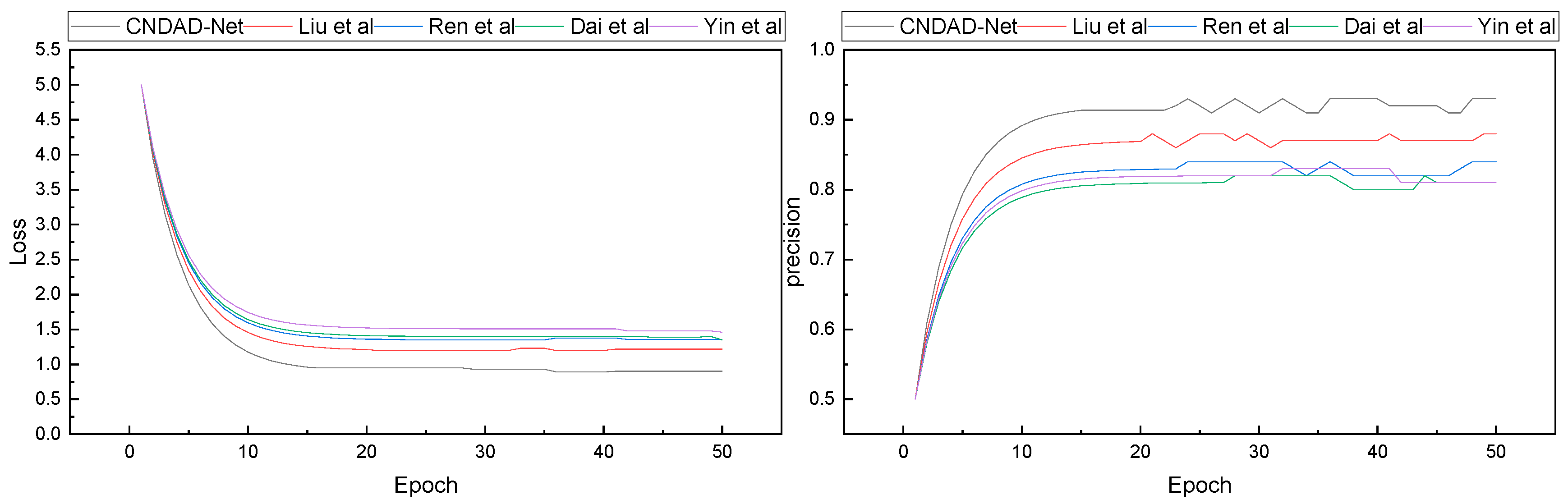
After finalizing the model construction and confirming the experimental environment, we proceeded with model training and testing. Following the 80%, 10%, and 10% split ratio specified in Algorithm 1 for training, validation, and testing, respectively, the training loss dynamics and corresponding accuracy progression during the training process were recorded. The results of these changes are illustrated in
Figure 3.
| Algorithm 1: Training process of CNDAD—Net. |
Input: training part of LUNA16 dataset
Output: Trained CNDAD—Net
Initialization: batch size, leaning rate, epochs, weights, bias.
Define CNDAD—Net and domain adaption module
Combine two frameworks
Generate the domain transferred data
while epoch < preset-epoch do
Sample a batch of image.
Feed data to the CNDAD—Net
Update model.
End
Parameters tuning
while epoch < preset-epoch do
Feed validation data to CNDAD—Net
Loss calculation.
Model updated.
Select the optimal model
Save CNDAD—Net
end |
In
Figure 3, it is evident that with continuous iterations, each model demonstrates progressively improved performance, achieving a final precision exceeding 0.8. During the loss convergence phase, the proposed CNDAD—Net exhibits superior results within approximately 15 iterations. Building upon these findings, we conducted a detailed statistical analysis of the final results, focusing on four evaluation metrics. A comparative analysis of these metrics is presented in
Figure 4.
