Qualitative and Quantitative Mass Spectrometry Approaches for the Analysis of Phenolic Compounds in Complex Natural Matrices
Featured Application
Abstract
1. Introduction
2. Phenolic Compounds
2.1. Phenolic Acids
2.1.1. Structural Classification
2.1.2. Phenolic Acids Fragmentation
2.2. Flavonoids
2.2.1. Structural Classification
2.2.2. Flavonoid Fragmentation
2.2.3. Flavonoid Glycosides Fragmentation
3. MS Analysis
3.1. Targeted Analysis
3.1.1. MRM and SRM
3.1.2. Precursor and Product Ion Scan
3.1.3. Neutral Loss
3.1.4. Absolute vs. Relative Concentration
3.2. Untargeted Analysis
3.2.1. Acquisition Modes and Reproducibility
Implications for Phenolic Chemistries
3.2.2. Direct Injection (DI) MS
3.2.3. Untargeted LC-MS/MS
| Matrix | Analyzed Compounds | Identification | Purpose | Ref. |
|---|---|---|---|---|
| LC-QTOF | ||||
| Plukenetia volubilis leaves | 16 compounds (Kaempferol trihexoside, Kaempferol-3-O- glucoside (astragalin), etc.) | Accurate mass measurements, fragmentation patterns, UV spectra, and comparison with reference standards and literature data | To characterize the phytochemical composition of Plukenetia volubilis leaves extract and evaluate its anti-Helicobacter pylori activity through in vitro assays and in silico docking, with focus on flavonoids such as astragalin. | [70] |
| Schima argentea | 30 compounds (Quercetin 3-arabinoside, Nodularin, Ricinoleic acid methyl ester, etc.) | Compound identification was performed using Progenesis QI’s online METLIN database and a custom library from Biomarker Technologies Co., Ltd., with theoretical fragment ions considered. The mass error was controlled within 100 ppm | To investigate the antioxidant and photoprotective effects of 3,4-dihydroxybenzoic acid and (+)-catechin, identified from Schima argentea extract, in UVB-irradiated HaCaT keratinocytes. | [72] |
| Stem, Roots, and Leaves of Syzygium cumini | 12 compounds (phenolic compounds and lignanas, for example: 3,4-O-Dimethylgallic acid, Scutellarein, etc.) | Compounds assigned by retention time (RT), isotopic pattern, and database/software matching (Agilent MassHunter) | To profile and compare phenolic constituents in stem, root, and leaf of Syzygium cumini and assess their antioxidant capacity across multiple assays, linking phenolic content to activity. | [76] |
| Symphorema polyandrum | 11 compounds (Aciculatinone (O-Methylated flavonoids, 2″-p-Coumaroylvitexin (Flavonoid glycosides), etc.) | Scientific literature and mass-spectra databases such as the METLIN database, Chemspider, Pub-chem, NIST MS–MS database, and npatlas database | Characterization and in vitro assessment of its antioxidant, anticancer, and anti-inflammatory potential | [77] |
| Litsea monopetala bark | 9 compounds (fraxetin, Kaempferol-3-O-alpha-L-rhamnoside, Kaempferol-3- neohesperidoside, etc.) | MassBank Europe Mass Spectral Database, the Human Metabolome Database, and relevant literature sources | Metabolic profiling and demonstration of hepatoprotective potential, supporting traditional use against jaundice and liver disorders | [78] |
| Gliricidia sepium leaves | 22 bioactive compounds (flavonoids, phenolic acids, triterpenoid saponins, fatty acid derivatives, and coumarins) | Molecular formula prediction and peak identification via dereplication using ChemCalc online, Dictionary of Natural Products (DNP) Database, Global Natural Product Social Molecular Networking (GNPS) Database, Human Metabolome Database (HMDB), MassBank of North America (MoNA) Database, MassBank Europe | Metabolomic profiling with in vivo renoprotective assessment in diabetic hamsters, supporting use against diabetic nephropathy. | [79] |
| Achillea ligustica | 45 phenolic compounds(caffeoylquinic and dicaffeoylquinic acid isomers, dihydroxybenzoic acid derivatives, coumarins, flavones, flavonols, lignin) | Accurate mass, MS/MS fragments and standards | Phytochemical profiling with evaluation of antioxidant activity (DPPH, ABTS, phenanthroline, reducing power), antimicrobial effects, acute toxicity, and analgesic activity, supporting traditional uses. | [71] |
| Strobilanthes sarcorrhiza root | 55 compounds (terpenoids, phenylethanol glycosides, fatty acid derivatives, chain/other glycosides, flavonoid glycosides, sterols, alkaloids, nucleosides, esters, alkylene oxides, organic acids; 34 first reported in plant | Accurate mass, MS/MS fragments, RT; differences assessed by chemometrics | Chemical profiling and part-differentiation showing phenylethanol glycosides enriched in underground parts and terpenoids in aboveground parts, supporting rhizomes as an alternative medicinal part to improve resource utilization | [80] |
| Ziziphus budhensis Leaves | 46 compounds (phenolic compounds, benzyl-isoquinolinic alkaloids, cyclopeptide alkaloids, triterpene aglycone and saponins) | Confirmation by diagnostic MS fragments and reference standards | Phytochemical profiling with evaluation of antioxidant, antibacterial, antifungal, cytotoxicity, and acute oral toxicity in mice to assess medicinal potential. | [81] |
| Lemon, lime, orange, and grapefruit juices | 57 compounds ranging from polar phenolic acids over flavonoid glycosides to a polar coumarins, psoralens, and polymethoxyflavones in lemon, lime, orange, and grapefruit juices | Assignments confirmed using UV spectra, accurate mass, diagnostic MS fragments and authentic standards | Phytochemical profiling and quantitation of phenolic compounds/PMFs/coumarins/psoralens for authentication of Citrus juices and identification of species-specific chemical markers | [82] |
| Fagonia arabica | 42 phenolic compounds (3 phenolic acids (cinnamic acid derivatives), 15 flavonols, 1 flavanol, 4 flavanones, 8 flavones, 2 isoflavones, 1 chalcone, 1 aurone O-glycosides, 1 stilbene and 6 anthocyanins) | ReSpect databases | Profiling and evaluation of cholinesterase inhibition potential through in vitro and in silico approaches | [83] |
