A Query-Based Progressive Aggregation Network for 3D Medical Image Segmentation
Abstract
1. Introduction
- (1)
- A Texture Attention (TA) mechanism is developed, which employs the Sobel operator to effectively capture local texture details, improving the structural representation capability of feature maps.
- (2)
- Structural Feature Query and Content Feature Query mechanisms are proposed to allow for precise interaction between structural and semantic information at various stages.
- (3)
- Mechanisms for forward cross-stage attention (FCA) and backward cross-stage attention (BCA) are introduced in order to achieve precise interaction and fusion of multi-scale features.
- (4)
- A Progressive Aggregation Strategy is developed to gradually integrate semantic features at various scales, effectively reducing semantic deviations in cross-scale fusion.
2. Related Work
2.1. Feature Fusion Paradigms of CNN and Transformer
2.2. Cross-Scale Semantic Alignment Methods
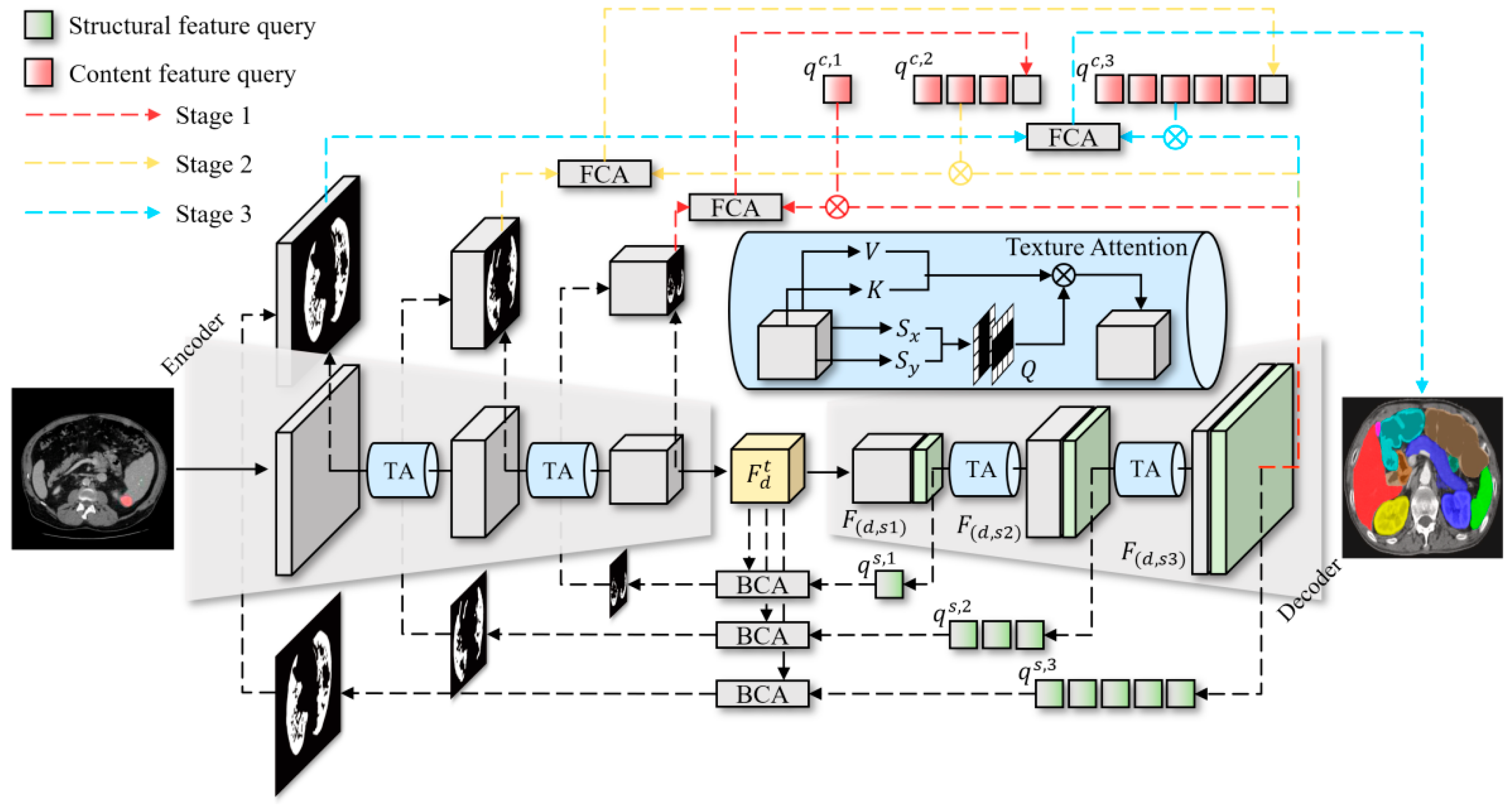
3. Proposed Method
3.1. Texture Attention and Structural Features
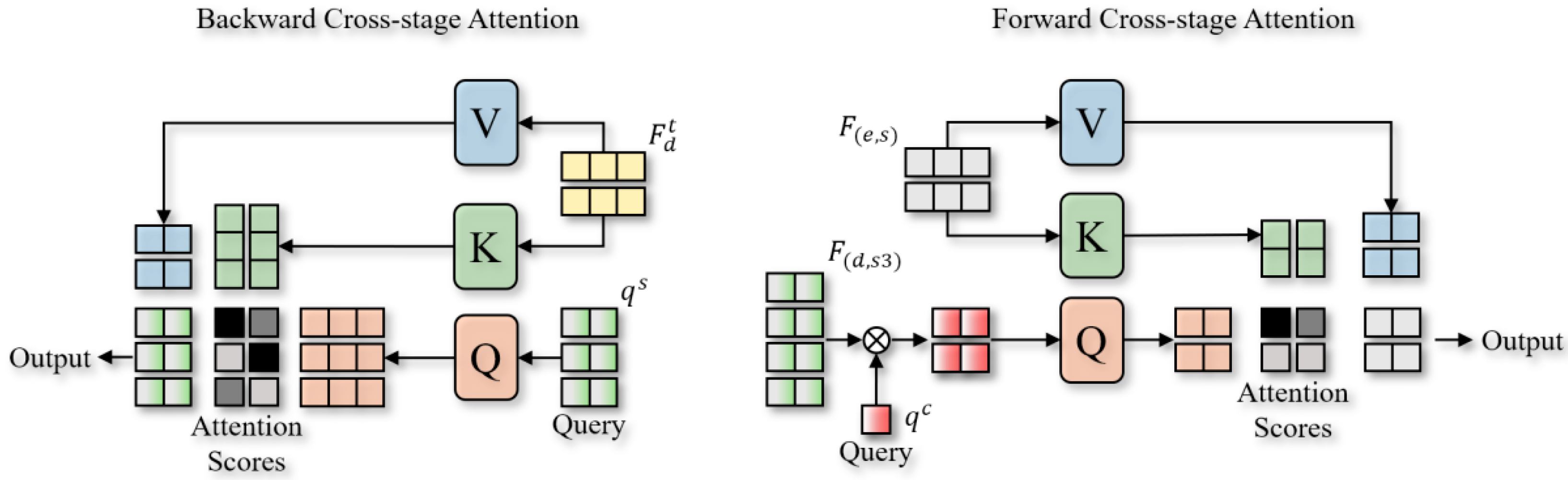
3.2. Cross-Stage Attention
3.3. Loss Function
4. Experiments
4.1. Experimental Environment and Setup
4.2. Datasets and Evaluation Metrics
4.3. Results and Analysis
4.3.1. Ablation Study
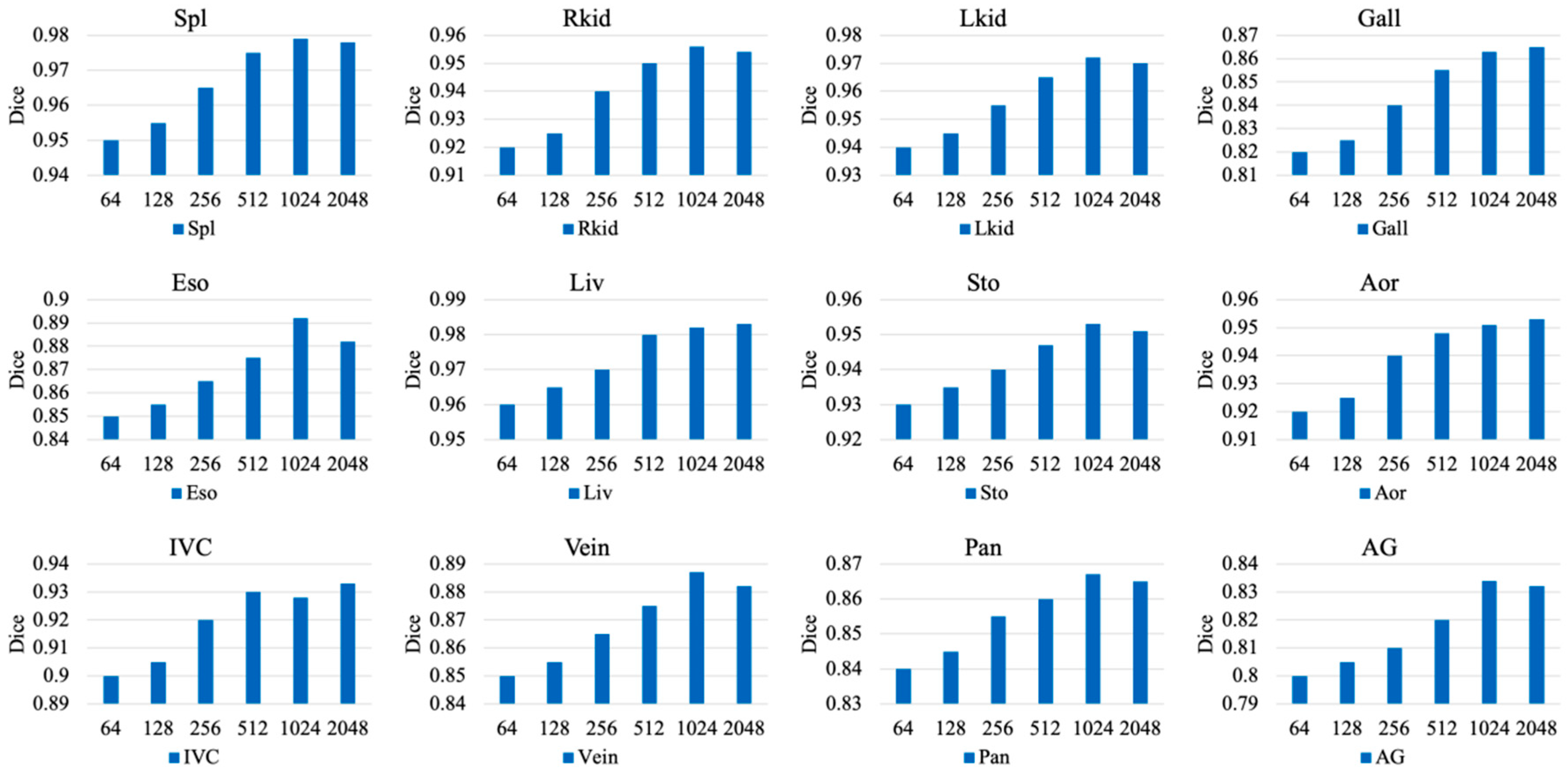
4.3.2. Hyperparameter Study
4.3.3. Comparative Study
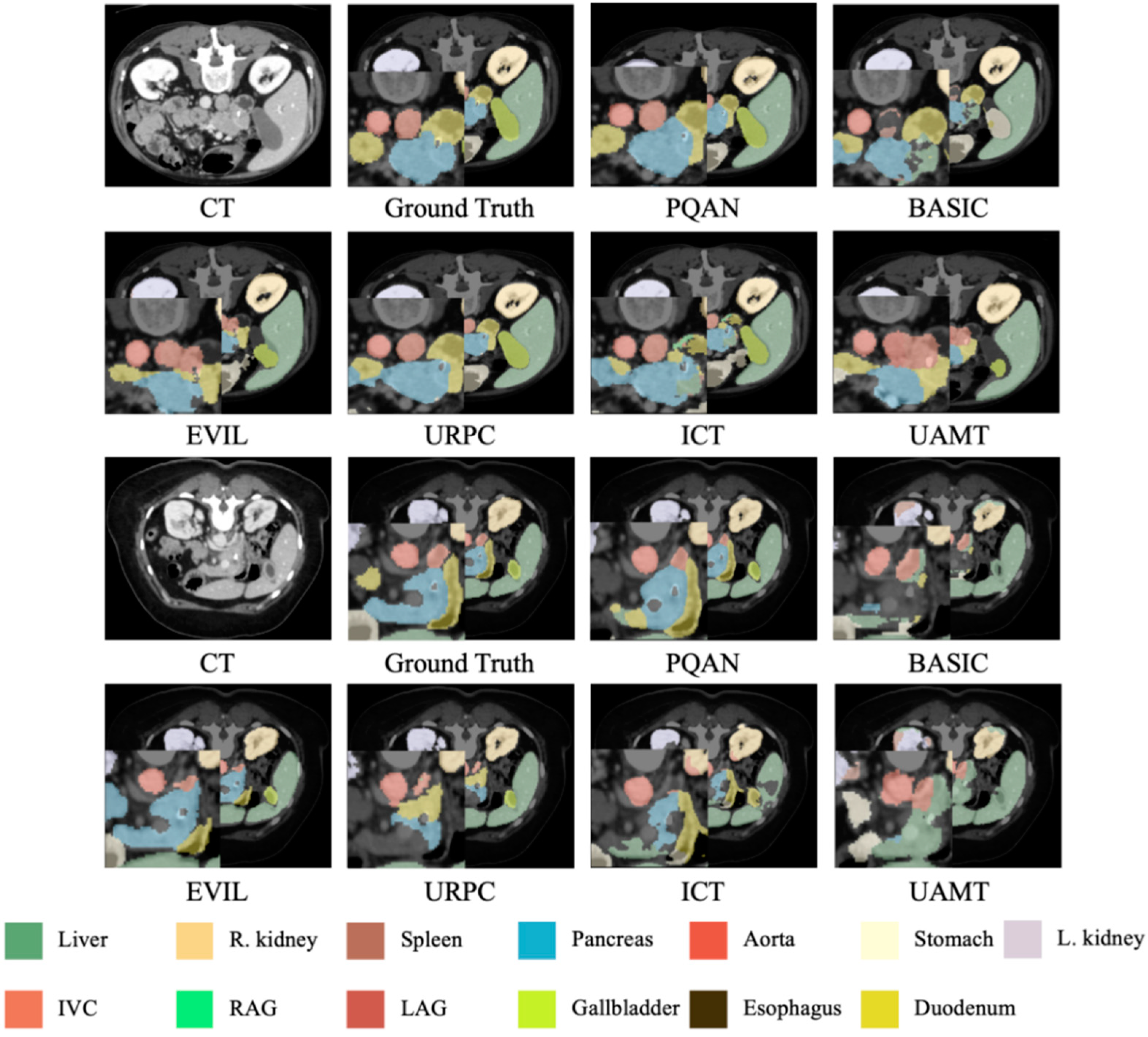
4.3.4. Visualization Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Y.; Wang, S.; Ren, Y.; Wang, J.; Wang, S.; Qiao, S.; Liu, T.; Pang, S. TPFIANet: Three path feature progressive interactive attention learning network for medical image segmentation. Knowl.-Based Syst. 2025, 323, 113778. [Google Scholar] [CrossRef]
- Rayed, M.E.; Islam, S.S.; Niha, S.I.; Jim, J.R.; Kabir, M.M.; Mridha, M.F. Deep learning for medical image segmentation: State-of-the-art advancements and challenges. Inform. Med. Unlocked 2024, 47, 101504. [Google Scholar] [CrossRef]
- Azad, R.; Aghdam, E.K.; Rauland, A.; Jia, Y.; Avval, A.H.; Bozorgpour, A.; Karimijafarbigloo, S.; Cohen, J.P.; Adeli, E.; Merhof, D. Medical image segmentation review: The success of u-net. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10076–10095. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Wang, J.; Zhang, A.; Shi, J.; Wu, X.J. CSCA U-Net: A channel and space compound attention CNN for medical image segmentation. Artif. Intell. Med. 2024, 150, 102800. [Google Scholar] [CrossRef] [PubMed]