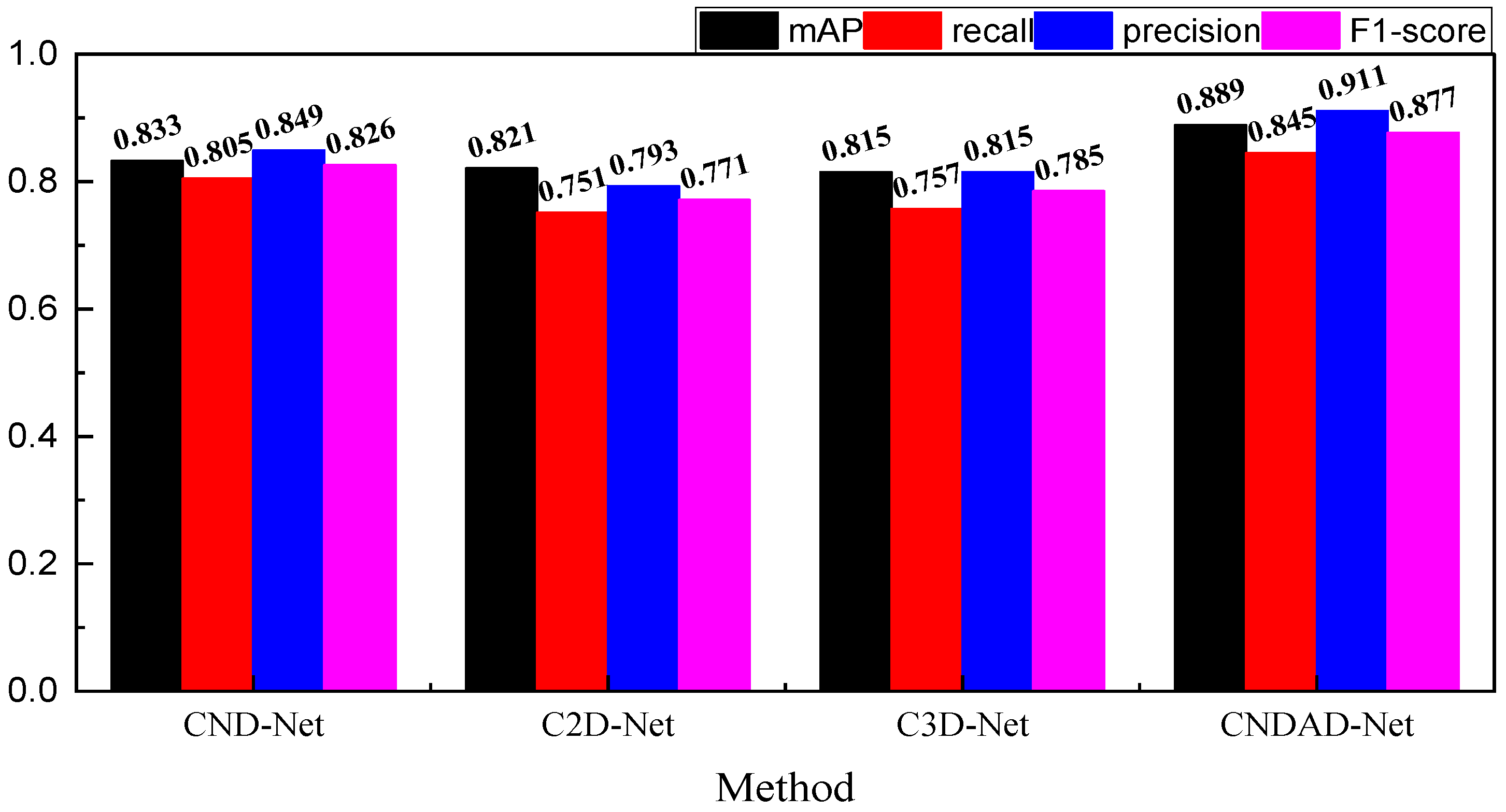
In
Figure 4, we give the specific results of the four types of evaluation metrics—mAP, recall, precision and F1-score, respectively—which are explained in detail below. For the mAP metric, mAP @ 0.5 is adpoted, the overall performance is generally better than other methods under this metric, and its value reaches 0.889, which is significantly better than traditional methods such as Faster-RCNN and slightly better than the method proposed by Liu et al. In addition, on the RECOLL indicator, the network proposed is 0.845, which indicates that it has a better performance in leakage detection compared to other methods, and can ensure more complete lesion detection. In addition, CNDAD—Net also has better performance in precision, which proves that it can detect relevant targets more accurately. In contrast, under