| LC-Orbitrap | ||||
| Shenhua Tablets | 183 compounds (64 flavonoids, 52 terpenoids, 37 organic acids, 6 phenylpropanoids, 5 phenols, and 19 other phytochemicals) | mzCloud and mzVault library | Characterization | [73] |
| Lagopsis supina | 114 compounds | opensource software, including GNPS web tools and MS-Dial, alongside public mass spectrometry databases (GNPS, HMDB, LipidMaps, KNApSAcK, and the American Mass Bank) | Chemical Composition and Antioxidant, Adipogenic, and Ani-Inflammatory activities | [84] |
| Rubi fructus | 47 components (10 organic acids, 15 flavonoids, 12 phenols, 2 alkaloids, 4 terpenoids, 1 miscellaneous compound, 1 stilbene, 1 steroid and its derivatives, and 1 diterpenoid) | databases and relevant literature | Phytochemical profiling and evaluation of anti-diabetic mechanism through network pharmacology and experimental validation | [74] |
| Ribes nigrum leaf | 24 compounds | Compound Discoverer (v. 2.1, Thermo, Waltham, MA, USA): accurate mass and mass fragmentation pattern spectra against customized database of different classes of phytochemicals created on the basis of literature data | Characterization and antioxidant and anti-inflammatory capabilities, concentrating on the influence of oxidative stress, gene expression, and enzymatic activity in microglial cells. | [75] |
| Thymbra spicata L. | 31 compounds | calibration curves, RT and MS/MS fragmentation patterns (Quan Peak and confirming ions) | Phytochemical profiling and anti-inflammatory mechanism | [85] |
Identification
3.2.4. Statistics of Untargeted MS Data
4. Extraction of Phenols from Complex Natural Matrices
4.1. Solvent
Acidification
4.2. Ultrasound (UAE) and Microwave Assisted Extractions (MAE)
4.3. Solid Phase Extraction (SPE)
4.4. Data-Driven Optimization of Phenolic Extraction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pereira, D.M.; Valentão, P.; Pereira, J.A.; Andrade, P.B. Phenolics: From Chemistry to Biology. Molecules 2009, 14, 2202–2211. [Google Scholar] [CrossRef]
- Sun, W.; Shahrajabian, M.H. Therapeutic Potential of Phenolic Compounds in Medicinal Plants-Natural Health Products for Human Health. Molecules 2023, 28, 1845. [Google Scholar] [CrossRef]
- Rahman, M.M.; Rahaman, M.S.; Islam, M.R.; Rahman, F.; Mithi, F.M.; Alqahtani, T.; Almikhlafi, M.A.; Alghamdi, S.Q.; Alruwaili, A.S.; Hossain, M.S.; et al. Role of Phenolic Compounds in Human Disease: Current Knowledge and Future Prospects. Molecules 2021, 27, 233. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional Control of Flavonoid Biosynthesis by MYB-bHLH-WDR Complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cai, P.; Cheng, G.; Zhang, Y. A Brief Review of Phenolic Compounds Identified from Plants: Their Extraction, Analysis, and Biological Activity. Nat. Prod. Commun. 2022, 17, 1934578X211069721. [Google Scholar] [CrossRef]
- López-Yerena, A.; Domínguez-López, I.; Vallverdú-Queralt, A.; Pérez, M.; Jáuregui, O.; Escribano-Ferrer, E.; Lamuela-Raventós, R.M. Metabolomics Technologies for the Identification and Quantification of Dietary Phenolic Compound Metabolites: An Overview. Antioxidants 2021, 10, 846. [Google Scholar] [CrossRef]
- Kumar, N.; Goel, N. Phenolic Acids: Natural Versatile Molecules with Promising Therapeutic Applications. Biotechnol. Rep. 2019, 24, e00370. [Google Scholar] [CrossRef]
- Awwad, S.; Issa, R.; Alnsour, L.; Albals, D.; Al-Momani, I. Quantification of Caffeine and Chlorogenic Acid in Green and Roasted Coffee Samples Using HPLC-DAD and Evaluation of the Effect of Degree of Roasting on Their Levels. Molecules 2021, 26, 7502. [Google Scholar] [CrossRef]
- Guan, H.; Luo, W.; Bao, B.; Cao, Y.; Cheng, F.; Yu, S.; Fan, Q.; Zhang, L.; Wu, Q.; Shan, M. A Comprehensive Review of Rosmarinic Acid: From Phytochemistry to Pharmacology and Its New Insight. Molecules 2022, 27, 3292. [Google Scholar] [CrossRef]
- González-Hernández, A.I.; Scalschi, L.; Vicedo, B.; Marcos-Barbero, E.L.; Morcuende, R.; Camañes, G. Putrescine: A Key Metabolite Involved in Plant Development, Tolerance and Resistance Responses to Stress. Int. J. Mol. Sci. 2022, 23, 2971. [Google Scholar] [CrossRef]
- Neilson, A.P.; Ferruzzi, M.G. Chapter 22—Bioavailability and Metabolism of Bioactive Compounds from Foods. In Nutrition in the Prevention and Treatment of Disease, 3rd ed.; Coulston, A.M., Boushey, C.J., Ferruzzi, M.G., Eds.; Academic Press: New York, NY, USA, 2013; pp. 407–423. ISBN 978-0-12-391884-0. [Google Scholar]
- Chandrasekara, A. Phenolic Acids. In Encyclopedia of Food Chemistry; Melton, L., Shahidi, F., Varelis, P., Eds.; Academic Press: Oxford, UK, 2019; pp. 535–545. ISBN 978-0-12-814045-1. [Google Scholar]
- Siquet, C.; Paiva-Martins, F.; Lima, J.L.F.C.; Reis, S.; Borges, F. Antioxidant Profile of Dihydroxy- and Trihydroxyphenolic Acids--a Structure-Activity Relationship Study. Free Radic. Res. 2006, 40, 433–442. [Google Scholar] [CrossRef]
- Sinosaki, N.B.M.; Tonin, A.P.P.; Ribeiro, M.A.S.; Poliseli, C.B.; Roberto, S.B.; Silveira, R.d.; Visentainer, J.V.; Santos, O.O.; Meurer, E.C. Structural Study of Phenolic Acids by Triple Quadrupole Mass Spectrometry with Electrospray Ionization in Negative Mode and H/D Isotopic Exchange. J. Braz. Chem. Soc. 2020, 31, 402–408. [Google Scholar] [CrossRef]
- Ostrowski, W.; Wojakowska, A.; Grajzer, M.; Stobiecki, M. Mass Spectrometric Behavior of Phenolic Acids Standards and Their Analysis in the Plant Samples with LC/ESI/MS System. J. Chromatogr. B 2014, 967, 21–27. [Google Scholar] [CrossRef]
- Chemicalize—Instant Cheminformatics Solutions. Available online: https://chemicalize.com (accessed on 24 October 2025).