- Allaoui, M.L.; Allili, M.S.; Belaid, A. HA-U3Net: A Modality-Agnostic Framework for 3D Medical Image Segmentation Using Nested V-Net Structure and Hybrid Attention. Knowl.-Based Syst. 2025, 327, 114127. [Google Scholar] [CrossRef]
- Chen, J.; Mei, J.; Li, X.; Lu, Y.; Yu, Q.; Wei, Q.; Luo, X.; Xie, Y.; Adeli, E.; Wang, Y.; et al. TransUNet: Rethinking the U-Net architecture design for medical image segmentation through the lens of transformers. Med. Image Anal. 2024, 97, 103280. [Google Scholar] [CrossRef]
- Yu, X.; Wu, T.; Zhang, D.; Zheng, J.; Wu, J. EDGE: Edge distillation and gap elimination for heterogeneous networks in 3D medical image segmentation. Knowl.-Based Syst. 2025, 314, 113234. [Google Scholar] [CrossRef]
- Xing, Z.; Ye, T.; Yang, Y.; Liu, G.; Zhu, L. Segmamba: Long-range sequential modeling mamba for 3d medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; pp. 578–588. [Google Scholar]
- Chen, C.; Miao, J.; Wu, D.; Zhong, A.; Yan, Z.; Kim, S.; Hu, J.; Liu, Z.; Sun, L.; Li, X.; et al. Ma-sam: Modality-agnostic sam adaptation for 3d medical image segmentation. Med. Image Anal. 2024, 98, 103310. [Google Scholar] [CrossRef]
- Isensee, F.; Wald, T.; Ulrich, C.; Baumgartner, M.; Roy, S.; Maier-Hein, K.; Jaeger, P.F. nnu-net revisited: A call for rigorous validation in 3d medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; pp. 488–498. [Google Scholar]
- Saifullah, S.; Dreżewski, R. Enhancing breast cancer diagnosis: A CNN-based approach for medical image segmentation and classification. In Proceedings of the International Conference on Computational Science, Malaga, Spain, 2–4 July 2024; pp. 155–162. [Google Scholar]
- Tamilmani, G.; Phaneendra Varma, C.H.; Devi, V.B.; Ramesh Babu, G. Medical image segmentation using grey wolf-based u-net with bi-directional convolutional LSTM. Int. J. Pattern Recognit. Artif. Intell. 2024, 38, 2354025. [Google Scholar] [CrossRef]
- Tang, H.; Chen, Y.; Wang, T.; Zhou, Y.; Zhao, L.; Gao, Q.; Du, M.; Tan, T.; Zhang, X.; Tong, T. HTC-Net: A hybrid CNN-transformer framework for medical image segmentation. Biomed. Signal Process. Control 2024, 88, 105605. [Google Scholar] [CrossRef]