the F1 metric, the framework of this paper is 0.886, which achieves a more balanced performance. In order to further test the characteristics of this paper’s method, we used a ten-fold validation form for the further validation of CNDAD—Net, the results of which are shown in
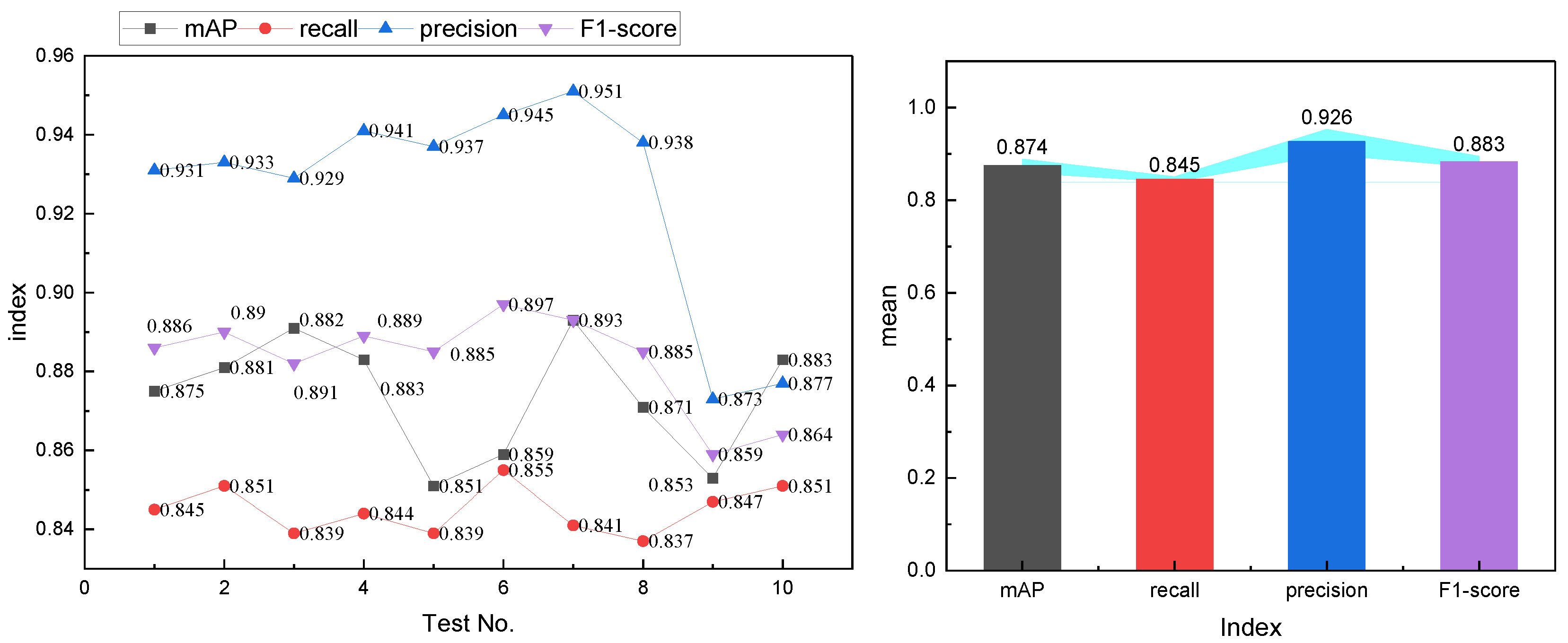
Figure 5.
In
Figure 5, it is observed that CNDAD—Net exhibits consistent stability across the first eight experiments for all evaluation metrics. However, in the final two validations, a slight deviation in precision is noted. This deviation is attributed to the imbalance in category distribution during these experiments, which impacts the model’s performance to a certain extent. Addressing this issue will require greater emphasis on balancing categories in future datasets to ensure more robust and reliable model performance.