- Zheng, X.; Zhang, X.; Zeng, F. Biological Functions and Health Benefits of Flavonoids in Fruits and Vegetables: A Contemporary Review. Foods 2025, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- Khlestkina, E. The Adaptive Role of Flavonoids: Emphasis on Cereals. Cereal Res. Commun. 2013, 41, 185–198. [Google Scholar] [CrossRef]
- He, H.-F.; Wei, K.; Yin, J.; Ye, Y. Insight into Tea Flavonoids: Composition and Chemistry. Food Rev. Int. 2021, 37, 812–823. [Google Scholar] [CrossRef]
- Fernandes, I.; Pérez-Gregorio, R.; Soares, S.; Mateus, N.; De Freitas, V. Wine Flavonoids in Health and Disease Prevention. Molecules 2017, 22, 292. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, X.; Qiu, H. Flavones and Flavonols: Phytochemistry and Biochemistry. In Natural Products; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1821–1847. ISBN 978-3-642-22144-6. [Google Scholar]
- Jomova, K.; Alomar, S.Y.; Valko, R.; Liska, J.; Nepovimova, E.; Kuca, K.; Valko, M. Flavonoids and Their Role in Oxidative Stress, Inflammation, and Human Diseases. Chem. Biol. Interact. 2025, 413, 111489. [Google Scholar] [CrossRef]
- Yang, B.; Liu, H.; Yang, J.; Gupta, V.K.; Jiang, Y. New Insights on Bioactivities and Biosynthesis of Flavonoid Glycosides. Trends Food Sci. Technol. 2018, 79, 116–124. [Google Scholar] [CrossRef]
- Gao, Y.; Xia, W.; Shao, P.; Wu, W.; Chen, H.; Fang, X.; Mu, H.; Xiao, J.; Gao, H. Impact of Thermal Processing on Dietary Flavonoids. Curr. Opin. Food Sci. 2022, 48, 100915. [Google Scholar] [CrossRef]
- Saftić, L.; Peršurić, Ž.; Fornal, E.; Pavlešić, T.; Kraljević Pavelić, S. Targeted and Untargeted LC-MS Polyphenolic Profiling and Chemometric Analysis of Propolis from Different Regions of Croatia. J. Pharm. Biomed. Anal. 2019, 165, 162–172. [Google Scholar] [CrossRef]
- Pal, S.; Saha, C. A Review on Structure-Affinity Relationship of Dietary Flavonoids with Serum Albumins. J. Biomol. Struct. Dyn. 2014, 32, 1132–1147. [Google Scholar] [CrossRef]
- Singh, P.; Arif, Y.; Bajguz, A.; Hayat, S. The Role of Quercetin in Plants. Plant Physiol. Biochem. 2021, 166, 10–19. [Google Scholar] [CrossRef]
- Semwal, D.K.; Semwal, R.B.; Combrinck, S.; Viljoen, A. Myricetin: A Dietary Molecule with Diverse Biological Activities. Nutrients 2016, 8, 90. [Google Scholar] [CrossRef]
- Rice-Evans, C. Flavonoids and Isoflavones: Absorption, Metabolism, and Bioactivity. Free Radic. Biol. Med. 2004, 36, 827–828. [Google Scholar] [CrossRef]
- Heim, K.E.; Tagliaferro, A.R.; Bobilya, D.J. Flavonoid Antioxidants: Chemistry, Metabolism and Structure-Activity Relationships. J. Nutr. Biochem. 2002, 13, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Kurek-Górecka, A.; Rzepecka-Stojko, A.; Górecki, M.; Stojko, J.; Sosada, M.; Świerczek-Zięba, G. Structure and Antioxidant Activity of Polyphenols Derived from Propolis. Molecules 2013, 19, 78–101. [Google Scholar] [CrossRef] [PubMed]
- Vukics, V.; Guttman, A. Structural Characterization of Flavonoid Glycosides by Multi-Stage Mass Spectrometry. Mass Spectrom. Rev. 2010, 29, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mabry, T.J.; Markham, K.R.; Thomas, M.B. The Aglycone and Sugar Analysis of Flavonoid Glycosides. In The Systematic Identification of Flavonoids; Mabry, T.J., Markham, K.R., Thomas, M.B., Eds.; Springer: Berlin/Heidelberg, Germany, 1970; pp. 23–32. ISBN 978-3-642-88458-0. [Google Scholar]
- Domon, B.; Costello, C.E. A Systematic Nomenclature for Carbohydrate Fragmentations in FAB-MS/MS Spectra of Glycoconjugates. Glycoconj. J. 1988, 5, 397–409. [Google Scholar] [CrossRef]
- Ferreres, F.; Gil-Izquierdo, A.; Andrade, P.B.; Valentão, P.; Tomás-Barberán, F.A. Characterization of C-Glycosyl Flavones O-Glycosylated by Liquid Chromatography–Tandem Mass Spectrometry. J. Chromatogr. A 2007, 1161, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Saftić, L.; Peršurić, Ž.; Kraljević Pavelić, S. LC–QQQ and LC–QTOF MS Methods for Comprehensive Detection of Potential Allergens in Various Propolis Extracts. Eur. Food Res. Technol. 2019, 245, 1981–1995. [Google Scholar] [CrossRef]
- Rodríguez, M.S.; Martínez León, A.J.; Porras, L.S.; Giraldo, I.A.; Rojas, E.; Lavao, F.E.; Martínez, K. Flavonoid and Phenolic Quantification from Açaí (Euterpe Oleracea Mart and Euterpe Precatoria Mart), Mirití (Mauritia flexuosa L.), and Cupuassu (Theobroma Grandiflorum (Wild. Ex Spreng.) Schum) from Vaupés, Colombia, Using LC-QqQ-MS. Plants 2025, 14, 2632. [Google Scholar] [CrossRef]
- Myo, H.; Nantarat, N.; Khat-Udomkiri, N. Changes in Bioactive Compounds of Coffee Pulp through Fermentation-Based Biotransformation Using Lactobacillus Plantarum TISTR 543 and Its Antioxidant Activities. Fermentation 2021, 7, 292. [Google Scholar] [CrossRef]
- Zhang, S.; Long, X.; Xing, Y.; Ee, K.-H.; Goh, R.M.V.; Huang, Y.; Pua, A.; Jublot, L.; Liu, S.Q.; Yu, B. A Systematic Approach to Analyzing Catechins and Catechin Derivatives in Ceylon Black Tea Using Liquid Chromatography Coupled with Triple Quadrupole Mass Spectrometry. Food Chem. X 2025, 28, 102621. [Google Scholar] [CrossRef]
- Lambert, M.; Meudec, E.; Verbaere, A.; Mazerolles, G.; Wirth, J.; Masson, G.; Cheynier, V.; Sommerer, N. A High-Throughput UHPLC-QqQ-MS Method for Polyphenol Profiling in Rosé Wines. Molecules 2015, 20, 7890–7914. [Google Scholar] [CrossRef]
- Barrales-Cureño, H.J.; Salgado-Garciglia, R.; López-Valdez, L.G.; Monribot-Villanueva, J.L.; Guerrero-Analco, J.A.; Lucho-Constantino, G.G.; Zaragoza-Martínez, F.; Herrera-Cabrera, B.E.; Reyes, C. Metabolomic Data of Phenolic Compounds from Acer Negundo Extracts. Data Brief 2020, 30, 105569. [Google Scholar] [CrossRef] [PubMed]