- Guo, X.; Lin, X.; Yang, X.; Yu, L.; Cheng, K.T.; Yan, Z. UCTNet: Uncertainty-guided CNN-Transformer hybrid networks for medical image segmentation. Pattern Recognit. 2024, 152, 110491. [Google Scholar] [CrossRef]
- Lan, L.; Cai, P.; Jiang, L.; Liu, X.; Li, Y.; Zhang, Y. Brau-net++: U-shaped hybrid cnn-transformer network for medical image segmentation. arXiv 2024, arXiv:2401.00722. [Google Scholar]
- Ao, Y.; Shi, W.; Ji, B.; Miao, Y.; He, W.; Jiang, Z. MS-TCNet: An effective Transformer–CNN combined network using multi-scale feature learning for 3D medical image segmentation. Comput. Biol. Med. 2024, 170, 108057. [Google Scholar] [CrossRef]
- Fu, B.; Peng, Y.; He, J.; Tian, C.; Sun, X.; Wang, R. HmsU-Net: A hybrid multi-scale U-net based on a CNN and transformer for medical image segmentation. Comput. Biol. Med. 2024, 170, 108013. [Google Scholar] [CrossRef]
- Wang, X.; Girshick, R.; Gupta, A.; He, K. Non-local neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7794–7803. [Google Scholar]
- Hu, J.; Chen, S.; Pan, Z.; Zeng, S.; Yang, W. Perspective+ Unet: Enhancing Segmentation with Bi-Path Fusion and Efficient Non-Local Attention for Superior Receptive Fields. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; pp. 499–509. [Google Scholar]
- Song, E.; Zhan, B.; Liu, H. Combining external-latent attention for medical image segmentation. Neural Netw. 2024, 170, 468–477. [Google Scholar] [CrossRef]
- Liu, X.; Nguyen, T.D.C. Medical Images Enhancement by Integrating CLAHE with Wavelet Transform and Non-Local Means Denoising. Acad. J. Comput. Inf. Sci. 2024, 7, 52–58. [Google Scholar] [CrossRef]
- Maria Nancy, A.; Sathyarajasekaran, K. SwinVNETR: Swin V-net Transformer with non-local block for volumetric MRI Brain Tumor Segmentation. Automatika Časopis za Automatiku Mjerenje Elektroniku Računarstvo i Komunikacije 2024, 65, 1350–1363. [Google Scholar]
- Zhan, S.; Yuan, Q.; Lei, X.; Huang, R.; Guo, L.; Liu, K.; Chen, R. BFNet: A full-encoder skip connect way for medical image segmentation. Front. Physiol. 2024, 15, 1412985. [Google Scholar] [CrossRef] [PubMed]
- Perera, S.; Navard, P.; Yilmaz, A. Segformer3d: An efficient transformer for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 4981–4988. [Google Scholar]