4.3. Ablation Experiment and Model Analysis
After completing the model comparison tests, an ablation experiment is conducted to evaluate the contributions of individual modules within the model. The decomposition and naming of each variant are described in detail in
Section 4.1. The results of these ablation experiments on both the training and test sets are presented in
Figure 6.
In
Figure 6, it is evident that the proposed model outperforms the CND-Net model without domain adaptation, achieving nearly a 10% improvement in overall performance. Furthermore, the detection performance significantly declines when using either 2D or 3D data alone, underscoring the importance of incorporating domain adaptation through transfer learning and leveraging multi-dimensional feature fusion. This approach, which integrates domain adaptation and multi-scale feature fusion, substantially enhances the detection accuracy of lung lesions.
Additionally, we conducted a detailed analysis of each evaluation metric under a 10-fold validation scheme, assessing the model’s performance across different test stages. The results, visualized through box plots, are presented in
Figure 7.
From
Figure 7, it is evident that the proposed CNDAD—Net achieves outstanding detection performance, demonstrating robust results across all four evaluation metrics. In the figure, the box is given with the mean value by the line and square. From observation, it is clear that the positioning of quartiles as well as the mean values of each indicator are ahead of those observed in other methods. It is further obvious that the incorporation of domain adaptation and the fusion of multi-level data have significantly added to the robustness and precision. These observations increase the model’s potential in practical applications developed for the reliable and accurate detection of lung lesion.
5. Discussion
In order to perform the task of lung lesion target detection with high performance, the CNDAD—Net framework proposed in this study adopted multi-scale feature fusion approaches and domain adaptation through transfer learning. Notably, the domain discriminator of the GAN differentiates the features from the source and target domains to force the main network to update its feature representations. Even in cases where data in the target domain may be sparse or even demonstrate significant distributional differences, this adversarial training is guaranteed to provide excellent performance in detection by enabling successful knowledge transfer from the source to the target domain. This would significantly enhance the model’s generalizing and flexibility capabilities toward an array of data conditions, surpassing any currently available conventional technique. A significant improvement in CNDAD—Net is the CFB, which improves the ability of the model to recognize complex lesion areas. The CFB allows for bidirectional feature flow across dimensions, fully utilizing potential correlations between features by combining 2D, 3D, and domain-adapted features across several scales. The multi-scale feature fusion technique, in contrast to direct feature map stacking, fully captures global structural information and specific features of lesion sites, improving the model’s discriminatory power, robustness and precision. This design enables CNDAD—Net to accurately recognize lesion targets with complicated morphologies and a range of sizes. CNDAD—Net outperforms traditional target detection algorithms like YOLO and Faster R-CNN when there are small targets or ones that are acquired against complex backgrounds. Even though Faster R-CNN shows a very high level of accuracy, it most often fails in adapting to cross-domain data situations. CNDAD—Net successfully balances speed and accuracy by fusing the cross-domain transfer capabilities of domain adaptation technology, the detailed feature expression of multi-scale fusion, and the global context capture capability of its encoding–decoding structure. Moreover, it shows excellent robustness in scenarios with complex data distributions, making it a very flexible and reliable framework for medical image analysis.