- Abouelenein, D.; Mustafa, A.M.; Nzekoue, F.K.; Caprioli, G.; Angeloni, S.; Tappi, S.; Castagnini, J.M.; Dalla Rosa, M.; Vittori, S. The Impact of Plasma Activated Water Treatment on the Phenolic Profile, Vitamins Content, Antioxidant and Enzymatic Activities of Rocket-Salad Leaves. Antioxidants 2023, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Irakli, M.; Skendi, A.; Bouloumpasi, E.; Chatzopoulou, P.; Biliaderis, C.G. LC-MS Identification and Quantification of Phenolic Compounds in Solid Residues from the Essential Oil Industry. Antioxidants 2021, 10, 2016. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yang, J.; Tong, X.; Zhao, C.; He, Y.; Wan, H. Precursor Ion Scan Enhanced Rapid Identification of the Chemical Constituents of Danhong Injection by Liquid Chromatography–Tandem Mass Spectrometry: An Integrated Strategy. J. Chromatogr. A 2019, 1602, 378–385. [Google Scholar] [CrossRef]
- Goh, R.M.V.; Ee, K.H.; Pua, A.; Huang, Y.; Liu, S.Q.; Lassabliere, B.; Yu, B. Neutral Loss Scan in Complement with High-Resolution MS/MS: Combination of Detection Methods for Flavonoid and Limonoid Glycosides Analysis. J. Mass Spectrom. 2022, 57, e4810. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, F.; Cao, Y.; Tong, C.; Wei, Q.; Shi, S.; Guo, Y. Rapid Profiling of Potential Antitumor Polymethoxylated Flavonoids in Natural Products by Integrating Cell Biospecific Extraction with Neutral Loss/Diagnostic Ion Filtering-Based High-Performance Liquid Chromatography-Quadrupole Time-of-Flight Tandem Mass Spectrometry. Phytochem. Anal. 2022, 33, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Tian, Y.; Xiong, M.; Yi, Y.; Qiao, X.; Yang, Y.; Ye, M. Characterization of Prenylated Phenolics in Glycyrrhiza Uralensis by Offline Two-Dimensional Liquid Chromatography/Mass Spectrometry Coupled with Mass Defect Filter. J. Pharm. Biomed. Anal. 2022, 220, 115009. [Google Scholar] [CrossRef]
- Peršurić, Ž.; Saftić, L. Phenolic Composition of Extra Virgin Olive Oil Samples from Istria (Croatia). Riv. Ital. Delle Sostanze Grasse 2019, 96, 231–239. [Google Scholar]
- Pavlešić, T.; Poljak, S.; Mišetić Ostojić, D.; Lučin, I.; Reynolds, C.A.; Kalafatovic, D.; Saftić Martinović, L. Mint (Mentha spp.) Honey: Analysis of the Phenolic Profile and Antioxidant Activity. Food Technol. Biotechnol. 2022, 60, 509–519. [Google Scholar] [CrossRef]
- Saftić Martinović, L.; Birkic, N.; Pavlešić, T.; Planinić, A.; Gobin, I.; Mišetić Ostojić, D.; Pedisić, S. Chemical Characterization of Rare Unifloral Honeys of Ailanthus (Ailanthus Altissima), Fennel (Foenicum Vulgare), and Raspberry (Rubus Idaeus) and Their Antimicrobial and Antioxidant Activity. Agric. Res. 2025, 14, 130–142. [Google Scholar] [CrossRef]
- Glauser, G.; Veyrat, N.; Rochat, B.; Wolfender, J.-L.; Turlings, T.C.J. Ultra-High Pressure Liquid Chromatography–Mass Spectrometry for Plant Metabolomics: A Systematic Comparison of High-Resolution Quadrupole-Time-of-Flight and Single Stage Orbitrap Mass Spectrometers. J. Chromatogr. A 2013, 1292, 151–159. [Google Scholar] [CrossRef] [PubMed]
- La Barbera, G.; Capriotti, A.L.; Cavaliere, C.; Montone, C.M.; Piovesana, S.; Samperi, R.; Zenezini Chiozzi, R.; Laganà, A. Liquid Chromatography-High Resolution Mass Spectrometry for the Analysis of Phytochemicals in Vegetal-Derived Food and Beverages. Food Res. Int. 2017, 100, 28–52. [Google Scholar] [CrossRef]
- Tang, H.; Wang, X.; Xu, L.; Ran, X.; Li, X.; Chen, L.; Zhao, X.; Deng, H.; Liu, X. Establishment of Local Searching Methods for Orbitrap-Based High Throughput Metabolomics Analysis. Talanta 2016, 156–157, 163–171. [Google Scholar] [CrossRef]
- El Boudlali, H.; Lehmicke, L.; Ceglarek, U. High-Resolution Accurate Mass- Mass Spectrometry Based- Untargeted Metabolomics: Reproducibility and Detection Power across Data- Dependent Acquisition, Data-Independent Acquisition, and AcquireX. Comput. Struct. Biotechnol. J. 2025, 27, 2412–2423. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Huan, T. Comparison of Full-Scan, Data-Dependent, and Data-Independent Acquisition Modes in Liquid Chromatography-Mass Spectrometry Based Untargeted Metabolomics. Anal. Chem. 2020, 92, 8072–8080. [Google Scholar] [CrossRef]
- Cooper, B.; Yang, R. An Assessment of AcquireX and Compound Discoverer Software 3.3 for Non-Targeted Metabolomics. Sci. Rep. 2024, 14, 4841. [Google Scholar] [CrossRef] [PubMed]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and Considerations for the Use of System Suitability and Quality Control Samples in Mass Spectrometry Assays Applied in Untargeted Clinical Metabolomic Studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for Large-Scale Metabolic Profiling of Serum and Plasma Using Gas Chromatography and Liquid Chromatography Coupled to Mass Spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Märtens, A.; Holle, J.; Mollenhauer, B.; Wegner, A.; Kirwan, J.; Hiller, K. Instrumental Drift in Untargeted Metabolomics: Optimizing Data Quality with Intrastudy QC Samples. Metabolites 2023, 13, 665. [Google Scholar] [CrossRef]
- Wang, R.-Q.; Ding, J.; Geng, Y.; Li, Y.-Z.; Mei, Y.-W.; Bao, K.; Yu, H.-D.; Feng, Y.-Q. CRB-SWATH: A Method for Enhancing Untargeted Precursor Ion Extraction and Automatically Constructing Their Tandem Mass Spectra from SWATH Datasets by Chromatographic Retention Behaviors. Anal. Chem. 2021, 93, 12273–12280. [Google Scholar] [CrossRef] [PubMed]
- Aigensberger, M.; Bueschl, C.; Castillo-Lopez, E.; Ricci, S.; Rivera-Chacon, R.; Zebeli, Q.; Berthiller, F.; Schwartz-Zimmermann, H.E. Modular Comparison of Untargeted Metabolomics Processing Steps. Anal. Chim. Acta 2025, 1336, 343491. [Google Scholar] [CrossRef]