- Fu, L.; Chen, Y.; Ji, W.; Yang, F. SSTrans-Net: Smart Swin Transformer Network for medical image segmentation. Biomed. Signal Process. Control 2024, 91, 106071. [Google Scholar] [CrossRef]
- Liu, X.; Gao, P.; Yu, T.; Wang, F.; Yuan, R.Y. CSWin-UNet: Transformer UNet with cross-shaped windows for medical image segmentation. Inf. Fusion 2025, 113, 102634. [Google Scholar] [CrossRef]
- Wu, J.; Ji, W.; Fu, H.; Xu, M.; Jin, Y.; Xu, Y. Medsegdiff-v2: Diffusion-based medical image segmentation with transformer. In Proceedings of the AAAI Conference on Artificial Intelligence, Philadelphia, PA, USA, 25 February–4 March 2025; Volume 38, pp. 6030–6038. [Google Scholar]
- Benabid, A.; Yuan, J.; Elhassan, M.A.; Benabid, D. CFNet: Cross-scale fusion network for medical image segmentation. J. King Saud Univ.-Comput. Inf. Sci. 2024, 36, 102123. [Google Scholar] [CrossRef]
- Sui, F.; Wang, H.; Zhang, F. Cross-scale informative priors network for medical image segmentation. Digit. Signal Process. 2025, 157, 104883. [Google Scholar] [CrossRef]
- Dai, W.; Wu, Z.; Liu, R.; Zhou, J.; Wang, M.; Wu, T.; Liu, J. Sosegformer: A Cross-Scale Feature Correlated Network For Small Medical Object Segmentation. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–4. [Google Scholar]
- Wang, Z.; Yu, L.; Tian, S.; Huo, X. CRMEFNet: A coupled refinement, multiscale exploration and fusion network for medical image segmentation. Comput. Biol. Med. 2024, 171, 108202. [Google Scholar] [CrossRef]
- Sun, J.; Zheng, X.; Wu, X.; Tang, C.; Wang, S.; Zhang, Y. CasUNeXt: A Cascaded Transformer With Intra-and Inter-Scale Information for Medical Image Segmentation. Int. J. Imaging Syst. Technol. 2024, 34, e23184. [Google Scholar] [CrossRef]
- Kervadec, H.; Bouchtiba, J.; Desrosiers, C.; Granger, E.; Dolz, J.; Ben Ayed, I. Boundary loss for highly unbalanced segmentation. In Proceedings of the International Conference on Medical Imaging with Deep Learning, London, UK, 8–10 July 2019; pp. 285–296. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. “V-net: Fully convolutional neural networks for volumetric medical image segmentation”. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, UK, 2016; Volume 1. [Google Scholar]
- Landman, B.; Xu, Z.; Igelsias, J.; Styner, M.; Langerak, T.; Klein, A. Multi-Atlas Labeling Beyond the Cranial Vault. Available online: https://www.synapse.org (accessed on 5 December 2025).