CNDAD—Net has significant importance in the lung lesion target detection domain. It addresses the problem of cross-domain feature distribution disparities and the varied morphology of lesions, which are quite prevalent in complex medical image analysis. By allowing precise lesion target detection, this framework offers critical technical support for the early diagnosis of lung diseases, enhancing the efficiency and accuracy of clinical diagnosis and treatment. CNDAD—Net is the new technique that can help more effectively screen early lung nodules and other lesions and assists clinicians in identifying them early. Early detection enhances the probability of improving the prognosis for severe diseases, such as lung cancer. High precision with minimal error rates reduces risks in traditional manual readings. Thus, missed diagnoses decrease and patients are treated within timely manners. This framework has an amazing adaptability with diverse and inconsistent datasets. The capability of the framework is extremely useful in remote or resource-poor regions where medical data are usually sparse or not uniformly distributed. CNDAD—Net provides stable detection performance over data coming from different medical institutions or imaging devices, allowing for universal target detection and support for hierarchical diagnosis and treatment systems. Furthermore, the multi-scale feature fusion incorporated within the framework makes it robust to the complexity of lesion morphology. Lung lesions have wide variances in shape and size; traditional approaches find it challenging to maintain the subtle detail extraction of features as well as global context comprehension. The mechanism of CNDAD—Net’s fusion highly enhances the adaptive model capability, establishing a paradigm for subsequent medical image analysis frameworks, though several considerations must be taken into account for practical deployments. This shows that success in domain adaptation strongly relies on the target domain data distribution, and the minimum requirement to train the model is that the target data must exhibit some form of basic consistency. Adversarial training also introduces instability that may be difficult to stabilize. In return, it reinforces multi-scale feature fusion in improving model performance; however, computational complexity increases dramatically during deployment. With careful optimization and deployment strategies, CNDAD—Net has the potential to realize a significant advancement in lung lesion detection and provide a basic framework for other applications in medical image analysis.
6. Conclusions
In this study, we deeply explored the challenge of lung lesion target detection and proposed the CNDAD—Net, which integrates domain adaptation techniques with multi-scale feature fusion. This framework addresses the limitations of traditional detection methods in handling cross-domain data distribution disparities and complex lesion morphologies. First, the input data are preprocessed by a classical encoding–decoding structure to provide a robust foundation for subsequent feature learning. Utilizing the domain discriminator in GAN aligns feature distributions between the source and target domains, and significantly improves the generalization of the model in cross-domain scenarios. Finally, CFB is integrated into the model effectively to fuse 2D, 3D, and domain-adapted features. This module also enhances the capability of the model to sense and detect regions of lesion areas for good performance. Experimental tests that were carried out on the LUNA16 dataset result in an exceptional performance from CNDAD—Net with accuracy near 0.9 for the main medical metrics such as mAP and recall of the target detection task. These results form a very good basis for its usage in clinical applications in the future.
Despite the strong performance of our model in lung nodule detection, several limitations remain. First, deep learning-based detection methods have high computational costs, particularly 3D CNNs and domain adaptation techniques, which require substantial GPU resources. This poses challenges for deployment in low-resource medical settings or real-time applications. Second, the model’s generalization ability in real-world clinical applications is still a concern. Domain shifts caused by differences in imaging protocols, scanner types, and patient demographics can lead to performance degradation when applied in diverse hospital environments. To address these limitations, future research will focus on optimizing computational efficiency through model compression, lightweight architectures, and cloud-based inference solutions, making real-time applications more feasible. Additionally, we plan to extend the model’s capabilities by integrating multimodal medical imaging data, such as X-ray and MRI scans, to enhance its adaptability across different imaging modalities. Further exploration of advanced feature extraction techniques, including Transformer-based architectures and more efficient convolutional networks, is expected to improve detection performance. Lastly, improving model robustness in clinical settings through self-supervised learning, continual learning, and federated learning will be essential in mitigating domain shift issues and enhancing generalization across medical institutions. These improvements will contribute to the development of a more scalable, efficient, and clinically applicable AI-driven lung nodule detection system.