- Wartmann, Y.; Boxler, M.I.; Kraemer, T.; Steuer, A.E. Impact of Three Different Peak Picking Software Tools on the Quality of Untargeted Metabolomics Data. J. Pharm. Biomed. Anal. 2024, 248, 116302. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Alseekh, S.; Scossa, F.; Fernie, A.R. Ultra-High-Performance Liquid Chromatography High-Resolution Mass Spectrometry Variants for Metabolomics Research. Nat. Methods 2021, 18, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.-L.; Nuzillard, J.-M.; van der Hooft, J.J.J.; Renault, J.-H.; Bertrand, S. Accelerating Metabolite Identification in Natural Product Research: Toward an Ideal Combination of Liquid Chromatography-High-Resolution Tandem Mass Spectrometry and NMR Profiling, in Silico Databases, and Chemometrics. Anal. Chem. 2019, 91, 704–742. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Peršurić, Ž.; Saftić Martinović, L.; Malenica, M.; Gobin, I.; Pedisić, S.; Dragović-Uzelac, V.; Kraljević Pavelić, S. Assessment of the Biological Activity and Phenolic Composition of Ethanol Extracts of Pomegranate (Punica granatum L.) Peels. Molecules 2020, 25, 5916. [Google Scholar] [CrossRef]
- Pachulicz, R.J.; Yu, L.; Jovcevski, B.; Bulone, V.; Pukala, T.L. Structural Analysis and Identity Confirmation of Anthocyanins in Brassica Oleracea Extracts by Direct Injection Ion Mobility-Mass Spectrometry. ACS Meas. Sci. Au 2023, 3, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Masike, K.; de Villiers, A.; de Beer, D.; Joubert, E.; Stander, M.A. Application of Direct Injection-Ion Mobility Spectrometry-Mass Spectrometry (DI-IMS-MS) for the Analysis of Phenolics in Honeybush and Rooibos Tea Samples. J. Food Compos. Anal. 2022, 106, 104308. [Google Scholar] [CrossRef]
- Tan, A.; Scortecci, K.C.; Cabral De Medeiros, N.M.; Kukula-Koch, W.; Butler, T.J.; Smith, S.M.; Boylan, F. Plukenetia Volubilis Leaves as Source of Anti-Helicobacter Pylori Agents. Front. Pharmacol. 2024, 15, 1461447. [Google Scholar] [CrossRef]
- Boubertakh, H.; Kabouche, Z.; Boudechicha, A.; Madi, A.; Khalfallah, A.; Kabouche, A. RP-UHPLC-ESI-QTOF-MSn Analyses, Antioxidant, Antimicrobial, Analgesic Activities and Toxicity of Achillea Ligustica All. Nat. Prod. Res. 2025, 39, 3316–3321. [Google Scholar] [CrossRef]
- He, Q.; Chen, Y.-P.; Li, J.; Wu, H.; Chen, F.; Li, M.; Wu, C. Antioxidant and Photoprotective Activities of 3,4-Dihydroxybenzoic Acid and (+)-Catechin, Identified from Schima Argentea Extract, in UVB-Irradiated HaCaT Cells. Antioxidants 2025, 14, 241. [Google Scholar] [CrossRef]
- Wang, M.; Li, P.; Yin, F.; Liu, H.; Sun, H.; Zheng, Y.; Liu, C.; Wang, Y.; Chen, X.; Guan, S.; et al. Rapid Characterization of the Constituents in Shenhua Tablets by UHPLC-Q-Orbitrap-LTQ-MS. J. Sep. Sci. 2024, 47, e70030. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Liao, Q.; Rui, X.; Wang, R. Study on the Mechanism of Raspberry (Rubi Fructus) in Treating Type 2 Diabetes Based on UPLC-Q-Exactive Orbitrap MS, Network Pharmacology, and Experimental Validation. Phytochem. Anal. 2025, 36, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Minasyan, A.; Pires, V.; Gondcaille, C.; Ginovyan, M.; Mróz, M.; Savary, S.; Cherkaoui-Malki, M.; Kusznierewicz, B.; Bartoszek, A.; Andreoletti, P.; et al. Ribes Nigrum Leaf Extract Downregulates Pro-Inflammatory Gene Expression and Regulates Redox Balance in Microglial Cells. BMC Complement Med. Ther. 2025, 25, 49. [Google Scholar] [CrossRef]
- Imran, A.; Ye, S.; Li, J.A.; Ajaj, R.; Rauf, A.; Ahmad, Z.; Hemeg, H.A.; Al-Awthan, Y.S.M.; Bahattab, O.S.; Quradha, M.M.; et al. LC-ESI-QTOF-MS/MS Characterization of Phenolic Compounds in the Stem, Roots, and Leaves of Syzygium Cumini and Their Antioxidant Potential. Food Sci. Nutr. 2025, 13, e70112. [Google Scholar] [CrossRef]
- Sahoo, D.R.; Babu, S.K.; Naik, B.B.; Hota, S.S.; Bhoi, N.; Sarkar, B.; Ali, S.K.M.; Naik, P.K. UPLC-QToF-MS/MS Screening and Characterization of Symphorema Polyandrum Wight and in Vitro Assessment of Its Antioxidant, Anticancer, and Anti-Inflammatory Potential. 3 Biotech 2024, 14, 298. [Google Scholar] [CrossRef]
- Sahoo, S.; Khan, H.A.; Kar, D.M.; Pattanaik, S.; Rath, D. LC-QTOF-MS/MS Metabolic Profiling and Hepatoprotective Effects of Litsea Monopetala Bark Methanol Extract against Liver Injury in Rats and HepG2 Cells. Cell Mol. Biol. 2024, 70, 156–169. [Google Scholar] [CrossRef]
- Wafaey, A.A.; El-Hawary, S.S.; El Raey, M.A.; Abdelrahman, S.S.; Ali, A.M.; Mandour, Y.M.; Fayek, F.H.; Mohamed, S.S.; El-Rashedy, A.A.; Mohamed, O.G.; et al. UHPLC-QTOF-MS/MS Metabolomic Profiling and in Silico-in vIvo Evaluation of Gliricidia Sepium Leaves for Renoprotection in Diabetic Nephropathy. Fitoterapia 2025, 186, 106875. [Google Scholar] [CrossRef]
- Chen, R.; Fan, J.; Wu, Y.; Huang, X.; Zhang, W.; Xu, Y.; Zhang, Y.; Li, L.; Wang, C.; Yu, M.; et al. Strobilanthes Sarcorrhiza Root Phenolic Extract Prevent Diabetic Nephropathy in Mice by Regulating NF-κB/IL-1β Signaling and Glycerophospholipid Metabolism. J. Pharm. Biomed. Anal. 2025, 253, 116534. [Google Scholar] [CrossRef]
- Bharati, S.; Maharjan, B.; Shrestha, T.; Shrestha, S.S.; Sut, S.; Devkota, H.P.; Shrestha, R.L.S.; Dall’Acqua, S. LC-DAD-MSn and HR-LC-QTOF Analysis of Ziziphus Budhensis Leaves and Evaluation of Their In Vitro and In Vivo Biological Activities. Chem. Biodivers. 2025, 22, e202402835. [Google Scholar] [CrossRef]
- Gilcher, C.; Jungen, M.; Schweiggert, R.; Steingass, C.B. Comparative HPLC-DAD-ESI-QTOF-HR-MS/MS Analyses of Phenolic Compounds of Authentic Lemon, Lime, Orange, and Grapefruit Juices. Food Chem. 2025, 485, 144416. [Google Scholar] [CrossRef]