- Ma, J.; Zhang, Y.; Gu, S.; Ge, C.; Wang, E.; Zhou, Q.; Huang, Z.; Lyu, P.; He, J.; Wang, B. Automatic organ and pan-cancer segmentation in abdomen ct: The flare 2023 challenge. arXiv 2024, arXiv:2408.12534. [Google Scholar] [CrossRef]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H.S.; et al. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 6881–6890. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Caron, M.; Touvron, H.; Misra, I.; Jégou, H.; Mairal, J.; Bojanowski, P.; Joulin, A. Emerging properties in self-supervised vision transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 9650–9660. [Google Scholar]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. Cotr: Efficiently bridging cnn and transformer for 3d medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Volume 3, pp. 171–180. [Google Scholar]
- Tang, Y.; Gao, R.; Lee, H.H.; Han, S.; Chen, Y.; Gao, D.; Nath, V.; Bermudez, C.; Savona, M.R.; Abramson, R.G.; et al. High-resolution 3D abdominal segmentation with random patch network fusion. Med. Image Anal. 2021, 69, 101894. [Google Scholar] [CrossRef]
- Chen, C.; Deng, M.; Zhong, Y.; Cai, J.; Chan, K.K.W.; Dou, Q.; Chong, K.K.L.; Heng, P.-A.; Chu, W.C.-W. Multi-organ Segmentation from Partially Labeled and Unaligned Multi-modal MRI in Thyroid-associated Orbitopathy. IEEE J. Biomed. Health Inform. 2025, 29, 4161–4172. [Google Scholar] [CrossRef]
- Huang, Z.; Deng, Z.; Ye, J.; Wang, H.; Su, Y.; Li, T.; Sun, H.; Cheng, J.; Chen, J.; He, J.; et al. A-Eval: A benchmark for cross-dataset and cross-modality evaluation of abdominal multi-organ segmentation. Med. Image Anal. 2025, 101, 103499. [Google Scholar] [CrossRef] [PubMed]
- Gillot, M.; Baquero, B.; Le, C.; Deleat-Besson, R.; Bianchi, J.; Ruellas, A.; Gurgel, M.; Yatabe, M.; Al Turkestani, N.; Najarian, K.; et al. Automatic multi-anatomical skull structure segmentation of cone-beam computed tomography scans using 3D UNETR. PLoS ONE 2022, 17, e0275033. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ding, L.; Zhang, D.; Wu, J.; Liang, M.; Zheng, J.; Pang, W. Decoupled pixel-wise correction for abdominal multi-organ segmentation. Complex Intell. Syst. 2025, 11, 203. [Google Scholar] [CrossRef]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Sutskever, I.; et al. Learning transferable visual models from natural language supervision. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 8748–8763. [Google Scholar]