Author Contributions
Conceptualization, X.L. and W.L.; methodology, X.L.; software, W.L.; validation, X.L., W.L. and A.W.; formal analysis, X.L.; investigation, W.L.; resources, X.L.; data curation, A.W.; writing—original draft preparation, A.W.; writing—review and editing, A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Acknowledgments
We thank the anonymous reviewers whose comments and suggestions helped to improve the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jian, M.; Chen, H.; Zhang, Z.; Yang, N.; Zhang, H.; Ma, L.; Xu, W.; Zhi, H. A Lung Nodule Dataset with Histopathology-based Cancer type annotation. Sci. Data 2024, 11, 824. [Google Scholar] [CrossRef]
- Miyamoto, A.; Michimae, H.; Nakahara, Y.; Akagawa, S.; Nakagawa, K.; Minegishi, Y.; Ogura, T.; Hontsu, S.; Date, H.; Takahashi, K.; et al. Acute exacerbation predicting poor outcomes in idiopathic interstitial pneumonia and advanced lung cancer patients undergoing cytotoxic chemotherapy. Sci. Rep. 2024, 14, 10162. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Tian, S.; Chen, H.; Li, X.; Pu, X.; Zhang, X.; Zheng, Y.; Li, Y.; Huang, H.; Bai, C. Transbronchial lung cryobiopsy for peripheral pulmonary lesions. A narrative review. Pulmonology 2023, 30, 475–484. [Google Scholar] [CrossRef]
- Häder, A.; Köse-Vogel, N.; Schulz, L.; Mlynska, L.; Hornung, F.; Hagel, S.; Teichgräber, U.; Lang, S.M.; Pletz, M.W.; Saux, C.J.L.; et al. Respiratory infections in the aging lung: Implications for diagnosis, therapy, and prevention. Aging Dis. 2023, 14, 1091. [Google Scholar] [PubMed]
- Peng, L.; Liao, Y.; Zhou, R.; Zhong, Y.; Jiang, H.; Wang, J.; Fu, Y.; Xue, L.; Zhang, X.; Sun, M.; et al. [18F] FDG PET/MRI combined with chest HRCT in early cancer detection: A retrospective study of 3020 asymptomatic subjects. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 3723–3734. [Google Scholar] [CrossRef]
- Huang, H.; Wu, N.; Tian, S.; Shi, D.; Wang, C.; Wang, G.; Jin, F.; Li, S.; Dong, Y.; Simoff, M.J.; et al. Application of bronchoscopy in the diagnosis and treatment of peripheral pulmonary lesions in China: A national cross-sectional study. J. Cancer 2023, 14, 1398. [Google Scholar] [CrossRef] [PubMed]
- Asif, S.; Wenhui, Y.; Ur-Rehman, S.; Ul-Ain, Q.; Amjad, K.; Yueyang, Y.; Jinhai, S.; Awais, M. Advancements and Prospects of Machine Learning in Medical Diagnostics: Unveiling the Future of Diagnostic Precision. Arch. Comput. Methods Eng. 2024, 1–31. [Google Scholar] [CrossRef]
- Li, R.; Xiao, C.; Huang, Y.; Hassan, H.; Huang, B. Deep learning applications in computed tomography images for pulmonary nodule detection and diagnosis: A review. Diagnostics 2022, 12, 298. [Google Scholar] [CrossRef]
- Barnes, H.; Humphries, S.M.; George, P.M.; Assayag, D.; Glaspole, I.; Mackintosh, J.A.; Corte, T.J.; Glassberg, M.; Johannson, K.A.; Calandriello, L.; et al. Machine learning in radiology: The new frontier in interstitial lung diseases. Lancet Digit. Health 2023, 5, e41–e50. [Google Scholar] [CrossRef]
- Venkadesh, K.V.; Setio, A.A.A.; Schreuder, A.; Scholten, E.T.; Chung, K.; Wille, M.M.W.; Saghir, Z.; van Ginneken, B.; Prokop, M.; Jacobs, C. Deep learning for malignancy risk estimation of pulmonary nodules detected at low-dose screening CT. Radiology 2021, 300, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Adji, W.A.K.; Amalia, A.; Herriyance, H.; Elizar, E. Abnormal Object Detection in Thoracic X-Ray Using You Only Look Once (YOLO). In Proceedings of the 2021 International Conference on Computer System, Information Technology, and Electrical Engineering (COSITE), Banda Aceh, Indonesia, 20–21 October 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 118–123. [Google Scholar]
- Yang, Y.; Zhang, H.; Gichoya, J.W.; Katabi, D.; Ghassemi, M. The limits of fair medical imaging AI in real-world generalization. Nat. Med. 2024, 30, 2838–2848. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Li, Y.; He, K.; Sun, J. R-fcn: Object detection via region-based fully convolutional networks. Adv. Neural Inf. Process. Syst. 2016, 29. [Google Scholar]