- Badawy, S.A.; Hassan, A.R.; Abu Bakr, M.S.; Mohammed, A.E.-S.I. UPLC-qTOF-MS/MS Profiling of Phenolic Compounds in Fagonia Arabica L. and Evaluation of Their Cholinesterase Inhibition Potential through in-Vitro and in-Silico Approaches. Sci. Rep. 2025, 15, 5244. [Google Scholar] [CrossRef]
- Choi, J.; Le, D.D.; Roh, N.; Lee, J.; Shrestha, D.; Dinh, T.; Truong, V.; Bazarragchaa, B.; Kim, S.-Y.; Suh, S.-S.; et al. Chemical Composition and Biological Activities of Lagopsis Supina Extract: Antioxidant, Adipogenic, and Ani-Inflammatory Effects. Pharmaceuticals 2025, 18, 150. [Google Scholar] [CrossRef]
- Unsal, V.; Ercan, L.; Calıskan, C.G. Determination of Bioactive and Anti-Inflammatory Molecules of Thymbra Spicata L. from Mardin by GC-MS and LC-Orbitrap HRMS: A DFT, Molecular Docking, ADMET, Biological Target and Activity Study. BMC Complement Med. Ther. 2025, 25, 358. [Google Scholar] [CrossRef]
- Reisdorph, N.A.; Walmsley, S.; Reisdorph, R. A Perspective and Framework for Developing Sample Type Specific Databases for LC/MS-Based Clinical Metabolomics. Metabolites 2019, 10, 8. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Rotation and Interpretation of Principal Components. In Principal Component Analysis; Jolliffe, I.T., Ed.; Springer: New York, NY, USA, 2002; pp. 269–298. ISBN 978-0-387-22440-4. [Google Scholar]
- Jolliffe, I.T. Graphical Representation of Data Using Principal Components. In Principal Component Analysis; Jolliffe, I.T., Ed.; Springer: New York, NY, USA, 2002; pp. 78–110. ISBN 978-0-387-22440-4. [Google Scholar]
- Kumar, K. Partial Least Square (PLS) Analysis. Resonance 2021, 26, 429–442. [Google Scholar] [CrossRef]
- Pomerantsev, A.L.; Rodionova, O.Y. Multiclass Partial Least Squares Discriminant Analysis: Taking the Right Way—A Critical Tutorial. J. Chemom. 2018, 32, e3030. [Google Scholar] [CrossRef]
- Sulaiman, S.F.; Sajak, A.A.B.; Ooi, K.L.; Supriatno; Seow, E.M. Effect of Solvents in Extracting Polyphenols and Antioxidants of Selected Raw Vegetables. J. Food Compos. Anal. 2011, 24, 506–515. [Google Scholar] [CrossRef]
- Boeing, J.S.; Barizão, E.O.; E Silva, B.C.; Montanher, P.F.; de Cinque Almeida, V.; Visentainer, J.V. Evaluation of Solvent Effect on the Extraction of Phenolic Compounds and Antioxidant Capacities from the Berries: Application of Principal Component Analysis. Chem. Cent. J. 2014, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Meneses, N.G.T.; Martins, S.; Teixeira, J.A.; Mussatto, S.I. Influence of Extraction Solvents on the Recovery of Antioxidant Phenolic Compounds from Brewer’s Spent Grains. Sep. Purif. Technol. 2013, 108, 152–158. [Google Scholar] [CrossRef]
- Alara, O.R.; Abdurahman, N.H.; Ukaegbu, C.I. Extraction of Phenolic Compounds: A Review. Curr. Res. Food Sci. 2021, 4, 200–214. [Google Scholar] [CrossRef]
- Barros, H.D.F.Q.; Baseggio, A.M.; Angolini, C.F.F.; Pastore, G.M.; Cazarin, C.B.B.; Marostica-Junior, M.R. Influence of Different Types of Acids and pH in the Recovery of Bioactive Compounds in Jabuticaba Peel (Plinia Cauliflora). Food Res. Int. 2019, 124, 16–26. [Google Scholar] [CrossRef]
- Nuutila, A.M.; Kammiovirta, K.; Oksman-Caldentey, K.-M. Comparison of Methods for the Hydrolysis of Flavonoids and Phenolic Acids from Onion and Spinach for HPLC Analysis. Food Chem. 2002, 76, 519–525. [Google Scholar] [CrossRef]
- Chemat, F.; Rombaut, N.; Sicaire, A.-G.; Meullemiestre, A.; Fabiano-Tixier, A.-S.; Abert-Vian, M. Ultrasound Assisted Extraction of Food and Natural Products. Mechanisms, Techniques, Combinations, Protocols and Applications. A Review. Ultrason. sonochem. 2017, 34, 540–560. [Google Scholar] [CrossRef]
- Malenica, M.; Biesaga, M.; Pedisić, S.; Martinović, L.S. Stability of Propolis Phenolics during Ultrasound-Assisted Extraction Procedures. Foods 2024, 13, 2020. [Google Scholar] [CrossRef]
- Pellati, F.; Prencipe, F.P.; Bertelli, D.; Benvenuti, S. An Efficient Chemical Analysis of Phenolic Acids and Flavonoids in Raw Propolis by Microwave-Assisted Extraction Combined with High-Performance Liquid Chromatography Using the Fused-Core Technology. J. Pharm. Biomed. Anal. 2013, 81–82, 126–132. [Google Scholar] [CrossRef]
- Setyaningsih, W.; Saputro, I.E.; Palma, M.; Barroso, C.G. Optimisation and Validation of the Microwave-Assisted Extraction of Phenolic Compounds from Rice Grains. Food Chem. 2015, 169, 141–149. [Google Scholar] [CrossRef] [PubMed]
- V. González-de-Peredo, A.; Vázquez-Espinosa, M.; Espada-Bellido, E.; Ferreiro-González, M.; F. Barbero, G.; Palma, M.; Carrera, C. Optimization of a Microwave Assisted Extraction Method for Maximum Flavonols and Antioxidant Activity of Onion Extracts. Antioxidants 2022, 11, 2393. [Google Scholar] [CrossRef]
- Biesaga, M.; Pyrzyńska, K. Stability of Bioactive Polyphenols from Honey during Different Extraction Methods. Food Chem. 2013, 136, 46–54. [Google Scholar] [CrossRef]
- Pedisić, S.; Čulina, P.; Pavlešić, T.; Vahčić, N.; Elez Garofulić, I.; Zorić, Z.; Dragović-Uzelac, V.; Repajić, M. Efficiency of Microwave and Ultrasound-Assisted Extraction as a Green Tool for Polyphenolic Isolation from Monofloral Honeys. Processes 2023, 11, 3141. [Google Scholar] [CrossRef]
- Maisl, C.; Schuhmacher, R.; Bueschl, C. Accuracy, Linearity, and Statistical Differences in Comparative Quantification in Untargeted Plant Metabolomics Using LC-ESI-Orbitrap-MS. Anal. Bioanal. Chem. 2025, 417, 2293–2309. [Google Scholar] [CrossRef]