- Rao, Y.; Zhao, W.; Chen, G.; Tang, Y.; Zhu, Z.; Huang, G.; Zhou, J.; Lu, J. Denseclip: Language-guided dense prediction with context-aware prompting. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 18082–18091. [Google Scholar]
- Eslami, S.; de Melo, G.; Meinel, C. Does clip benefit visual question answering in the medical domain as much as it does in the general domain? arXiv 2021, arXiv:2112.13906. [Google Scholar] [CrossRef]
- Tian, W.; Huang, X.; Hou, J.; Ren, C.; Jiang, L.; Zhao, R.-W.; Jin, G.; Zhang, Y.; Geng, D. MOSMOS: Multi-organ segmentation facilitated by medical report supervision. Biomed. Signal Process. Control 2025, 106, 107743. [Google Scholar] [CrossRef]
- Springenberg, J.T. Unsupervised and semi-supervised learning with categorical generative adversarial networks. arXiv 2015, arXiv:1511.06390. [Google Scholar]
- Tarvainen, A.; Valpola, H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. arXiv 2017, arXiv:1703.01780. [Google Scholar]
- Yu, L.; Wang, S.; Li, X.; Fu, C.W.; Heng, P.A. Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Volume 22, pp. 605–613. [Google Scholar]
- Verma, V.; Kawaguchi, K.; Lamb, A.; Kannala, J.; Solin, A.; Bengio, Y.; Lopez-Paz, D. Interpolation consistency training for semi-supervised learning. Neural Netw. 2022, 145, 90–106. [Google Scholar] [CrossRef]
- Luo, X.; Wang, G.; Liao, W.; Chen, J.; Song, T.; Chen, Y.; Zhang, S.; Metaxas, D.N.; Zhang, S. Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency. Med. Image Anal. 2022, 80, 102517. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Z.; Shen, C.; Wang, Z.; Qin, Y.; Zhang, Y. EVIL: Evidential inference learning for trustworthy semi-supervised medical image segmentation. In Proceedings of the 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 18–21 April 2023. [Google Scholar]
- Feng, Z.; Wen, L.; Xu, Y.; Yan, B.; Wu, X.; Zhou, J.; Wang, Y. BASIC: Semi-supervised Multi-organ Segmentation with Balanced Subclass Regularization and Semantic-conflict Penalty. arXiv 2025, arXiv:2501.03580. [Google Scholar]
- Wang, S.; Safari, M.; Li, Q.; Chang, C.W.; Qiu, R.L.; Roper, J.; Yu, D.S.; Yang, X. Triad: Vision Foundation Model for 3D Magnetic Resonance Imaging. arXiv 2025, arXiv:2502.14064. [Google Scholar] [CrossRef]
- Dong, G.; Wang, Z.; Chen, Y.; Sun, Y.; Song, H.; Liu, L.; Cui, H. An efficient segment anything model for the segmentation of medical images. Sci. Rep. 2024, 14, 19425. [Google Scholar] [CrossRef]
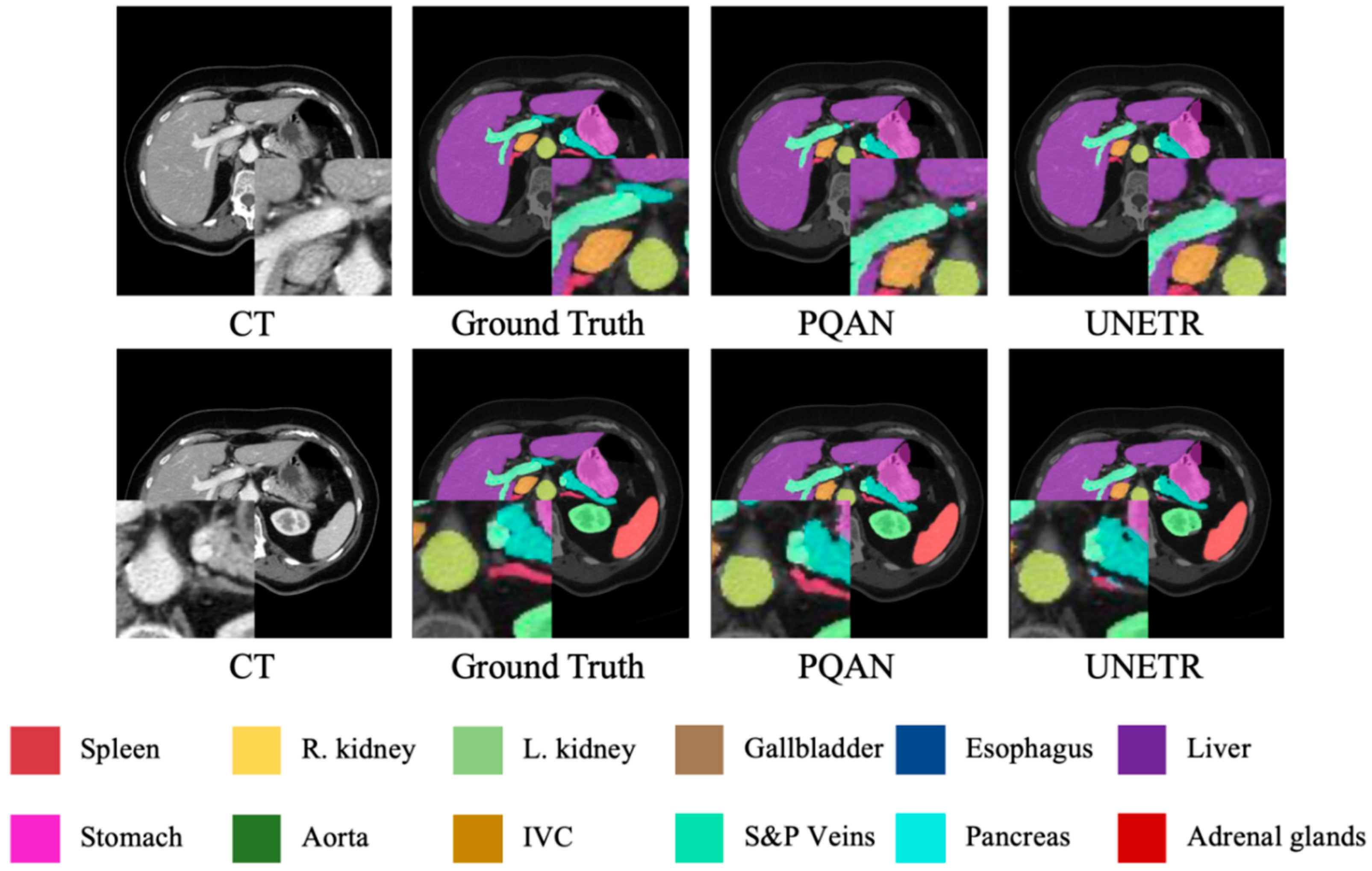
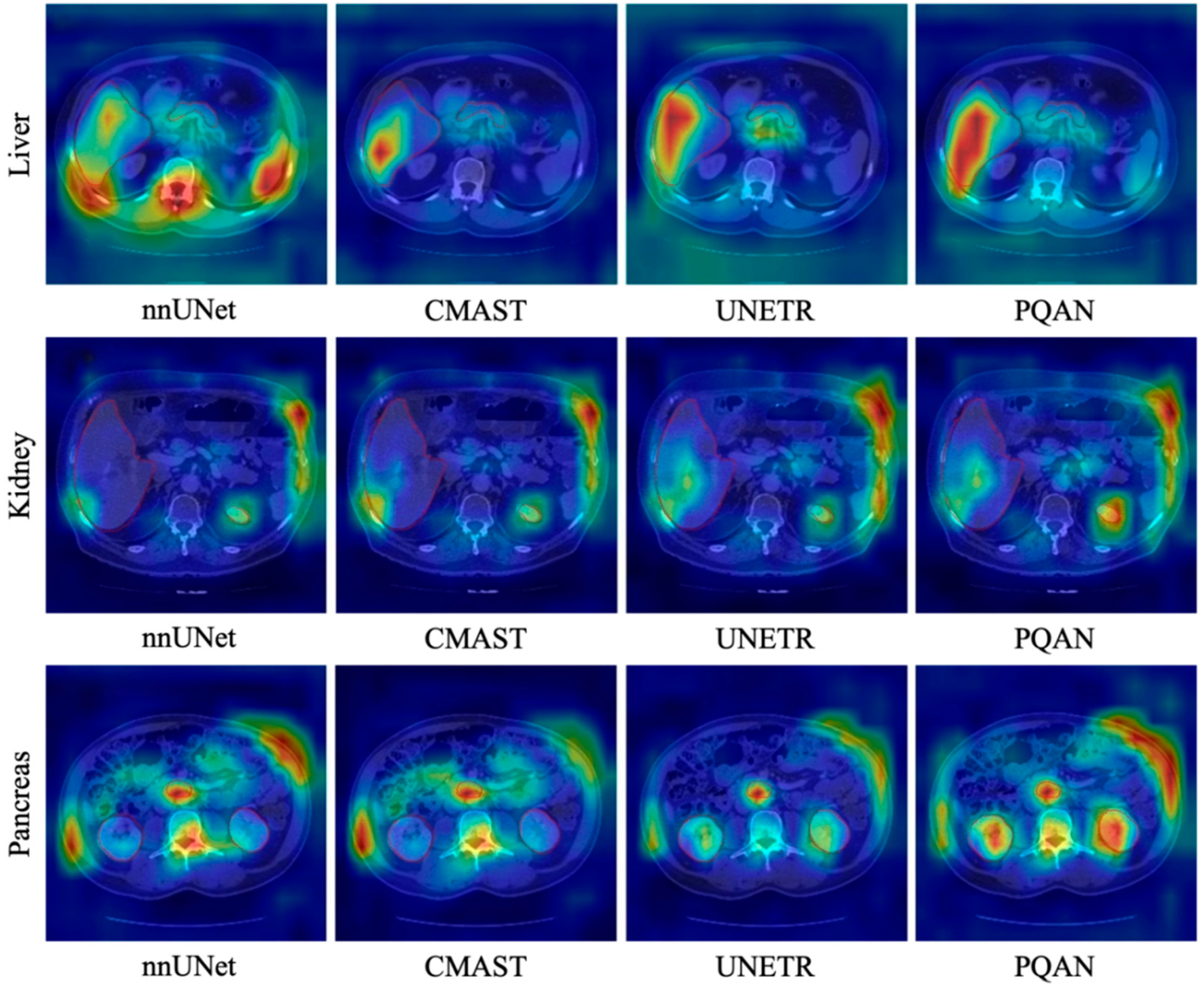
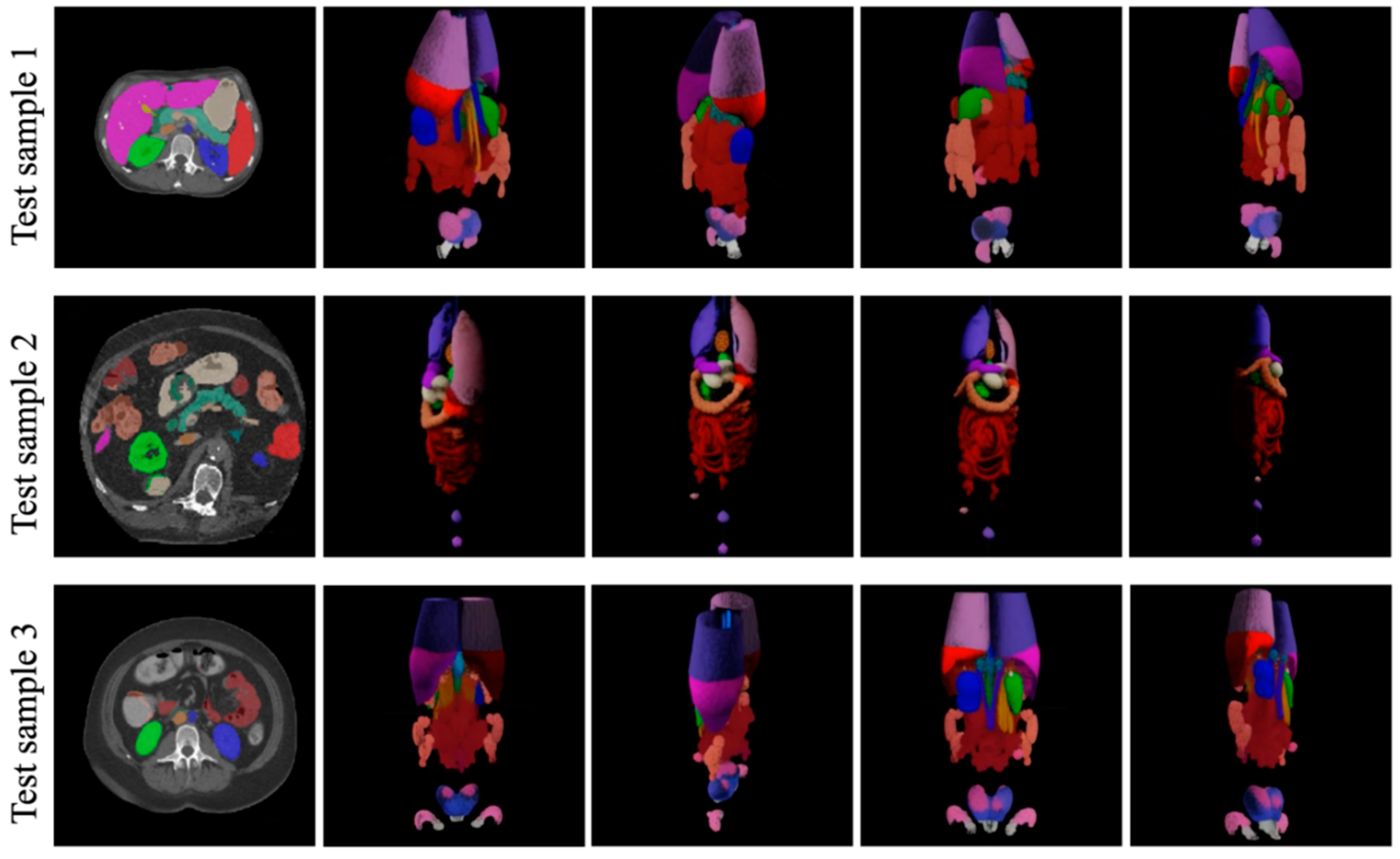