- Redmon, J. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. Ssd: Single shot multibox detector. In Proceedings of the Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; Proceedings, Part I 14; Springer: Berlin/Heidelberg, Germany, 2016; pp. 21–37. [Google Scholar]
- Wang, Y.; Wang, C.; Zhang, H.; Dong, Y.; Wei, S. Automatic ship detection based on RetinaNet using multi-resolution Gaofen-3 imagery. Remote Sens. 2019, 11, 531. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Dai, J.; Qi, H.; Xiong, Y.; Li, Y.; Zhang, G.; Hu, H.; Wei, Y. Deformable convolutional networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 764–773. [Google Scholar]
- Zhang, P.; Zhang, B.; Chen, D.; Yuan, L.; Wen, F. Cross-domain correspondence learning for exemplar-based image translation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 5143–5153. [Google Scholar]
- Oza, P.; Sindagi, V.A.; Sharmini, V.V.; Patel, V.M. Unsupervised domain adaptation of object detectors: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 46, 4018–4040. [Google Scholar] [CrossRef]
- Li, J.; Chen, E.; Ding, Z.; Zhu, L.; Lu, K.; Shen, H.T. Maximum density divergence for domain adaptation. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 3918–3930. [Google Scholar] [CrossRef]
- Kang, G.; Jiang, L.; Yang, Y.; Hauptmann, A.G. Contrastive adaptation network for unsupervised domain adaptation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 4893–4902. [Google Scholar]
- Yang, S.; Wang, Y.; Van De Weijer, J.; Herranz, L.; Jui, S. Generalized source-free domain adaptation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 8978–8987. [Google Scholar]
- Li, S.; Liu, C.; Lin, Q.; Xie, B.; Ding, Z.; Huang, G.; Tang, J. Domain conditioned adaptation network. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; pp. 11386–11393. [Google Scholar]
- Zhuo, J.; Wang, S.; Zhang, W.; Huang, Q. Deep unsupervised convolutional domain adaptation. In Proceedings of the 25th ACM International Conference on Multimedia, Mountain View, CA, USA, 23–27 October 2017; pp. 261–269. [Google Scholar]
- Ma, X.; Gao, J.; Xu, C. Active universal domain adaptation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 8968–8977. [Google Scholar]
- Saito, K.; Saenko, K. Ovanet: One-vs-all network for universal domain adaptation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 9000–9009. [Google Scholar]
- Khodaee, P.; Esfahanipour, A.; Taheri, H.M. Forecasting turning points in stock price by applying a novel hybrid CNN-LSTM-ResNet model fed by 2D segmented images. Eng. Appl. Artif. Intell. 2022, 116, 105464. [Google Scholar] [CrossRef]
- Muhathir, M.; Ryandra, F.D.; Syah, R.B.Y.; Khairina, N.; Muliono, R. Convolutional Neural Network (CNN) of Resnet-50 with Inceptionv3 Architecture in Classification on X-Ray Image. In Computer Science On-line Conference; Springer: Berlin/Heidelberg, Germany, 2023; pp. 208–221. [Google Scholar]
- Zhang, L.; Gao, X. Transfer adaptation learning: A decade survey. IEEE Trans. Neural Netw. Learn. Syst. 2022, 35, 23–44. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Sasaki, Y.; Wang, W.; Shimizu, K. 3D Object Detection Method Based on YOLO and K-Means for Image and Point Clouds. arXiv 2020, arXiv2005.02132. [Google Scholar] [CrossRef]
- Ivanov, M.A.; Head, J.W.; Hiesinger, H. New insights into the regional and local geological context of the Luna 16 landing site. Icarus 2023, 400, 115579. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Hu, F.; Feng, L.; Zhou, T.; Zheng, C. LDNNET: Towards robust classification of lung nodule and cancer using lung dense neural network. IEEE Access 2021, 9, 50301–50320. [Google Scholar] [CrossRef]
- Dai, X.; Chen, Y.; Yang, J.; Zhang, P.; Yuan, L.; Zhang, L. Dynamic detr: End-to-end object detection with dynamic attention. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 2988–2997. [Google Scholar]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).