- Michalkiewicz, A.; Biesaga, M.; Pyrzynska, K. Solid-Phase Extraction Procedure for Determination of Phenolic Acids and Some Flavonols in Honey. J. Chromatogr. A 2008, 1187, 18–24. [Google Scholar] [CrossRef]
- Ma, M.; Lu, X.; Wang, L.; Guo, Y.; Ding, H.; Wang, S.; Liang, X. A Stable Core-Shell Metal-Organic Framework@covalent Organic Framework Composite as Solid-Phase Extraction Adsorbent for Selective Enrichment and Determination of Flavonoids. J. Chromatogr. A 2023, 1707, 464324. [Google Scholar] [CrossRef] [PubMed]
- Klongdee, S.; Klinkesorn, U. Optimization of Accelerated Aqueous Ethanol Extraction to Obtain a Polyphenol-Rich Crude Extract from Rambutan (Nephelium lappaceum L.) Peel as Natural Antioxidant. Sci. Rep. 2022, 12, 21153. [Google Scholar] [CrossRef]
- Brzezińska, R.; Górska, A.; Wirkowska-Wojdyła, M.; Piasecka, I. Response Surface Methodology as a Tool for Optimization of Extraction Process of Bioactive Compounds from Spent Coffee Grounds. Appl. Sci. 2023, 13, 7634. [Google Scholar] [CrossRef]
- Dahmen-Ben Moussa, I.; Masmoudi, M.A.; Choura, S.; Chamkha, M.; Sayadi, S. Extraction Optimization Using Response Surface Methodology and Evaluation of the Antioxidant and Antimicrobial Potential of Polyphenols in Scenedesmus sp. and Chlorella sp. Biomass Conv. Bioref. 2023, 13, 7185–7198. [Google Scholar] [CrossRef]
- Mobasheri, F.; Khajeh, M.; Ghaffari-Moghaddam, M.; Piri, J.; Bohlooli, M. Machine Learning Optimization of Microwave-Assisted Extraction of Phenolics and Tannins from Pomegranate Peel. Sci. Rep. 2025, 15, 19439. [Google Scholar] [CrossRef] [PubMed]
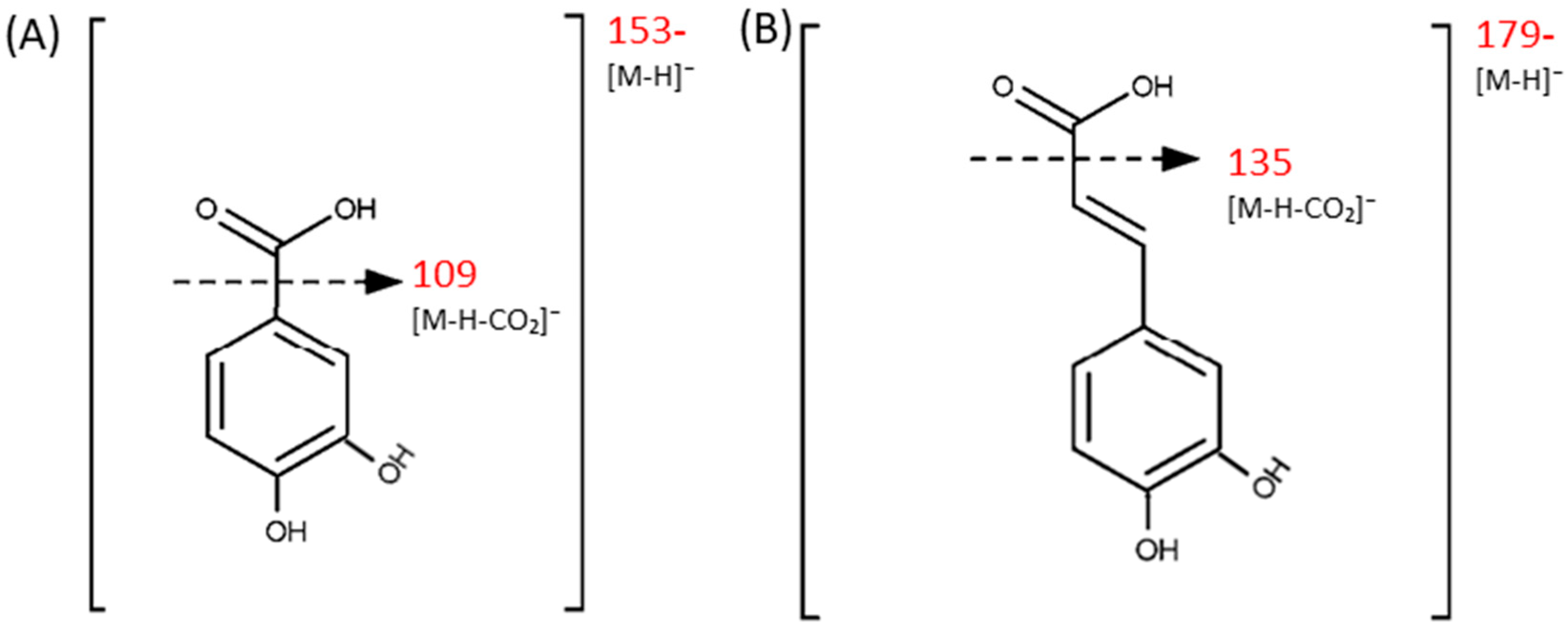


| Compound | pKa (Lowest) | Log S (at pH = 2) | Log D (at pH = 2) | Molecular Formula | Exact Mass/Da | Precursor Ion | Fragment Ions |
|---|---|---|---|---|---|---|---|
| 3,4-DHBA (protocatechuic acid) | 2.56 | −0.579 | 1.570 | C7H6O4 | 154.0266 | 153 (-) | 109 (-), 108 (-), 81 (-), 53 (-) |
| 2,5-DHBA (gentisic acid) | 2.53 | −0.572 | 1.561 | C7H6O4 | 154.0266 | 153 (-) | 109 (-), 108 (-), 81 (-), 53 (-) |
| o-coumaric acid | 4.42 | −1.675 | 1.831 | C9H8O3 | 164.0473 | 163 (-) | 145 (-), 119 (-), 93 (-) |
| m-coumaric acid | 4.17 | −1.674 | 1.830 | C9H8O3 | 164.0473 | 163 (-) | 145 (-), 119 (-), 93 (-) |
| p-coumaric acid | 4.20 | −1.674 | 1.829 | C9H8O3 | 164.0473 | 163 (-) | 145 (-), 119 (-), 93 (-) |
| Specific Fragment | Hexoses | Deoxyhexoses | Pentoses |
|---|---|---|---|
| Mass/Da | |||
| 0,1X | 150 | 134 | 120 |
| 0,2X | 120 | 104 | 90 |
| 0,3X | 90 | 74 | 60 |
| 1,5X | 134 | 120 | 104 |
| 2,3X-2H2O | 66 | 66 | |
| 0,4X-2H2O | 96 | 80 | 66 |
| 0,2X-H2O | 138 | 122 | 108 |
| 0,2X-2H2O | 156 | 140 | 126 |
| 2,3X-3H2O | 84 | 84 | |
| Yi | 162 | 146 | 132 |
| Xi | 180 | 164 | 150 |
| Phenolic Compound | Class | pKa (Lowest) | Molecular Mass/Da | |
|---|---|---|---|---|
| p-hydroxybenzoic acid | Phenolic acid; hydroxybenzoic acid | 4.38 | 138.0317 | 100% in neutral form when pH below 2 |
| p-coumaric acid | Phenolic Acid; hydroxycinnamic acid | 4.20 | 164.0473 | |
| Quercetin | Flavonoid; flavonol | 7.58 | 302.0426 | 100% in neutral form when pH below 6 |
| Rutin | Glycosylated flavonoid; Quercetin 3-rutinoside | 6.37 | 610.1534 | 100% in neutral form when pH below 4 |
| Matrix | Mobile Phase A | Mobile Phase B | Column | Gradient | Flow Rate/mL/min | Ref. |
|---|---|---|---|---|---|---|
| Propolis | 0.1% formic acid in Milli-Q water | 0.1% formic acid in acetonitrile | Zorbax SB-C18, (2.1 mm × 50 mm I.D, 1.8) | 0.00–0.90 min: 1% → 10% B 0.90–3.00 min: 10% → 20% B 3.00–4.50 min: 20% → 25% B 4.50–6.00 min: 25% → 30% B 6.00–7.50 min: 30% B (isocratic) 7.50–9.00 min: 30% → 90% B 9.00–9.30 min: 90% B (isocratic) 9.30–9.60 min: 90% → 10% B 9.60–15.00 min: 10% → 1% B 15.00–17.00 min (re-equilibration) | 0.33 | [37] |
| Plants | 0.1% formic acid in Milli-Q water | 0.1% formic acid in acetonitrile | C18 column (InfinityLab Poroshell 120 EC-C18, 2.1 × 150 mm, 2.7 μm) | 0.00–6.00 min: 20% B (isocratic) 6.00–16.00 min: 20% → 80% B 16.00–20.00 min: 80% B (isocratic) 20.00–25.00 min: 80% → 20% B (return to initial) 25.00–30.00 min: 20% B (re-equilibration) | 0.40 | [38] |
| Coffee pulp | 0.2% formic acid in Milli-Q water | Acetonitrile | C18 reversed-phase Avantor® ACE® Excel® C18-PFP (100 mm × 2.1 mm, 1.7 μm) | 0.00–0.30 min: 10% B (isocratic) 0.30–2.40 min: 10% → 15% B 2.40–3.25 min: 15% → 20% B 3.25–3.60 min: 20% B (isocratic) 3.60–6.20 min: 20% → 95% B 6.20–7.00 min: 95% B (isocratic) 7.00–7.50 min: 95% → 10% B 7.50–11.00 min: 10% B (re-equilibration) | 0.30 | [39] |
| Ceylon black tea | 0.1% formic acid in Milli-Q water | Acetonitrile | InfinityLab Poroshell 120 EC-C18 column (2.1 mm ×150 mm, 1.9 μm) | 0.00–1.50 min: 5% B (isocratic) 1.50–11.00 min: 5% → 15% B 11.00–18.00 min: 15% → 35% B 18.00–25.00 min: 35% → 95% B 25.00–27.00 min: 95% B (isocratic) 27.00–27.10 min: 95% → 5% B 27.10–29.00 min: 5% B (re-equilibration) 29.00+ min: column washing and re-equilibration | 0.28 | [40] |
| Rosé Wines | 0.1% formic acid in Milli-Q water | 0.1% formic acid in methanol | reversed-phase Acquity HSS T3 1.8 µm 1.0 × 100 mm | 0.00–2.00 min: 1% B (isocratic) 2.00–2.10 min: 1% → 5% B 2.10–8.00 min: 5% → 10% B 8.00–12.00 min: 10% → 28% B 12.00–18.00 min: 28% B (isocratic) 18.00–22.00 min: 28% → 45% B 22.00–23.50 min: 45% → 99% B 23.50–26.50 min: 99% B (isocratic) 26.50–27.00 min: 99% → 1% B 27.00–30.00 min: 1% B (re-equilibration) | 0.17 | [41] |
| Acer negundo tree | 0.1% formic acid in Milli-Q water | 0.1% formic acid in acetonitrile | Waters, BEH, 2.1 mm × 50 mm, 1.7 Microns | 0.00–30.00 min: 1% → 50% B 30.00–35.00 min: 50% → 99% B 35.00–39.00 min: 99% B (isocratic) 39.00–40.00 min: 99% → 1% B 40.00–45.00 min: 1% B (re-equilibration) | 0.30 | [42] |