| Methods | Spl | Rkid | Lkid | Gall | Eso | Liv | Sto | Aor | IVC | Vein | Pan | AG | Avg. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| w/o TA | 0.977 | 0.954 | 0.969 | 0.857 | 0.888 | 0.979 | 0.951 | 0.947 | 0.925 | 0.884 | 0.863 | 0.830 | 0.918 |
| w/o Query | 0.975 | 0.952 | 0.967 | 0.852 | 0.885 | 0.978 | 0.949 | 0.945 | 0.922 | 0.881 | 0.862 | 0.828 | 0.912 |
| w/o FCA/BCA | 0.974 | 0.950 | 0.966 | 0.850 | 0.882 | 0.977 | 0.948 | 0.943 | 0.920 | 0.878 | 0.860 | 0.826 | 0.910 |
| w/o Prog. Agg | 0.972 | 0.949 | 0.964 | 0.848 | 0.880 | 0.975 | 0.947 | 0.941 | 0.918 | 0.876 | 0.858 | 0.824 | 0.908 |
| PQAN | 0.979 | 0.956 | 0.972 | 0.863 | 0.892 | 0.982 | 0.953 | 0.951 | 0.928 | 0.887 | 0.867 | 0.834 | 0.926 |
| Methods | Spl | Rkid | Lkid | Gall | Eso | Liv | Sto | Aor | IVC | Vein | Pan | AG | Avg. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SETR NUP [38] | 0.931 | 0.890 | 0.897 | 0.652 | 0.760 | 0.952 | 0.809 | 0.867 | 0.745 | 0.717 | 0.719 | 0.620 | 0.796 |
| SETR PUP [38] | 0.929 | 0.893 | 0.892 | 0.649 | 0.764 | 0.954 | 0.822 | 0.869 | 0.742 | 0.715 | 0.714 | 0.618 | 0.797 |
| SETR MLA [38] | 0.930 | 0.889 | 0.894 | 0.650 | 0.762 | 0.953 | 0.819 | 0.872 | 0.739 | 0.720 | 0.716 | 0.614 | 0.796 |
| nnUNet [39] | 0.942 | 0.894 | 0.910 | 0.704 | 0.723 | 0.948 | 0.824 | 0.877 | 0.782 | 0.720 | 0.680 | 0.616 | 0.802 |
| ASPP [40] | 0.935 | 0.892 | 0.914 | 0.689 | 0.760 | 0.953 | 0.812 | 0.918 | 0.807 | 0.695 | 0.720 | 0.629 | 0.811 |
| TransUNet [41] | 0.952 | 0.927 | 0.929 | 0.662 | 0.757 | 0.969 | 0.889 | 0.920 | 0.833 | 0.791 | 0.775 | 0.637 | 0.838 |
| CoTr [42] | 0.958 | 0.921 | 0.936 | 0.700 | 0.764 | 0.963 | 0.854 | 0.920 | 0.838 | 0.787 | 0.775 | 0.694 | 0.844 |
| RandomPatch [43] | 0.963 | 0.912 | 0.921 | 0.749 | 0.760 | 0.962 | 0.870 | 0.889 | 0.846 | 0.786 | 0.762 | 0.712 | 0.844 |
| CMAST [44] | 0.966 | 0.927 | 0.952 | 0.732 | 0.791 | 0.973 | 0.891 | 0.914 | 0.850 | 0.805 | 0.802 | 0.652 | 0.854 |
| A-Eval [45] | 0.972 | 0.924 | 0.958 | 0.780 | 0.841 | 0.976 | 0.922 | 0.921 | 0.872 | 0.831 | 0.842 | 0.775 | 0.884 |
| UNETR [46] | 0.972 | 0.942 | 0.954 | 0.825 | 0.864 | 0.983 | 0.945 | 0.948 | 0.890 | 0.858 | 0.799 | 0.812 | 0.891 |
| DPC-Net [47] | 0.958 | 0.908 | 0.911 | 0.695 | 0.781 | 0.955 | 0.842 | 0.880 | 0.833 | 0.786 | 0.762 | 0.729 | 0.840 |
| CLIP [48] | 0.958 | 0.914 | 0.924 | 0.706 | 0.766 | 0.963 | 0.874 | 0.889 | 0.855 | 0.825 | 0.826 | 0.720 | 0.854 |
| DenseCLIP [49] | 0.904 | 0.899 | 0.917 | 0.793 | 0.785 | 0.929 | 0.851 | 0.892 | 0.848 | 0.843 | 0.788 | 0.726 | 0.852 |
| PubMedCLIP [50] | 0.954 | 0.909 | 0.917 | 0.729 | 0.766 | 0.960 | 0.874 | 0.887 | 0.846 | 0.822 | 0.825 | 0.720 | 0.852 |
| MOSMOS [51] | 0.959 | 0.913 | 0.920 | 0.816 | 0.789 | 0.963 | 0.899 | 0.896 | 0.853 | 0.849 | 0.842 | 0.694 | 0.866 |
| PQAN | 0.979 | 0.956 | 0.972 | 0.863 | 0.892 | 0.982 | 0.953 | 0.951 | 0.928 | 0.887 | 0.867 | 0.834 | 0.926 |
| Methods | Liv | Spl | Lkid | Rkid | Sto | Gall | Eso | Pan | Duo | Col | Int | Adr | Rec | Avg. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UNet [52] | 0.890 | 0.575 | 0.708 | 0.455 | 0.809 | 0.688 | 0.588 | 0.583 | 0.620 | 0.625 | 0.679 | 0.391 | 0.644 | 0.635 |
| MT [53] | 0.936 | 0.777 | 0.805 | 0.525 | 0.872 | 0.761 | 0.557 | 0.526 | 0.414 | 0.688 | 0.719 | 0.366 | 0.831 | 0.675 |
| UAMT [54] | 0.915 | 0.732 | 0.858 | 0.455 | 0.864 | 0.748 | 0.630 | 0.588 | 0.608 | 0.672 | 0.662 | 0.347 | 0.771 | 0.681 |
| ICT [55] | 0.922 | 0.738 | 0.894 | 0.513 | 0.867 | 0.762 | 0.630 | 0.608 | 0.576 | 0.688 | 0.745 | 0.389 | 0.771 | 0.700 |
| URPC [56] | 0.924 | 0.735 | 0.863 | 0.484 | 0.883 | 0.749 | 0.619 | 0.622 | 0.683 | 0.695 | 0.632 | 0.413 | 0.789 | 0.699 |
| EVIL [57] | 0.940 | 0.834 | 0.889 | 0.501 | 0.852 | 0.764 | 0.678 | 0.595 | 0.666 | 0.693 | 0.700 | 0.416 | 0.860 | 0.722 |
| BASIC [58] | 0.963 | 0.944 | 0.951 | 0.622 | 0.936 | 0.827 | 0.710 | 0.619 | 0.730 | 0.723 | 0.745 | 0.502 | 0.913 | 0.783 |
| Triad [59] | 0.970 | 0.949 | 0.957 | 0.655 | 0.944 | 0.845 | 0.732 | 0.642 | 0.739 | 0.729 | 0.757 | 0.519 | 0.924 | 0.797 |
| EMedSAM [60] | 0.969 | 0.943 | 0.952 | 0.634 | 0.941 | 0.831 | 0.725 | 0.620 | 0.731 | 0.722 | 0.736 | 0.510 | 0.919 | 0.785 |
| PQAN | 0.976 | 0.953 | 0.962 | 0.687 | 0.951 | 0.863 | 0.753 | 0.664 | 0.748 | 0.735 | 0.769 | 0.535 | 0.936 | 0.801 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, W.; Hu, G.; Li, J.; Lyu, C. A Query-Based Progressive Aggregation Network for 3D Medical Image Segmentation. Appl. Sci. 2025, 15, 13153. https://doi.org/10.3390/app152413153
Peng W, Hu G, Li J, Lyu C. A Query-Based Progressive Aggregation Network for 3D Medical Image Segmentation. Applied Sciences. 2025; 15(24):13153. https://doi.org/10.3390/app152413153
Chicago/Turabian StylePeng, Wei, Guoqing Hu, Ji Li, and Chengzhi Lyu. 2025. "A Query-Based Progressive Aggregation Network for 3D Medical Image Segmentation" Applied Sciences 15, no. 24: 13153. https://doi.org/10.3390/app152413153
APA StylePeng, W., Hu, G., Li, J., & Lyu, C. (2025). A Query-Based Progressive Aggregation Network for 3D Medical Image Segmentation. Applied Sciences, 15(24), 13153. https://doi.org/10.3390/app152413153