| Rocket-Salad Leaves | 0.1% formic acid in Milli-Q water | 0.1% formic acid in methanol | Synergi Polar–RP C18 (250 mm × 4.6 mm, 4 µm) | 0.00–1.00 min: isocratic (initial composition; %B not specified) 1.00–25.00 min: 20% B (isocratic) 25.00–26.00 min: 20% → 85% B (linear ramp) (brief) isocratic hold at 85% B—duration not reported 26.00–32.00 min: 85% → 20% B (linear return) | 0.20 | [43] |
| Solid Residues from the Essential Oil Industry | 0.1% formic acid in Milli-Q water | Acetonitrile | Poroshell 120 EC-C18 (4.6 × 150 mm, 4 μm) | 0.00–5.00 min: 15% → 25% B (linear) 5.00–10.00 min: 25% → 35% B 10.00–28.00 min: 35% → 60% B 28.00–28.01 min: 60% → 15% B (fast return) 28.01–35.00 min: 15% B (isocratic/re-equilibration) | 0.50 | [44] |
| Level | Description | Minimum Evidence to Claim the Level | What You May Report |
|---|---|---|---|
| 1—Confirmed identification | Confirmed structure | In-house authentic standard measured in the same method with matching retention time (RT) window, exact mass, isotope pattern, and MS/MS (key fragments and ratios). | Definitive identity and quantitative data. |
| 2—putatively annotated compounds | Library/in silico match with orthogonal support | High-quality library or in silico MS/MS match. No in-house standard. | Probable identity (report as “putatively annotated”). |
| 3—Putative compound class | Substructure/class only | Diagnostic fragments/neutral losses define a class, but isomeric structures are unresolved; MS/MS present but not unique to a single structure. | Class-level assignment only. |
| 4—Unknown feature | Reproducible signal | Reproducible, alignable feature (m/z–RT; acceptable mass accuracy; clean peak shape); no reliable structural evidence. | Report as feature ID (m/z, RT) for statistics; do not name a compound. |
| Matrix | Mode | Fragmentor/V | Collision Energies/V | Ref. |
|---|---|---|---|---|
| Plukenetia volubilis leaves | negative | 110 | 10 and 20 | [70] |
| Schima argentea | positive and negative | NP * | NP | [72] |
| Stem, Roots, and Leaves of Syzygium cumini | positive and negative | NP * | NP | [76] |
| Symphorema polyandrum | positive and negative | NP | 10–40 (ramp) (for positive mode) and 10–30 (ramp) (for negative mode) | [77] |
| Litsea monopetala bark | positive | NP | 20 | [78] |
| Achillea ligustica | NP | NP | NP | [71] |
| Ziziphus budhensis Leaves | positive | NP | 30 | [81] |
| Lemon, lime, orange, and grapefruit juices | positive and negative | NP | 25–50 eV (stepping, negative and positive ion mode) and 40 eV (positive) | [82] |
| Fagonia arabica | positive and negative | NP | NP | [83] |
| Method | Extraction Principle and Typical Conditions | Strengths (Target Classes/Matrices) | Limitations and Risks | LC–MS(/MS) Implications for the Resulting Extract |
|---|---|---|---|---|
| UAE | Cavitation → rapid desorption/diffusion 50–80% aqueous alcohol; mild acid ≤40–50 °C; 10–30 min; controlled amplitude/duty | Fast; low thermal load Strong for cinnamates, flavanones/flavones Good in sugar-rich matrices when parameters constrained | Hot-spot oxidation if aggressive Matrix-specific tuning (pH/T/amplitude) needed | Align solvent with initial LC phase; favor negative ESI ([M–H]−) Often fewer sugar-borne co-extracts than MAE → lower ion suppression; light cleanup if waxes/lipids present (propolis) |
| MAE | Dielectric/volumetric heating; minutes-scale 60–100% MeOH/EtOH; mild acid Tight control of power/hold/temperature | Very rapid; high throughput Efficient for hydroxybenzoic and hydroxycinnamic acids Effective on robust plant residues | Over-power/time → glycoside cleavage, oxidation In sugar-rich matrices aglycones can degrade Tends to co-extract interferents without cleanup | Plan SPE to remove sugars/organic acids and stabilize adducts Tune polarity/pH to preserve glycosides when needed Useful for flavonols/benzoates with constrained dielectric input |
| SPE | Adsorptive cleanup/enrichment Acidified load; LC-compatible MeOH/ACN elution Sorbent selection: C18, polymeric, mixed-mode | Removes sugars/salts/lipids → ↑S/N, reproducibility Stabilizes electrospray; matrix-agnostic cleanup | Polar acids can break through C18 without strong acidification Glycosides show variable recovery; risk of over-retention | Essential to curb ion suppression and adduct variability Standardize loading/elution pH Consider polymeric/mixed-mode phases when C18 under-retains (honey/propolis/juices) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saftić Martinović, L.; Barbarić, A.; Gobin, I. Qualitative and Quantitative Mass Spectrometry Approaches for the Analysis of Phenolic Compounds in Complex Natural Matrices. Appl. Sci. 2025, 15, 12529. https://doi.org/10.3390/app152312529
Saftić Martinović L, Barbarić A, Gobin I. Qualitative and Quantitative Mass Spectrometry Approaches for the Analysis of Phenolic Compounds in Complex Natural Matrices. Applied Sciences. 2025; 15(23):12529. https://doi.org/10.3390/app152312529
Chicago/Turabian StyleSaftić Martinović, Lara, Ana Barbarić, and Ivana Gobin. 2025. "Qualitative and Quantitative Mass Spectrometry Approaches for the Analysis of Phenolic Compounds in Complex Natural Matrices" Applied Sciences 15, no. 23: 12529. https://doi.org/10.3390/app152312529
APA StyleSaftić Martinović, L., Barbarić, A., & Gobin, I. (2025). Qualitative and Quantitative Mass Spectrometry Approaches for the Analysis of Phenolic Compounds in Complex Natural Matrices. Applied Sciences, 15(23), 12529. https://doi.org/10.3390/app152312529









